Abstract
Genes involved in iron (Fe) acquisition often are regulated in response to the local availability of Fe. In many bacteria, Fe-dependent responsiveness is mediated by Fur, a global Fe-dependent transcriptional repressor. Tighter regulatory control of Fur-responsive genes is afforded by incorporating additional regulators into Fur-dependent regulatory cascades. RhuI, a Fur-dependent extracytoplasmic function sigma factor of Bordetella avium, in response to the dual stimulation of Fe starvation and the presence of heme (or hemoproteins), regulates PbhuR, a heme-responsive promoter which directs expression of the bhuRSTUV heme utilization operon. While BhuR, the outer membrane heme receptor, and RhuI have been shown to be indispensable for heme-dependent activation of PbhuR, collateral components of the regulatory cascade have not been described. In this investigation, RhuR, an integral cytoplasmic membrane protein with homology to anti-sigma factors, is shown to be an essential activator of PbhuR expression. The functional domain of RhuR required for heme-dependent activation of PbhuR expression was mapped to the N-terminal 97 amino acids of the protein by use of a chimeric RhuR-BlaM fusion. Expression of the chimera in a rhuR mutant rendered PbhuR constitutive, thereby decoupling the promoter from heme dependency. Growth studies confirmed that B. avium requires RhuR for optimal utilization of hemoglobin, but not hemin, as a sole source of nutrient Fe. These data imply that B. avium expresses, in addition to the BhuR heme/hemoprotein utilization system, an alternative RhuR-independent heme utilization mechanism. A model is proposed in which RhuR is the functional bridge between BhuR and RhuI in a heme-dependent regulatory cascade.
Iron (Fe), an essential element for nearly all pathogenic bacteria, must be derived from the host. In hosts, however, free Fe is usually sequestered by high-affinity molecules such as hemopexin, haptoglobin, and ferritin. Fe is also found coordinately bound with molecules such as hemoglobin, myoglobin, and transferrin. Efficient scavenging of Fe and Fe-containing molecules by the sequestering complexes reduces the concentration of the metal in the tissues and fluids of vertebrates well below the critical concentrations required for bacterial growth, rendering these compartments as inhospitable environments for colonization. To counteract this blockade to successful colonization, bacteria have evolved sophisticated Fe acquisition systems for removing the metal from the diverse sequestering molecules (4). Bacteria express a variety of Fe acquisition systems, each specific for one or more of the various host-specific Fe-containing molecules (4).
Expression of genes encoding the various Fe acquisition systems is rigidly regulated in response to environmental Fe availability and to the intracellular Fe concentration. When Fe is abundant, expression of the uptake systems is repressed. The capacity to downregulate expression of these systems when unneeded is an adaptive response that conserves metabolic resources. Tight control of Fe uptake also evolved to prevent accumulation of lethal concentrations of the metal in the bacterial cytoplasm. Bacteria have a variety of intracellular Fe sequestering systems for detoxification of the metal. Yet, if these systems are overwhelmed by uncontrolled Fe uptake, the result can be lethal. Fe is a potent oxidizer and has the capacity to damage DNA and a variety of other indispensable biomolecules. Extensive conservation of certain Fe-dependent regulators across genera suggests that subtle yet rigid control of expression of genes encoding Fe acquisition systems is crucial to most bacteria. For example, Fur (Fe uptake regulator), a global Fe-dependent transcriptional repressor which controls a variety of Fe acquisition systems, is highly conserved in virtually all bacteria. The capacity of Fur to respond to Fe stress is derived from the molecule's innate ability to reversibly bind Fe. When intracellular Fe is abundant, Fur is complexed with Fe. Binding of Fe stimulates binding of the protein to a 19-bp nucleotide sequence that is located proximal to Fur-dependent promoters. Binding of Fur to the “Fur box” blocks transcription of the Fur-regulated cistron (or polycistron). Upon Fe starvation, the metal dissociates from Fur, which decreases the repressor's affinity for the Fur box. Release of Fur from the Fur box derepresses transcription of the regulated gene (7).
In many cases, Fur is sufficient to adequately control expression of a specific gene. In other cases, genes encoding a particular Fur-dependent Fe acquisition system may require an additional level of regulation. These more complicated regulatory systems often operate in concert with Fur to activate specific genes under more limited environmental circumstances. Secondary regulatory systems have been described in several bacteria: siderophore expression in Pseudomonas aeruginosa is controlled by PchR, an AraC-type regulator which responds to extracellular pyochelin (12); genes for heme biosynthesis in Bradyrhizobium japonicum are controlled by Irr in response to internal Fe and heme stores (11); and irgA synthesis in Vibrio cholerae is induced by the LysR-type regulator IrgB (42). Recently, it was demonstrated that extracytoplasmic function (ECF) sigma factors are essential ancillary components in some Fur-dependent regulatory cascades (4).
The ECF class of σ70-type sigma factors (24) are small, functionally modifiable proteins that provide promoter specificity to core RNA polymerase (RNAP) (44). ECF sigma factors have been found to regulate a wide variety of cellular activities that mediate adaptive responses of bacteria to the local environment. These adaptive responses include the regulation of motility and carotenogenesis in Myxococcus xanthus (10, 41), toxin production in Clostridium difficile (26), and mycelium formation in Streptomyces coelicolor (3). Unlike σ70, the housekeeping sigma factor, the transcriptional activities of ECF sigma factors are modulated as a result of signal transduction cascades activated by an extracytoplasmic inducer molecule (24). Evidence is accumulating that bacterial pathogens have evolutionarily recruited ECF sigma factors for regulation of various virulence molecules (20, 45). Fur-dependent ECF sigma factors have been described which control genes for siderophore biosynthesis (22, 33, 40) and genes encoding proteins essential for uptake of exogenous Fe complexes (18, 19, 38, 39).
Interactions of ECF sigma factors with their affiliated regulators may be simple, complicated, or absent. While expression of the siderophore biosynthesis regulators PfrI of Pseudomonas putida and PbrA of Pseudomonas fluorescens is Fur dependent, no other regulatory partners for these ECF sigma factors have been identified (33, 40). In contrast, the ECF sigma factors FecI of Escherichia coli (39), PupI of P. putida (19), PvdS of P. aeruginosa (21), and HurI of Bordetella pertussis (38), which are associated with regulation of systems for uptake of various Fe sources, are dependent upon secondary transmembrane regulators for their functions. A useful paradigm to demonstrate the interdependence of the ECF sigma factor and its cognate transmembrane regulator is the FecI/FecR-regulated ferric dicitrate acquisition system of E. coli. Conditional transcription of the fec-encoded genes for ferric dicitrate transport is controlled by FecI, a modulated Fur-dependent sigma factor (2). Binding of ferric dicitrate to FecA, the outer membrane ferric dicitrate receptor, stimulates FecR, an integral cytoplasmic membrane protein (17) which transduces a signal to FecI, a cytoplasmic ECF sigma factor. FecI, in association with RNAP, upregulates transcription of fecABCDE (6, 25, 30). Although FecR is necessary to activate FecI, the precise molecular mechanism by which the dicitrate-dependent signal is transduced from FecR to FecI has yet to be elucidated. A similar ECF-based regulatory system for pseudobactin-dependent induction of PupI (ECF sigma factor) by PupR (cytoplasmic membrane PupI regulator) has also been described in P. putida (19).
Bordetella avium, a gram-negative coccobacillus, is the causative agent of avian coryza (bordetellosis). Since the pathology of coryza exhibits a strong similarity to whooping cough, an upper respiratory disease caused by the obligate human pathogen B. pertussis, B. avium has been employed as a surrogate pathogen to study B. pertussis pathogenesis (28). Previously, our laboratory described a heme and hemoprotein acquisition system (bhuRSTUV) in B. avium that is regulated by RhuI, a Fur-dependent ECF sigma factor, in concert with BhuR, the outer membrane heme receptor (18, 28). A similar locus (hurIRbhuRSTUV) was recently described in B. pertussis and Bordetella bronchiseptica (37). Using B. bronchiseptica as a model system, Vanderpool and Armstrong (38) showed that the B. pertussis bhuR and hurI genes are required for heme-dependent induction of bhuR expression (38). In this study, we describe the functional role of RhuR, a third component of the heme-dependent regulatory cascade, in the regulation of heme acquisition by B. avium. Database searches revealed that RhuR shares homology with anti-sigma factors FecR of E. coli (30) and PupR of P. putida (19), which are intimately involved in the regulatory activities of the Fe-dependent ECF sigma factors FecI and PupI, respectively. Based on these findings, it was postulated that RhuR functionally bridges the regulatory cascade from BhuR to RhuI (18). In this report, cell fractionation studies mapped the protein to the cytoplasmic membrane. In addition, experiments using fusion proteins revealed that the heme-dependent RhuI activation domain of RhuR is located within the N-terminal portion of this membrane protein. The critical significance of RhuR in the efficient utilization of hemoglobin, but not of hemin, as a source of nutrient Fe was also established.
