Abstract
Apical membrane antigen 1 (AMA1) is expressed on the surfaces of Plasmodium falciparum merozoites and is thought to play an important role in the invasion of erythrocytes by malaria parasites. To select for peptides that mimic conformational B-cell epitopes on AMA1, we screened a phage display library of >108 individual peptides for peptides bound by a monoclonal anti-AMA1 antibody, 4G2dc1, known to inhibit P. falciparum invasion of erythrocytes. The most reactive peptides, J1, J3, and J7, elicited antibody responses in rabbits that recognized the peptide immunogen and both recombinant and parasite AMA1. Human antibodies in plasma samples from individuals exposed to chronic malaria reacted with J1 and J7 peptides and were isolated using immobilized peptide immunoadsorbents. Both rabbit and human antibodies specific for J1 and J7 peptides were able to inhibit the invasion of erythrocytes by P. falciparum merozoites. This is the first example of phage-derived peptides that mimic an important epitope of a blood-stage malaria vaccine candidate, inducing and isolating functional protective antibodies. Our data support the use of J1 and J7 peptide mimics as in vitro correlates of protective immunity in future AMA1 vaccine trials.
Apical membrane antigen 1 (AMA1) is a leading malarial vaccine candidate, since many studies have demonstrated that recombinant AMA1 induces antibodies that protect against plasmodial infection in simian and rodent models of the human disease (6, 22). AMA1 appears to have an essential role in the erythrocyte invasion process, and anti-AMA1 monoclonal, polyclonal, and Fab′ fragment antibodies have been shown to inhibit this process in vitro (10, 23). AMA1 is a type I integral protein with eight intramolecular disulfide bonds in the ectodomain, which suggests a three-domain substructure (9). The constrained nature of AMA1 was found to be critical, since antibodies generated toward reduced and alkylated (R-A) AMA1 failed to inhibit invasion (2). This evidence strongly suggests that conformation stabilized by disulfide bonds is important for the generation of a protective immune response.
The monoclonal antibody (MAb) 4G2dc1, which binds to correctly folded AMA1 but not to the R-A protein, is a useful reagent for monitoring correct disulfide bonding of recombinant AMA1 (12). In previous studies, 4G2dc1 was shown to react with recombinant AMA1 in 10 different isolates of Plasmodium falciparum from diverse geographical locations, and it consistently inhibited the invasion of P. falciparum merozoites into erythrocytes in vitro by 60 to 70% (12). 4G2dc1 also inhibited invasion by Plasmodium reichenowi parasites in vitro by 44% at 1 mg/ml (13). The exact location of the 4G2dc1 conformational epitope is not known, but clearly it has significance with respect to the generation of protective antibodies that can inhibit merozoite invasion (14).
In this study, we have used phage display technology to isolate peptide sequences that mimic the conformation of the 4G2dc1 epitope. Immunization of rabbits with the peptide mimotopes induced high titers of peptide antibodies, which were reactive with native AMA1. Human antibodies were affinity purified from plasma samples from individuals living in regions of Papua New Guinea (PNG) where malaria is endemic, using immobilized peptide immunoadsorbents. Both rabbit and affinity-purified human antibodies inhibited the invasion of human erythrocytes by P. falciparum merozoites in vitro. Our data support the use of 4G2dc1 peptide mimics as in vitro correlates of protective immunity in future AMA1 vaccine trials.
MATERIALS AND METHODS
Phage library and selection.
We constructed a linear peptide library of 20 random amino acids displayed as N-terminal fusions to protein III of filamentous phage M13, using the Fuse 5 vector (20). The library of >5 × 108 random peptides was screened for MAb 4G2dc1-binding peptides using microtiter plates as described previously (1), with the following modifications. 4G2dc1-coated wells were blocked with 5% BLOTTO (milk powder diluted in phosphate-buffered saline [PBS]), and the 20-mer phage peptide library was preincubated in 1% BLOTTO for 15 min to remove any milk-binding phage before being added to the 4G2dc1-coated wells. Phage was amplified and titrated, and DNA sequencing was performed using procedures similar to those described previously (1).
Recombinant antigens.
The ectodomain of AMA1 from the 3D7 strain of P. falciparum was expressed in Escherichia coli with an N-terminal six-His tag to allow purification by Ni chelate chromatography using methods detailed elsewhere (10). The 3D7 strains of merozoite surface antigen 2 (MSP2) and MSP3 were expressed in E. coli and prepared using similar methods (25). The recombinant antigen designated Ag1505H, corresponding to ∼70% of the C terminus of the ring-stage parasite-infected erythrocyte surface antigen (RESA) polypeptide, was also used as a control antigen (25).
ELISAs.
Phage enzyme-linked immunosorbent assays (ELISAs) were performed by coating a microtiter plate (Nunc Maxisorp) with 5 μg of antibody/ml overnight at 4°C and subsequently blocking it with 10% skim milk