MATERIALS AND METHODS
Strains, media, reagents, and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. B. avium strains were maintained on brain heart infusion (BHI) agar or broth (Difco Laboratories, Detroit, Mich.). E. coli strains were cultured on Luria-Bertani agar. BHI was supplemented with 150 μM FeSO4 for Fe-replete growth conditions. Fe-stressed growth conditions were established by addition of ethylenediamine di-o-hydroxy-phenylacetic acid (EDDHA; Sigma Biochemicals, St. Louis, Mo.) to BHI at a final concentration of 100 μM. EDDHA at a concentration of 300 to 400 μM was used to inhibit growth of the bacterium in the absence of hemin or other hemoproteins. Unless otherwise noted, ampicillin was used at a concentration of 200 μg/ml, rifampin was used at 10 μg/ml, streptomycin was used at 100 μg/ml, tetracycline was used at 10 μg/ml, and gentamicin was used at 10 μg/ml. Antibiotics were obtained from Sigma Biochemicals and Amresco (Solon, Ohio). Biochemical reagents were purchased from Sigma Biochemicals. Restriction enzymes, DNA-modifying enzymes, and Taq polymerase were obtained from Fermentas, Inc. (Hanover, Md.) and Invitrogen (Carlsbad, Calif.). Deionized water with an electrical resistance of >18 MΩ was used for all solutions.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| B. avium strains | ||
| 4169rif | wt strain; Stpr Rifr | 28 |
| 4169rifΔrhuR | Chromosomal rhuR deletion of 4169rif; Stpr Rifr | This study |
| 4169rifbhuR::kan | Insertion of Kanr cassette into bhuR of 4169rif; Stpr Rifr | 28 |
| E. coli strains | ||
| DH5αF′tet | φ80dlacZM15 Δ(lacZYA-argF)U169 deoR recA1 phoA hsdR17(rK− mK+) supE44λ−thi-1 gyrA96 relA1 [F′ proA B lacqZΔM15 Tn10(Tetr)] | Invitrogen |
| HB101 | F− Δ(gpt-proA)62 leuB6 glnV44 ara-14 galK2 lacY1 Δ(mcr-mrr) rpsL20 xyl-5 mtl-1 recA13; Stpr | New England Biolabs |
| MC4100λpir | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR (λpir) | 34 |
| Plasmids | ||
| pBluescript KS(+) | Cloning vector; Ampr | Stratagene |
| pET21a | Protein expression vector; PT7; Ampr | Novagen |
| pMECA | Cloning vector; Ampr | 36 |
| pCVD442tet | Allelic exchange vector; λpir dependent; sacB; Ampr | 28 |
| pRK415 | Conjugative expression shuttle vector for B. avium and E. coli; Plac; Tetr | 16 |
| pRK2013 | Conjugative helper plasmid; Kanr | 8 |
| pAEK26 | rhuR ligated into pRK415 at the XbaI/EcoRI sites | This study |
| pAEK26.3 | rhuR-His6 ligated into pRK415 at the XbaI/EcoRI sites | This study |
| pAEK26.1 | rhuR ligated into pET21a at the NdeI/EcoRI sites | This study |
| pAEK21 | 5′ 291 bp of rhuR fused to blaM ligated into pRK415 at the BamHI/HindIII sites | This study |
| pAEK23 | 291 bp of 5′ end of rhuR ligated into pRK415 at the BamHI/EcoRI sites | This study |
| pAD3 | pBluescript KS(+) encoding the 5′ terminus of bhuR and 7.4 kbp of upstream sequence | 28 |
| pERM1 | 3-kbp EcoRI fragment encoding rhuIR-PbhuR-bhuR, in pMECA | 28 |
| pAEK16 | rhuR deletion allelic exchange clone in pCVD442tet | This study |
| pAEK20.1 | Mutated rhuR allele (G569A) in pET21a | This study |
| pAEK25 | rhuIR locus in pBluescript KS(+) at the EcoRI/BamHI sites | This study |
| pDJM41 | PbhuR-lacZYA reporter; Ampr Genr | 18 |
Construction of rhuR deletion strain.
rhuR was deleted from the B. avium 4169rif chromosome by use of allelic exchange. The mutant gene used in allelic exchange was engineered by PCR splicing by overlap extension (SOEing). Sequences flanking rhuR in the 4169rif chromosome were amplified by PCR using primers designed to amplify the 631 bp located immediately 5′ of the ATG start codon of rhuR (ΔrhuR-a, 5′-TCCGAGCTCTGACCTCGCCTGAGCCT-3′ [SacI site underlined]; ΔrhuR-b, 5′-CTATTTAGTAACAGGAATCATTTATCATGACGCATCCATCAC-3′) and the 451 bp immediately 3′ of the TGA stop codon (ΔrhuR-c, 5′-TAAATGATTCCTGTTACTAAATAGATT-3′; ΔrhuR-d, 5′-GCTCTAGACACATAGGTGTTGTCGCC-3′ [XbaI site underlined]). ΔrhuR-b was synthesized with a 5′ extension (indicated in bold) complementary to ΔrhuR-c, which enabled the two PCR products to anneal. Primary amplicons were produced using Taq polymerase (1 to 10 U) from the template pAD3 under the following conditions: 1× PCRx buffer (Promega, Madison, Wis.), 10% dimethyl sulfoxide, 150 μM deoxynucleoside triphosphates (dNTPs), 1 μM concentration of each oligonucleotide primer; 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min; 25 cycles. PCR products were purified using the GFX purification kit (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). The SOEing reaction was performed as follows: 1× PCRx buffer, 10% dimethyl sulfoxide, 250 μM dNTPs, 1 μM ΔrhuR-a, 1 μM ΔrhuR-d, 2 μl of each primary product (10 to 50 ng); Taq polymerase (1 to 10 U); 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min; 25 cycles. The 1,086-bp product was ligated into the allelic exchange vector pCVD442tet at the XbaI and SacI sites to produce pAEK16, which was conjugated into B. avium 4169rif. Transconjugants were selected on BHI agar containing tetracycline. To select for the secondary recombination event, transconjugants were plated on BHI agar containing 20% sucrose. Sucrose-resistant clones were screened by colony lift hybridization, Southern hybridization, and colony PCR to assure that rhuR had been deleted without inadvertent disruption of nucleotide sequences proximal to rhuR in the rhu-bhu locus.
Cloning wt rhuR.
The rhuR open reading frame (ORF) was amplified by PCR using primers rhuR-5N-2 (5′-GGAATTCCATATGAGCGCAGCCGGCGT-3′ [NdeI site underlined]) and rhuR-3E (5′-TCTATTTAGTAACAGGAATTCATTTA-3′ [EcoRI site underlined]). The PCR conditions using pERM1 as a template were as follows: 1× ThermalAce buffer, 1× dNTP mix, 240 nM each primer, 4 U of ThermalAce polymerase; 95°C for 30 s, 45°C for 30 s, and 72°C for 1.25 min; 30 cycles. The 958-bp amplification DNA fragment was ligated into pET21a at the NdeI and EcoRI sites to produce pAEK20.1, in which the ORF was oriented appropriately downstream of a vector-encoded ribosomal binding site. Sequencing revealed the presence of a PCR-derived nucleotide mutation in the rhuR coding sequence. To engineer pAEK26.1, a plasmid encoding a wild-type (wt) rhuR, pAEK20.1, was digested with ClaI and NcoI and the 657-bp fragment containing the point mutation was replaced with the corresponding ClaI/NcoI fragment from pAEK25. A 1,002-bp DNA fragment obtained by digestion of pAEK26.1 with EcoRI and XbaI was ligated into the equivalent restriction sites of the conjugal shuttle vector pRK415. The ligation properly oriented the rhuR gene proximal to the lac promoter for isopropyl-β-d-thiogalactopyranoside-inducible expression in E. coli and constitutive expression in B. avium. This shuttle plasmid was designated pAEK26.
β-Galactosidase assay.
Expression of the lacZYA reporter gene in pDJM41 was determined by measuring β-galactosidase activity (18). Briefly, overnight cultures of Fe-stressed cells were used to inoculate secondary cultures in desired culture media. Secondary cultures were incubated at 37°C for 16 to 20 h. Bacteria in 1.0 ml of culture were pelleted by centrifugation for 5 min at 5,000 × g. Cells were resuspended in Z buffer (60 mM Na2HPO4 · H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4 · H2O, 38 mM β-mercaptoethanol) and the optical density at 600 nm (OD600) of the cell suspension was adjusted to 0.28 to 0.70. After diluting 400 μl of the suspension with 400 μl of Z buffer, cells were permeabilized by addition of 45 μl of 0.1% sodium dodecyl sulfate (SDS) and 90 μl of chloroform. Cell suspensions were vortexed for 10 s, followed by a 15-min incubation at 30°C. Enzymatic reactions were initiated by addition of 160 μl of a 4-mg/ml solution of o-nitrophenyl-β-d-galactopyranoside. Following incubation at 30°C, samples were observed for development of a yellow color, at which point 400 μl of 1 M Na2CO3 was added to terminate the reaction. After a brief centrifugation to pellet debris and chloroform, the OD420 and OD550 of each reaction were recorded and the relative β-galactosidase activity was calculated by use of the following formula (27): {1,000 · [OD420 − 1.75(OD550)]}/(t)(0.4)(OD600), with t = time of reaction in minutes.
Relative enzymatic activities are reported as the mean of triplicate assays and were derived from at least two independent experiments.
Endpoint growth assay.
All strains were inoculated into BHI broth containing 100 μM EDDHA and appropriate antibiotics. After overnight incubation at 37°C, these primary Fe-starved cultures were used to inoculate triplicate 3-ml BHI broth cultures under the particular growth conditions to be investigated. All cultures were supplemented with 400 μM EDDHA, a concentration at which growth of B. avium is inhibited (28). An Fe-replete environment was produced by supplementing the culture with 300 μM FeSO4. Hemin or turkey hemoglobin was added to the cultures at desired concentrations. Endpoint culture densities after 24 h of incubation at 37°C were ascertained by use of a Beckman DU640 spectrophotometer (Fullerton, Calif.) using the average culture density obtained from three independent cultures under a given culture condition.
Cell fractionation.
For cell fractionation experiments, 5 ml of bacteria obtained from overnight Fe-limited BHI broth (BHI plus 100 μM EDDHA) cultures were used to inoculate 500 ml of Fe-replete (BHI plus 150 μM FeSO4), Fe-stressed (BHI plus 100 μM EDDHA), or heme-induced (BHI plus 100 μM EDDHA plus 1 μM hemin) broth. After overnight incubation at 37°C, cells from 1.0 ml of culture were pelleted by centrifugation and resuspended in 1× SDS-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. These samples represented the whole cell fraction. Total membrane, outer membrane, and cytoplasmic membrane (sarcosine extract) fractions were prepared from the remaining culture by the differential centrifugation and lauroyl sarcosine extraction method of Leyh and Griffith (23). Final membrane fractions were resuspended in sterile deionized H2O and stored at −70°C. The total protein concentration of each fraction was obtained using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.) and a bovine serum albumin standard. Solubilized proteins were separated by SDS-PAGE using 12% polyacrylamide gels. To visualize resolved proteins in the cell fractions, gels were stained with colloidal Coomassie brilliant blue (29). For Western immunoblotting for RhuR-His6 polypeptides, cell fractions were electrotransferred to BioTraceNT nitrocellulose filters (Pall Corporation, Ann Arbor, Mich.), which were subsequently probed with INDIA HisProbe-horseradish peroxidase (HRP) (Pierce, Rockford, Ill.) using the manufacturer's protocol. Rabbit polyclonal anti-β-lactamase antibodies (5 Prime-3 Prime, Inc., Boulder, Colo.) and peroxidase-conjugated anti-rabbit immunoglobulin G (whole molecule; Sigma Biochemicals) antibodies were used to detect RhuR-BlaM fusion protein. INDIA-HisProbe-reactive and anti-BlaM-immunoreactive proteins on the filters were visualized using the Renaissance Western blot chemiluminescence reagent (DuPont NEN Research Products, Boston, Mass.) and blue-sensitive autoradiographic film (Marsh Bioproducts, Rochester, N.Y.).
Nucleotide sequencing and analysis.
Nucleotide sequencing was performed by the Biopolymer Facility at Roswell Park Cancer Institute (Buffalo, N.Y.). B. pertussis, Bordetella parapertussis, and B. bronchiseptica sequences were obtained from the Sanger Center database (Bordetella Sequencing Group, Sanger Center; http://www.sanger.ac.uk/Projects). Sequence analysis was performed using the Wisconsin Package version 9.0 software package (Genetics Computer Group, Madison, Wis.) and ClustalW (http://www.ebi.ac.uk/clustalw).
RESULTS
RhuR has homology to anti-sigma factors.
RhuI, an ECF sigma factor located in the cytoplasm of B. avium (GenBank accession number AY095952) has been shown to induce PbhuR, a heme-responsive promoter controlling expression of BhuR, the outer membrane heme receptor of the bacterium (18). Subsequently, we found that heme-dependent induction of PbhuR expression was absent in a ΔrhuI mutant strain of B. avium (data not shown). Heme-dependent induction of PbhuR was restored when a plasmid copy of rhuI was introduced into the mutant. These results provided evidence that heme-dependent induction of PbhuR in B. avium requires expression of RhuI. Since BhuR and RhuI are separated by the plasma membrane, a third component to functionally bridge these two polypeptides in the regulatory cascade was hypothesized. As genes encoding accessory regulatory components are often genetically linked, it was predicted that this bridging protein would most likely be encoded by a gene located proximal to bhuR and rhuI in the B. avium chromosome (18). Nucleotide sequencing of the region of the B. avium chromosome between the 3′ end of rhuI and the 5′ end of bhuR revealed a 921-bp ORF with a coding capacity for a polypeptide of 307 amino acids with a predicted molecular mass of 32.8 kDa (18). A 29-amino-acid sequence located at the N terminus of this predicted polypeptide containing an Ala-X-Ala motif (amino acid positions 27 to 29) was consistent in structure with a typical signal peptide commonly observed in proteins destined for sec-dependent transport to the cytoplasmic membrane. Signal peptidase-dependent cleavage of this putative signal peptide would produce a mature protein of 29.7 kDa. From one to three putative transmembrane domains were predicted by computer analysis of the polypeptide sequence. Collectively, these characteristics were consistent with those expected for a cytoplasmic membrane-bound transducer that would functionally link BhuR and RhuI. For the sake of consistency, this ORF was designated rhuR.
Protein similarity searches of the database revealed that RhuR has homology to a family of proteins which regulate the activity of ECF sigma factors (Fig. 1), including the HurR proteins of B. bronchiseptica and B. pertussis (39% identity and 51% similarity). RhuR also exhibited significant homology to PupR (31% identity and 46% similarity), a predicted cytoplasmic membrane protein which is required for proper regulation of PupI, the pseudobactin-dependent ECF sigma factor of P. putida (19). FecR, which regulates the activity of FecI, the ECF sigma factor of E. coli essential for uptake of ferric dicitrate, exhibits slightly less similarity to RhuR (28% identity and 42% similarity) (30). Three Trp residues (Trp18, Trp38, and Trp49) located near the N terminus of RhuR are conserved in HurR of B. pertussis, HurR of B. bronchiseptica, PupR, and FecR (Fig. 1). In FecR, these tryptophan residues have been shown to be required for ferric dicitrate-dependent induction of the fecABCDE-encoded ferric dicitrate acquisition system (35). Experiments to determine the importance of the three tryptophan residues in RhuR in heme-dependent regulation of bhuR expression in B. avium are ongoing.
FIG. 1.
Homology alignment of the RhuR protein of B. avium with HurR of B. pertussis (38), HurR of B. bronchiseptica (38), FecR of E. coli (39), and PupR of P. putida (19). Amino acids are denoted using the one-letter code. Ba, B. avium; Bp, B. pertussis; Bb, B. bronchiseptica; Ec, E. coli; Pp, P. putida. *, amino acids which are conserved in all five proteins; colon, amino acids with similar side chains; -, gaps in the amino acid sequences in comparison to the other four proteins. Tryptophans which are conserved in all five proteins and which have been shown to be essential for induction in E. coli FecA (35) are denoted in boldface and larger font. Amino acid sequences of HurR from B. pertussis and B. bronchiseptica were taken from the National Center for Biotechnology Information website (accession numbers NP879221 and NP891186, respectively).
RhuR positively regulates PbhuR expression.
To control expression of a specific gene, anti-sigma factors commonly bind to their cognate sigma factors, thereby denying productive interactions between the cognate sigma factors and RNAP (15). Genetic deletion of the anti-sigma factor gene releases the sigma factor from anti-sigma factor sequestration, enabling it to interact in an uncontrolled manner with RNAP and thus promoting constitutive expression of the regulated gene (9, 10, 19, 21, 32). To determine the means by which RhuR is involved in the heme induction cascade of B. avium and to determine if the polypeptide behaves as a typical sequestering type of anti-sigma factor, a chromosomal deletion of the rhuR ORF was engineered. Southern hybridization and colony PCR of 4169rifΔrhuR confirmed that the deletion was confined to rhuR and that the nucleotide sequences encoding rhuI, bhuR, and the intergenic region located between rhuR and bhuR, which contains PbhuR, were undisturbed (data not shown). The RhuR-dependent induction capacity of the wt and mutant strains of B. avium on PbhuR was evaluated by introducing pDJM41, a reporter plasmid encoding a transcriptional fusion between PbhuR and a promoterless lacZ gene, into 4169rif and 4169rifΔrhuR (18). As shown previously by Kirby et al. (18), 4169rif(pDJM41, pRK415) exhibited minimal β-galactosidase activity when cultured in Fe-replete BHI broth and only slightly elevated β-galactosidase activity when cultured in Fe-limiting BHI broth (Fig. 2). Addition of 1 μM hemin to the Fe-limited BHI broth, however, elicited a dramatic increase in β-galactosidase reporter activity in 4169rif(pDJM41, pRK415). In contrast, when cultured under Fe-limited conditions regardless of the presence or absence of hemin supplementation, 4169rifΔrhuR(pDJM41, pRK415) elaborated only low amounts of β-galactosidase activity (Fig. 2). When grown in Fe-replete or in Fe-stressed BHI broth, neither of the vector control strains [4169rif(pDJM41, pRK415) and 4169rifΔrhuR (pDJM41, pRK415)] demonstrated significant differences in β-galactosidase reporter activities. These observations strongly indicated that 4169rif required RhuR for heme-dependent induction of PbhuR.
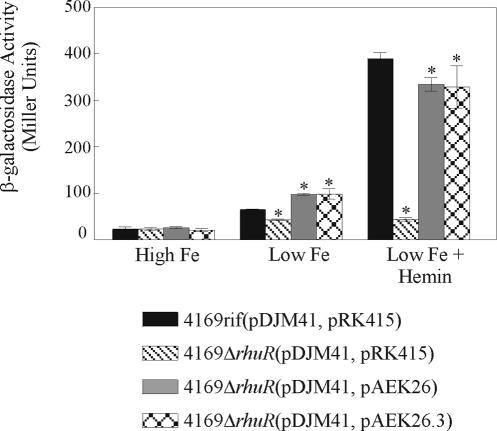
FIG. 2.
RhuR is a positive regulator of the BhuR-dependent heme induction cascade of B. avium. 4169rif(pDJM41, pRK415), 4169rifΔrhuR(pDJM41, pRK415), 4169rifΔrhuR(pDJM41, pAEK26), and 4169rifΔrhuR(pDJM41, pAEK26.3) were cultured in BHI broth supplemented with 100 μM EDDHA to deplete the internal Fe stores. Fe-stressed cultures were used to inoculate high-Fe (BHI plus 150 μM FeSO4), low-Fe (BHI plus 100 μM EDDHA) and low-Fe plus hemin (BHI plus 100 μM EDDHA plus 1 μM hemin) broth cultures. After 16 h of incubation, the β-galactosidase activity expressed from pDJM41 was determined using a modified Miller assay (27). Results shown represent the average of three experiments. Error bars indicate ±1 standard deviation. An asterisk indicates a significant difference (P < 0.05) from 4169rif(pDJM41, pRK415) under the same condition, as determined by Student's t test.
To confirm that the induction deficit of PbhuR was a consequence of the absence of rhuR, pAEK26, a recombinant plasmid that constitutively expresses wt rhuR, was conjugated into 4169rifΔrhuR. While introduction of pAEK26 restored heme-dependent induction of PbhuR in 4169rifΔrhuR(pDJM41, pAEK26), the level of induction was not equivalent to the level of induction observed in the wt background [4169rif(pDJM41, pRK415)] (Fig. 2). Observations in other systems suggested a possible explanation for this conundrum. Proper functioning of the CarQ/CarR sigma factor system in M. xanthus is exquisitely sensitive to differences in stoichiometric expression of CarQ and CarR (10). Several factors suggest that the expression of RhuI and RhuR are similarly coupled: rhuI and rhuR are encoded on a polycistronic message and are likely to be transcriptionally linked (A. E. Kirby and T. D. Connell, data not shown); rhuI and rhuR have a single codon overlap, a motif that is commonly observed in genes which are coupled at the level of translation (10). Thus, it is not unreasonable to hypothesize that, similar to CarQ and CarR, optimal functioning of RhuR and RhuI required close stoichiometric expression of both proteins. If that model is correct, the failure of pAEK26 to fully complement the rhuR mutant could be explained as a consequence of stoichiometric imbalance brought about by overexpression of the plasmid-encoded rhuR relative to the endogenous expression of the chromosomally encoded rhuI. To test this model, pAEK26 was introduced into 4169rif(pDJM41), a strain in which the chromosomal copies of rhuI and rhuR were undisturbed. Introduction of pAEK26 did not significantly alter the inductive capacity of PbhuR in the wt strain (data not shown). Thus, it is doubtful that the inability of pAEK26 to reestablish equivalent expression levels in the rhuR mutant was a result of a disparity in gene dosage between rhuI and rhuR.
Nevertheless, it was evident that efficient heme-dependent induction of PbhuR in B. avium required RhuR. Furthermore, as the deletion of rhuR did not elicit constitutive expression from the RhuI-dependent PbhuR in 4169rifΔrhuR, these experiments strongly implied that RhuR was not a sequestering type of anti-sigma factor. Rather, RhuR appeared to be a positive activator of PbhuR expression.
RhuR is a cytoplasmic membrane protein.
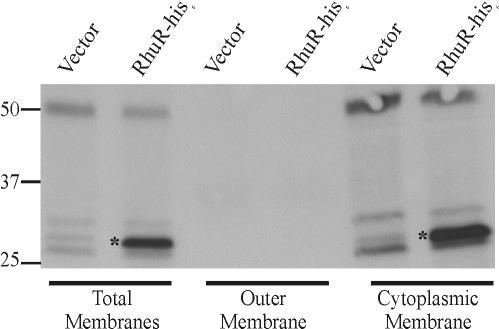
To functionally bridge the outer membrane protein BhuR and the cytoplasmic protein RhuI, RhuR was predicted to be localized either to the cytoplasmic membrane or to the periplasm. Evidence of a putative signal peptide in RhuR supported this model. Additional evidence to support a cytoplasmic membrane localization of RhuR was provided by computer-based polypeptide analysis (TMHMM [http://www.cbs.dtu.dk/services/TMHMM-2.0/], TMpred [http://www.ch.embnet.org/software/TMPRED_form.html], TopPred2 [http://www.sbc.su.se/∼erikw/toppred2/], and DAS [http://www.sbc.su.se/∼miklos/DAS]). The presence of a single membrane-spanning region (amino acids 81 to 100) with hydrophobic character was predicted by TMHMM. Three additional membrane-spanning regions (amino acids 199 to 219, 232 to 252, and 257 to 276) were predicted by TMpred, and two putative membrane-spanning regions were predicted by TopPred2 (amino acids 121 to 141 and 198 to 218). Somewhat paradoxically, analysis by DAS indicated that RhuR lacked motifs consistent with strong transmembrane domains. To resolve these contradictory predictions, fractionation experiments were conducted to directly establish the location of RhuR in the cell. Since antiserum to the wt RhuR was not available for use in these experiments, an alternative strategy was employed in which the cellular location of a His-tagged version of RhuR was ascertained. To verify that fusion of the His6 tag to the carboxyl terminus of RhuR did not interfere with proper localization of the protein, pAEK26.3, a plasmid encoding a His6-tagged version of RhuR, was introduced into 4169rifΔrhuR(pDJM41). Genetic complementation experiments confirmed that introduction of pAEK26.3 into 4169rifΔrhuR(pDJM41) restored heme-dependent expression from PbhuR to a level which was essentially equivalent to that observed in the mutant strain complemented with pAEK26 encoding wt rhuR [Fig. 2, 4169rifΔrhuR(pDJM41, pAEK26) versus 4169rifΔrhuR(pDJM41, pAEK26.3)]. Since it is likely that proper localization of the protein in the cell is essential for regulatory activity, this experiment indicated that the His6-tagged protein was transported to its active cellular location.
To localize RhuR-His6 in the cell, proteins in total membranes, outer membranes, and cytoplasmic membranes isolated from hemin-induced cultures of 4169rifΔrhuR(pDJM41, pRK415) and 4169rifΔrhuR(pDJM41, pAEK26.3) were resolved by SDS-PAGE and immunoblotted for poly-His epitopes. Western blotting with INDIA HisProbe-HRP (Pierce) revealed a strongly reactive polypeptide with an apparent molecular mass of 30 kDa in total membranes obtained from 4169rifΔrhuR(pDJM41, pAEK26.3) which was absent in total membranes isolated from the vector control (Fig. 3). Total membranes were further fractionated by extraction with 1% lauroyl sarcosine, which dissolves the cytoplasmic membrane but not the outer membrane. INDIA HisProbe-reactive protein was observed only in the cytoplasmic membrane fraction and not in the outer membrane fraction from 4169rifΔrhuR(pDJM41, pAEK26.3) (Fig. 3). Polymyxin B sulfate extracts of whole cells analyzed using similar blotting strategies demonstrated that the INDIA HisProbe-reactive protein of 4169rifΔrhuR(pDJM41, pAEK26.3) was not a component of the periplasm (data not shown). These cell fractionation experiments provided strong evidence that RhuR is either an integral cytoplasmic membrane protein or is tightly associated with the cytoplasmic membrane. These observations were consistent with the structural predictions obtained from the computer analysis of the predicted RhuR polypeptide.
FIG. 3.
RhuR-His6 is localized to the inner membrane. Fe-stressed cultures of 4169rifΔrhuR(pDJM41, pRK415) (Vector) and 4169rifΔrhuR(pDJM41, pAEK26.3) (RhuR-his6) were used to inoculate 500 ml BHI plus 100 μM EDDHA plus 1 μM hemin. At stationary phase, cells were isolated for preparation of total membranes, outer membranes, and cytoplasmic membranes. Twenty micrograms of total protein from each membrane preparation was resolved by SDS-PAGE using SDS-12% polyacrylamide gels. Proteins were transferred to nitrocellulose, and His-tagged RhuR was detected by Western blotting by use of the INDIA HisProbe-HRP. The positions of the molecular mass markers (in kilodaltons) are indicated. Asterisks indicate the position of RhuR-his6.
The N terminus of RhuR contains the activation domain.
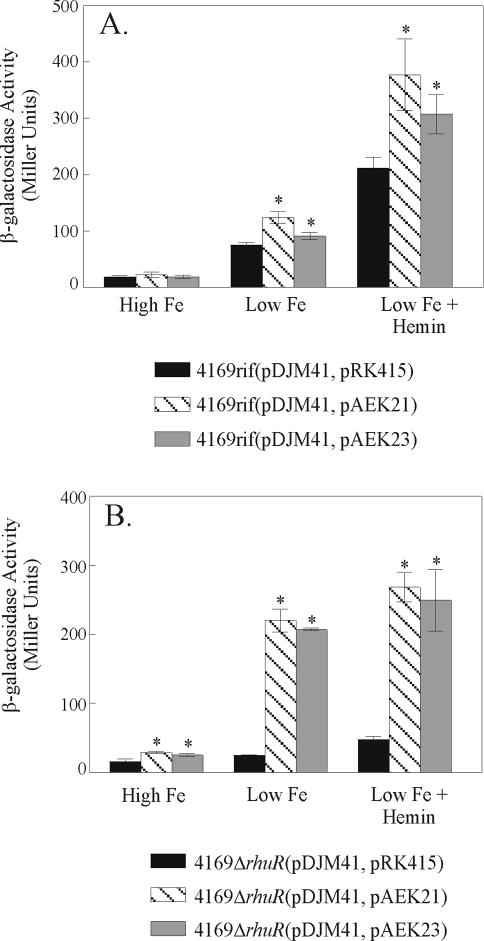
To understand the precise molecular interactions which mediate RhuR-dependent regulation of RhuI, it will be essential that the functionally important domains in these proteins are mapped. To begin to map the region(s) of RhuR required for heme-dependent activation of PbhuR, a series of fusion proteins was engineered. pAEK21 encodes a chimera in which the N-terminal 97 amino acids of RhuR, inclusive of the native signal peptide, is genetically fused to BlaM. As expected, growth of 4169rifΔrhuR(pDJM41, pAEK21) in Fe-replete (150 μM FeSO4) culture broth induced only minimal β-galactosidase activity (Fig. 4B). Surprisingly, 4169rifΔrhuR(pDJM41, pAEK21) cultured under conditions of Fe stress, but in the absence of hemin, exhibited levels of PbhuR reporter activity similar to that observed for hemin-induced, Fe-stressed wt cells [4169rif(pDJM41, pRK415)] (Fig. 4A versus B). Addition of hemin to the Fe-stressed cultures of 4169rifΔrhuR(pDJM41, pAEK21) did not further increase PbhuR activity (Fig. 4B). These results are consistent with a model in which the RhuR-BlaM chimera constitutively activates the PbhuR promoter in Fe-stressed cells. In effect, the RhuR-BlaM fusion decoupled PbhuR from hemin-dependent induction. The observation that the effect could be elicited only under Fe-stressed conditions suggested that chimera-dependent decoupled induction required expression of Fe-repressible RhuI. To determine if the chimera exerted its activity in a wt background, pAEK21 was also introduced into 4169rif(pDJM41). Under both Fe-stressed and hemin-induced conditions, 4169rif(pDJM41, pAEK21) exhibited higher PbhuR activity than did the relevant vector control [Fig. 4A, 4169rif(pDJM41, pAEK21) versus 4169rif(pDJM41, pRK415)] under similar growth conditions. The effect was also hemin inducible. Thus, wt RhuR is dominant over the chimera in respect to RhuI-dependent, hemin-dependent induction of PbhuR.
FIG. 4.
The N terminus of RhuR activates PbhuR in the absence of hemin. Fe-stressed overnight cultures were used to inoculate high-Fe (BHI plus 150 μM FeSO4), low-Fe (BHI plus 100 μM EDDHA), and low-Fe plus hemin (BHI plus 100 μM EDDHA plus 1 μM hemin) broth cultures. After 16 h of incubation, the β-galactosidase activity of the cultures was determined using a modified Miller assay (27). (A) Activity of the RhuR fusions in wt B. avium. pAEK21 expresses a chimeric protein consisting of the first 97 amino acids of RhuR fused to β-lactamase (BlaM); pAEK23 encodes the same 97-amino-acid region of RhuR fused to 18 irrelevant amino acids encoded by the vector. (B) PbhuR promoter activity of the RhuR fusions in 4169rifΔrhuR. Numbers represent the average β-galactosidase activity of triplicate cultures. Error bars indicate ±1 standard deviation. Asterisks indicate a significant difference (P < 0.05) from the vector control cultured under the same condition, as determined by Student's t test.
As a control to demonstrate that the decoupling activity of the RhuR-BlaM fusion was a property of the N-terminal 97 amino acids of RhuR and not to the BlaM portion of the chimera, a second fusion was engineered. In this new recombinant plasmid (pAEK23), a genetic fusion was created between the N-terminal 97 amino acids of the polypeptide and 18 irrelevant amino acids encoded by plasmid vector sequences. As shown in Fig. 4, PbhuR activity of this 115-amino-acid chimera was similar to that of the RhuR-BlaM fusion, indicating that the decoupled regulatory activity was engendered solely by the N-terminal region of RhuR and was not influenced by the presence of BlaM.
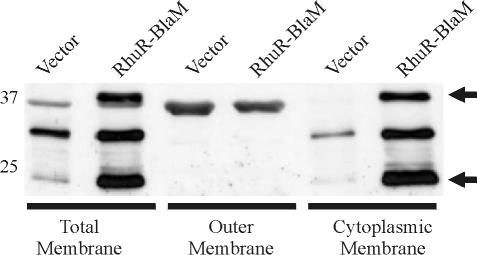
Prior cellular fractionation experiments indicated that wt RhuR localized to the cytoplasmic membrane. To investigate the cellular localization of the RhuR-BlaM chimera, cells expressing the chimera were fractionated into total membranes, outer membranes, and cytoplasmic membranes, each of which was analyzed by SDS-PAGE and immunoblotting using anti-BlaM antiserum. Anti-BlaM Western blotting of total membranes from the vector control, 4169rifΔrhuR(pDJM41, pRK415), revealed three cross-reactive polypeptides (Fig. 5). When equal amounts of protein from total membranes of 4169rifΔrhuR(pDJM41, pAEK21) were probed with anti-BlaM, two additional immunoreactive proteins that migrated to 37 and 25 kDa were apparent (Fig. 5). The 37-kDa species corresponded with the predicted molecular mass of RhuR-BlaM. As it was only seen in membrane fractions from 4169rifΔrhuR(pDJM41, pAEK21), the 25-kDa species was hypothesized to be a degradation product of RhuR-BlaM. Fractionations of the total membranes demonstrated that a set of immunoreactive proteins, which were identical in electrophoretic mobility to those in total membranes, were expressed in the cytoplasmic membrane fraction (Fig. 5). Only a vector-specific band was detected in the outer membrane (and in the periplasmic extract) (data not shown) of 4169rifΔrhuR(pDJM41, pAEK21). These experiments provided clear evidence that the RhuR-BlaM chimera, and also wt RhuR, localized to the cytoplasmic membrane of B. avium.
FIG. 5.
RhuR-BlaM is localized to the inner membrane of 4169rifΔrhuR. Fe-stressed cultures of 4169rifΔrhuR(pDJM41, pRK415) (vector) and 4169rifΔrhuR(pDJM41, pAEK21) (RhuR-BlaM) were used to inoculate BHI broth supplemented with 100 μM EDDHA and 1 μM hemin. At stationary phase, cells were collected for isolation of total membranes, outer membranes, and cytoplasmic membranes. Ten micrograms of total protein from each membrane preparation was separated by SDS-PAGE using SDS-12% polyacrylamide gels. Western immunoblotting using a rabbit polyclonal anti-BlaM antiserum was used to detect immunoreactive proteins. The positions of the molecular mass markers (in kilodaltons) are indicated. Arrows indicate the position of the 37-kDa RhuR-BlaM and its major 25-kDa degradation product.
The inductive capacity of the RhuR-BlaM chimera suggested a model for RhuR activity. Since expression of RhuR-BlaM activates PbhuR, it can be surmised that the N-terminal 97 amino acids of RhuR likely encode the residues which are required for activation of RhuI. Furthermore, since activation of the cytoplasmic RhuI likely requires interaction between RhuI and RhuR, at least a portion of the N-terminal 97 amino acids of RhuR are likely exposed to the cytoplasm. Extrapolative interpretation of that model implies that the C-terminal portion of RhuR (amino acids 98 to 307) is exposed to the periplasm and encodes the cis-acting heme-dependent (i.e., BhuR-dependent) domain(s) that regulates the N-terminal activation portion of the protein. Chimeras for expression in B. avium in which the carboxyl portion of RhuR is to be genetically fused to MalE are being engineered to experimentally test this model.
Expression of BhuR and growth of B. avium in hemin does not require RhuR.
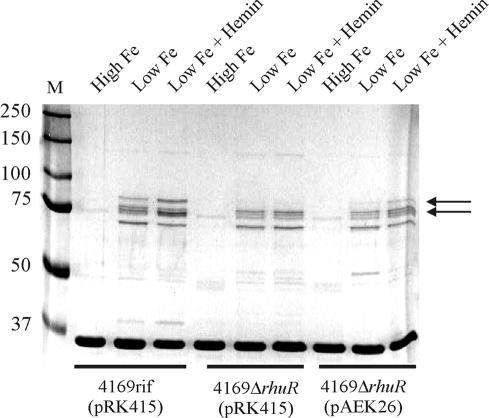
Expression of BhuR is essential for optimal growth of B. avium in culture medium in which hemin or hemoproteins are the sole sources of nutrient Fe (28). Having shown that RhuR was required to upregulate expression of PbhuR, it was hypothesized that RhuR was also required for efficient growth of B. avium in culture medium in which hemin was the Fe-limiting nutrient source. Previous endpoint growth assays demonstrated that growth of B. avium is inhibited when cultured in BHI broth depleted of available Fe by the addition of 400 μM EDDHA (18, 28). Supplementation of BHI plus 400 μM EDDHA broth with 300 μM FeSO4 rescued growth of the cells (28) (Fig. 6, FeSO4). To our surprise, when 4169rifΔrhuR was cultured in Fe-deficient BHI broth supplemented with either 5 μM hemin (data not shown) or 1.25 μM hemoglobin (Fig. 6), growth of the mutant was indistinguishable from growth of the isogenic parental strain (28). To examine this apparent conundrum, the outer membrane protein profile of 4169rifΔrhuR was examined to compare the expression of BhuR in the mutant strain with expression of BhuR in the wt parent strain. Contrary to expectations, expression of the two forms of BhuR (91 and 82 kDa) (18, 28) was not inhibited in the rhuR mutant strain [Fig. 7, 4169rifΔrhuR(pRK415), low Fe and low Fe plus hemin]. Expression of BhuR in 4169rifΔrhuR(pRK415), however, was somewhat lower than expression of the protein in 4169rif(pRK415) when both strains were cultured under conditions of Fe stress in the absence of hemin induction [Fig. 7, 4169rifΔrhuR(pRK415) versus 4169rif(pRK415), both in low Fe]. When hemin was added to the Fe-stressed cultures, expression of the two forms of BhuR was enhanced in the wt strain [Fig. 7, 4169rif(pRK415), low Fe, versus 4169rif(pRK415), low Fe plus hemin]. Expression of BhuR was unaffected in the mutant strain [Fig. 7, 4169rifΔrhuR(pRK415), low Fe versus 4169rifΔrhuR(pRK415), low Fe plus hemin] when cultured in low Fe plus hemin. Further analysis showed that, while 4169rifΔrhuR(pRK415), in comparison to the isogenic wt strain [4169rif(pRK415)], was incapable of hemin-dependent upregulation of BhuR expression, complementation of 4169rifΔrhuR with pAEK26 (i.e., with wt rhuR) restored the ability of the mutant to upregulate expression of BhuR in a hemin-dependent manner [Fig. 7, 4169rifΔrhuR(pAEK26), low Fe and low Fe plus hemin]. It should be noted that the low level expression of BhuR in wt B. avium in the absence of hemin is promoted, at least in part, by read-through transcription from the upstream Fur-dependent PrhuIR (unpublished results).
FIG. 6.
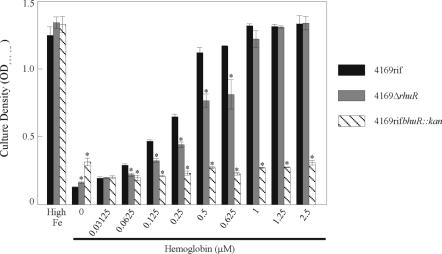
RhuR is required by B. avium for efficient utilization of hemoglobin as source of nutrient Fe. Fe-stressed cultures of 4169rif, 4169rifbhuR::kan, and 4169rifΔrhuR were used to inoculate 2 ml of BHI broth which was supplemented with 400 μM EDDHA and 300 μM FeSO4 (High Fe) or with turkey hemoglobin at the indicated concentrations. After 24 h of incubation at 37°C, the OD600 of the cultures was measured. Results represent the average cell densities of three cultures. The heme acquisition mutant 4169rifbhuR::kan was used as a negative control (28). Error bars indicate ±1 standard deviation. Asterisks indicate a statistically significant difference from 4169rif (Student's t test).
FIG. 7.
BhuR is expressed at low levels in the outer membrane of 4169rifΔrhuR. Fe-stressed (BHI plus 100 μM EDDHA), Fe-stressed plus heme (BHI plus 100 μM EDDHA plus 1 μM hemin), and Fe-replete (BHI plus 150 μM FeSO4) cultures of 4169rif(pRK415), 4169rifΔrhuR(pRK415), and 4169rifΔrhuR(pAEK26) were used for isolation of outer membranes. The proteins in the outer membrane preparations were resolved in an SDS-8.75% polyacrylamide gel. Protein profiles were visualized by staining with colloidal Coomassie brilliant blue. M, molecular mass markers (in kilodaltons). Arrows indicate the positions of the 91-kDa and 82-kDa forms of BhuR (28).
During a natural infection, the various sequestration and compartmentalization mechanisms operating in the host reduce the physiological concentrations of heme or hemoglobin to levels considerably below the 5 μM used in the prior in vitro growth experiments. With this point in mind, it was surmised that the failure of the rhuR mutant to demonstrate a detectible growth defect in comparison to wt B. avium was a result of the use of physiologically irrelevant amounts of hemin in the broth cultures. Experiments, therefore, were repeated using lower concentrations of hemin and hemoglobin to address this issue. As a control to establish a baseline for hemin- and hemoglobin-limited growth of the bacterium, 4169rifbhuR::kan, a mutant deficient in expression of bhuR, was included in these growth experiments. As expected, the growth of 4169rifbhuR::kan was significantly inhibited under all conditions with the exception of culture broth supplemented with FeSO4 (Fig. 6). Growth of 4169rifΔrhuR, however, was indistinguishable from that of 4169rif when each strain was cultured in BHI broth without hemin supplementation or in BHI broth supplemented with hemin. This effect was observable at all concentrations of hemin between 0.125 and 10 μM in a twofold dilution series (data not shown). In contrast, a significant difference in Fe substrate utilization was observed when hemoglobin was substituted for hemin in the broth cultures. While dose-dependent growth of 4169rif was evident in BHI broth containing hemoglobin at concentrations as low as 0.0625 μM, dose-dependent growth of 4169rifΔrhuR was not evident until the concentration of hemoglobin in the broth reached 0.125 μM (Fig. 6). Furthermore, the difference in dose-dependent hemoglobin utilization between the rhuR mutant and the wt strain remained significant from 0.0625 to 0.625 μM hemoglobin in the broth. At hemoglobin concentrations exceeding 1.0 μM, no significant difference was observed between the growth of the rhuR mutant and the wt strain.
DISCUSSION
During invasion of vertebrate hosts, pathogenic bacteria are normally confronted with a stringently Fe-limited environment. Without mechanisms to efficiently procure Fe from the cells, fluids, and tissues of the infected host, most bacteria are incapable of expressing full virulence (13, 28). Since Fe is toxic to the bacterial cell when intracellular concentrations are uncommonly high, expression of Fe uptake mechanisms must be rigidly controlled. It is critical, therefore, that bacteria balance nutritional needs against potential toxicity by tightly regulating the active uptake of Fe. While various Fe-dependent bacterial regulators are currently known, only Fur, a global Fe-dependent repressor of Fe uptake systems, has been extensively studied. In most cases, Fur represses transcription of Fur-dependent genes when the cellular concentration of Fe is optimal (7). When concentrations of Fe within the cell fall to critical levels, Fur-dependent Fe uptake genes are derepressed. This system of negative regulation ensures that Fur-regulated Fe acquisition systems are expressed only when the cell has a critical need for the metal.
Most bacteria exhibit multiple Fur-regulated Fe acquisition systems, each of which targets one or more forms of inorganic or organic Fe as a nutrient source. Expression by the bacterium of these systems solely in response to Fe stress in the absence of those specific Fe-containing substrates in the local environment potentially wastes precious energy and metabolic resources. In response to that selective pressure, bacteria evolved complicated regulatory systems to fine-tune the expression of some Fur-dependent Fe uptake mechanisms. The evolution or acquisition of the Fur- and heme-dependent Bhu uptake system in B. avium likely arose as an adaptive response to the abundance of heme and hemoproteins in the avian host (28). It is becoming increasingly evident that the regulation of the Bhu system of B. avium is a complicated regulatory cascade which responds both to Fe stress and the presence of heme and hemoglobin (18, 28). In Fe-replete cells, transcription of the bhuRSTUV operon is repressed by Fur (18). Upon Fe starvation, the upstream Fur-dependent PrhuIR is derepressed with the subsequent transcription of rhuI and rhuR. Concomitant with transcription of rhuI and rhuR is a low level expression of BhuR in the outer membrane of the Fe-stressed bacterium. This low level of expression of BhuR is a consequence of incomplete termination of rhuIR transcription, which gives rise to a rare polycistronic mRNA that encodes rhuI, rhuR, and bhuR (unpublished data). This system of read-through transcription ensures that Fe starvation of B. avium not only elicits optimal expression of RhuI and RhuR but also produces a low level of BhuR expression sufficient to prime the cell for up-regulation of the entire bhuRSTUV-encoded heme uptake system, should heme become locally available. Upregulation of the regulatory cascade initiates by binding of trace molecules of heme by BhuR. Binding to BhuR transduces a signal across the periplasm to RhuR. The signal is propagated through RhuR across the cytoplasmic membrane to RhuI, which is localized in the cytoplasm. Upon reception of the signal, RhuI directly or indirectly escorts RNAP to PbhuR, thus promoting high-level transcription of bhuR and the other four genes of the polycistron (bhuSTUV). This heme-dependent, three-component regulatory system parsimoniously ensures that B. avium expresses the heme utilization mechanism only when two conditions are met: (i) when the cell is starved for Fe and (ii) when either heme or hemoproteins are available in the local environment.
It is clear that the mechanism for heme uptake in B. avium is governed by a precise molecular interplay between the ECF sigma factor (RhuI) and the cytoplasmic membrane activator (RhuR). Many traditional anti-sigma factors control the function of their cognate sigma factor by sequestering the sigma factor from core RNAP (15). In those cases, genetic deletion of the anti-sigma factor typically releases the sigma factor from regulatory control, which manifests as constitutive expression of the sigma factor-dependent regulated gene (9, 10, 19, 21, 32). Other sigma factor regulators exert control of their cognate sigma factors via various molecular interactions, including phosphorylation-dependent partner switching (5), extracellular export of the anti-sigma factor (14), and a regulated degradation of the anti-sigma factor (1, 10). In this study, deletion of rhuR did not elicit a constitutive deregulation of the RhuI-dependent PbhuR. Rather, deletion of rhuR inhibited heme-dependent induction of PbhuR, thereby providing strong evidence that RhuR is not a traditional sequestering type of anti-sigma factor but is a positive regulator of RhuI activity. Similar sigma factor activation mechanisms have been reported in other bacteria. Genetic experiments revealed that FecR, an E. coli homologue of RhuR, is required for transcriptional activity of FecI, its cognate ECF sigma factor (30). Restoration of PfecA activity was not established when a fecIR-fecABCDE null strain was complemented solely with a plasmid encoding fecA (the outer membrane ferric citrate receptor) and fecI (the ECF sigma factor). Promoter activity was restored only when the fecIR-fecABCDE null strain was complemented with fecA, fecI, and fecR (30). While studies confirmed that FecR directly interacts with FecI (6, 25), the molecular mechanism by which FecI is activated, or the nature of the difference between the functional structures of activated and inactivated FecI, have not been well elucidated. The successful mapping of the activator domain of RhuR reported herein will likely facilitate the elucidation of the molecular mechanisms by which RhuI is biochemically or biophysically activated.
Using cells expressing chromosomally encoded and plasmid-encoded truncated FecR derivatives (30) and plasmid-encoded FecR′-BlaM hybrid proteins (43), it was demonstrated that truncated forms of FecR as small as 59 residues had the capacity to stimulate ferric citrate-independent expression of the fecA and fecB transport genes. It was later confirmed using a bacterial two-hybrid Lex-based system (6) that the N-terminal portion of RhuR directly interacts with RhuI. Expression of the N-terminal 97 amino acids of RhuR decouples the heme dependency of PbhuR from the heme induction cascade in B. avium. Fusions of these 97 amino acids to either BlaM or to 18 vector-encoded irrelevant amino acids constitutively stimulated PbhuR activity in cells cultured under Fe-stressed conditions in the absence of hemin to levels which were equivalent to those observed in Fe-starved cells cultured in the presence of hemin. These data support a model in which the N-terminal 97-amino-acid region of RhuR interacts productively with cytoplasmic RhuI. This region, therefore, has been designated as the activator domain of the protein. The fact that activation via RhuR requires heme-dependent transduction from BhuR suggests that RhuR must also harbor a controller domain for intercepting this transduction signal. Experiments in E. coli revealed that the C-terminal region of FecR is located in the periplasm (43) and interacts productively with FecA (6). Using the Fec system as a model, we hypothesize that the C-terminal region of RhuR is periplasmically exposed and that this region is the controller domain, which receives the heme-dependent transducing signals from BhuR. In general, we anticipate that interaction of heme-bound BhuR with the C-terminal region of RhuR elicits a conformational change in RhuR, which is translated across the cytoplasmic membrane to the N terminus of RhuR. A concomitant conformational change in the N-terminal region of RhuR elicits, directly or indirectly, a heme-dependent activation of RhuI. Future studies will investigate these putative interactions between BhuR and the C-terminal region of RhuR and between the N-terminal region of RhuR and RhuI and the structural changes in each protein that occur.
It should be noted that our domain model of RhuR is based upon the hypothesis that the decoupling activity of the RhuR chimera is due to the separation of its activator domain from its controller domain. There is, however, an alternative model that could explain the manner in which the chimeras decouple PbhuR from heme-dependent regulation. Decoupling could simply be a result of the overexpression of the chimera, which disturbs a regulatory equilibrium in the cell. We discount this alternative model, since overexpression of a plasmid-encoded wt rhuR in Fe-stressed wt cells does not elicit a detectible change in PbhuR activity (data not shown).
The establishment of the essential nature of BhuR for heme uptake and the corresponding dependence of RhuR for optimal BhuR expression provide solid evidence to support the hypothesis that RhuR is required for efficient growth in culture medium in which heme is the sole source of nutrient Fe. To our surprise, endpoint growth assays demonstrated that, regardless of hemin concentration, growth of B. avium in culture broth in which heme was the limiting Fe source did not require expression of RhuR. The capacity to use hemoglobin as a sole source of Fe, however, was strongly correlated with expression of RhuR. This response was particularly evident at lower hemoglobin concentrations, which may be indicative of the actual substrate likely encountered by B. avium during a natural infection. Free heme is rarely found in the fluids and tissues of infected hosts. Hemoglobin, however, becomes quite abundant as the bacterium degrades tissues in the upper respiratory system. It is not surprising, therefore, that B. avium evolved a system to rapidly access the concentration of hemoglobin, the most likely form of organic Fe, and used the sensing system to finely coordinate the regulated expression of the proteins involved in its acquisition. Based on the ability of B. avium to efficiently utilize hemoglobin as an Fe source, we predicted the existence of a molecule which can extract or release the heme moiety from hemoglobin and from other hemoproteins (28). A logical assumption would be that the bacterium produces a protease that degrades hemoglobin, thus releasing heme into the medium. In E. coli, for example, hemoglobin is degraded by a secreted protease (31). Using sensitive hemoglobin-specific immunoblotting, we have been unable to demonstrate a similar proteolytic activity in B. avium. The molecular mechanism by which heme is obtained from hemoglobin by B. avium warrants further investigation.
Comparisons between the rhuIR and bhuR genes and their respective protein products have been informative in substantiating the importance of the genes in Fe acquisition in the bordetellae. The rhuIR-bhuR locus is conserved at the nucleotide and amino acid levels and in genetic organization throughout the genus Bordetella. RhuI of B. avium is 65% similar to the HurI of B. pertussis and 83% similar to the HurI of B. bronchiseptica (38). Similarly, the BhuR of B. pertussis is 64% similar to the BhuR of B. avium. BhuRs from B. bronchiseptica and from B. parapertussis are 81% similar with the BhuR of B. avium. A very strong selective pressure is required to maintain these significant levels of similarity between RhuI and BhuR among the four virulent species of Bordetella. Complementation experiments are being performed to determine whether BhuR and RhuI of B. avium are true functional homologues in these species.
In contrast to the high degree of conservation of BhuR and RhuI, RhuR is more divergent among the Bordetella species. HurR (the RhuR homologue) of B. bronchiseptica and HurR of B. pertussis are both only 51% similar to RhuR of B. avium (Fig. 1). Perhaps the most interesting member of the RhuR family is encoded by B. parapertussis. While the first 83 amino acids of RhuR of B. parapertussis are 100% identical to the corresponding region of HurR of B. pertussis, the remaining 151 amino acids of the B. parapertussis protein have little or no homology with the RhuR polypeptides of B. avium or to either of the other two Bordetella species. Perhaps the heme-dependent induction system in B. parapertussis has been rendered nonfunctional. There is, however, another intriguing possibility. It is the N-terminal region of the B. parapertussis RhuR that is highly conserved. That region in RhuR of B. avium was shown in this study to harbor the RhuI activation domain. And, when that domain was separated from the putative C-terminal controller domain, the polypeptide stimulated expression of PbhuR in a heme-independent manner. Thus, it is possible that RhuR of B. parapertussis constitutively activates its cognate RhuI when exposed to Fe starvation, regardless of heme availability. Although eliminating heme-dependent control of heme uptake, such a decoupled system would, nevertheless, enable B. parapertussis to maximally express BhuR and presumably the other genes of the bhu locus. Whether such a decoupled system in B. parapertussis would be energy efficient or selectively adaptive is unclear.
Experiments detailed in this report provide strong evidence that expression of RhuR is essential for heme-dependent induction of BhuR and that the regulatory protein is required by B. avium for efficient utilization of hemoglobin as a source of nutrient Fe. Future studies will examine the function of RhuR throughout the Bordetella genus.
Acknowledgments
Funds to support this investigation were made available to T.D.C. from The School of Medicine and Biomedical Sciences, The University of Buffalo, The State University of New York. A.E.K. was supported by a National Institutes of Health training grant (AIO7614) awarded to the Witebsky Center for Microbial Pathogenesis and Immunology and by a Presidential Fellowship administered by The Office of the Provost, The University of Buffalo. N.K. was supported by an NIH training grant (DE007034-27) awarded to the Department of Oral Biology in the School of Dental Medicine at The University of Buffalo.
Editor: J. T. Barbieri
REFERENCES
- 1.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 13:2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angerer, A., S. Enz, M. Ochs, and V. Braun. 1995. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. Fecl belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol. Microbiol. 18:163-174. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor. J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V., and H. Killman. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109.10203757 [Google Scholar]
- 5.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escolar, L., J. Perez-Martin, and V. deLorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehring, A. M., N. J. Yoo, and R. Losick. 2001. RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J. Bacteriol. 183:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorham, H. C., S. J. McGowan, P. R. H. Robson, and D. A. Hodgson. 1996. Light-induced carotenogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by anti-sigma factor CarR. Mol. Microbiol. 19:171-186. [DOI] [PubMed] [Google Scholar]
- 11.Hamza, I., S. Chauhan, R. Hassett, and M. R. O'Brian. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273:21669-21674. [DOI] [PubMed] [Google Scholar]
- 12.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, D. P., and S. M. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsky. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231-286. [DOI] [PubMed] [Google Scholar]
- 16.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Kim, I., A. Stiefel, S. Plantor, A. Angerer, and V. Braun. 1997. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol. Microbiol. 23:333-344. [DOI] [PubMed] [Google Scholar]
- 18.Kirby, A. E., D. J. Metzger, E. R. Murphy, and T. D. Connell. 2001. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect. Immun. 69:6951-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koster, M., W. van Klompenburg, W. Bitter, J. Leong, and P. Weisbeek. 1994. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 13:2805-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacikova, G., and K. Skorupski. 2002. The alternative sigma factor σE plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leoni, L., N. Orsi, V. deLorenzo, and P. Visca. 2000. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J. Bacteriol. 182:1481-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leyh, R., and R. W. Griffith. 1992. Characterization of the outer membrane proteins of Bordetella avium. Infect. Immun. 60:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonetto, M., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahren, S., and V. Braun. 2003. The FecI extracytoplasmic-function sigma factor of Escherichia coli interacts with the β′ subunit of RNA polymerase. J. Bacteriol. 185:1796-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mani, N., and B. Dupuy. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. USA 98:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics, p. 72-74. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 28.Murphy, E. R., R. E. Sacco, A. Dickenson, D. J. Metzger, Y. Hu, P. E. Orndorff, and T. D. Connell. 2002. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect. Immun. 70:5390-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 30.Ochs, M., S. Veitinger, I. Kim, D. Welz, A. Angerer, and V. Braun. 1995. Regulation of citrate-dependent iron transport of Escherichia coli: FecR is required for transcription activation by FecI. Mol. Microbiol. 15:119-132. [DOI] [PubMed] [Google Scholar]
- 31.Otto, B. R., J. M. van Dooren, J. H. Nuijens, J. Luirink, and B. Oudega. 1998. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 188:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paget, M. S., J. B. Bae, M. Y. Hahn, W. Li, C. Kleanthous, J. H. Roe, and M. J. Buttner. 2001. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol. Microbiol. 39:1036-1047. [DOI] [PubMed] [Google Scholar]
- 33.Sexton, R., P. R. Gill, M. J. Callanan, D. J. O'Sullivan, D. N. Dowling, and F. O'Gara. 1995. Iron-responsive gene expression in Pseudomonas fluorescens M114: cloning and characterization of a transcription-activating factor, PbrA. Mol. Microbiol. 15:297-306. [DOI] [PubMed] [Google Scholar]
- 34.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 35.Stiefel, A., S. Mahren, M. Ochs, P. T. Schindler, S. Enz, and V. Braun. 2001. Control of the ferric citrate transport system of Escherichia coli: mutations in region 2.1 of the FecI extracytoplasmic-function sigma factor suppress mutations in the FecR transmembrane regulatory protein. J. Bacteriol. 183:162-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson, J. M., and W. A. Parrott. 1998. pMECA: a cloning plasmid with 44 unique restriction sites that allows selection of recombinants based on colony size. BioTechniques 24:922-924. [DOI] [PubMed] [Google Scholar]
- 37.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanderpool, C. K., and S. K. Armstrong. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Hove, B., H. Staudenmaier, and V. Braun. 1990. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J. Bacteriol. 172:6749-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venturi, V., C. Ottewanger, J. Leong, and P. J. Weisbeek. 1995. Iron regulation of siderophore biosynthesis and transport in Pseudomonas putida WCS358: involvement of a transcriptional activator and of the Fur protein. Mol. Microbiol. 15:1081-1093. [DOI] [PubMed] [Google Scholar]
- 41.Ward, M. J., H. Lew, A. Treuner-Lange, and D. R. Zusman. 1998. Regulation of motility behavior in Myxococcus xanthus may require an extracytoplasmic function sigma factor. J. Bacteriol. 180:5668-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watnick, P. I., J. R. Butterton, and S. B. Calderwood. 1998. The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes, irgA and irgB. Gene 209:65-70. [DOI] [PubMed] [Google Scholar]
- 43.Welz, D., and V. Braun. 1998. Ferric citrate transport of Escherichia coli: functional regions of the FecR transmembrane regulatory protein. J. Bacteriol. 180:2387-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wosten, M. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 45.Xiong, Y., M. L. Vasil, Z. Johnson, U. A. Ochsner, and A. S. Bayer. 2000. The oxygen- and iron-dependent sigma factor pvdS of Pseudomonas aeruginosa is an important virulence factor in experimental infective endocarditis. J. Infect. Dis. 181:1020-1026. [DOI] [PubMed] [Google Scholar]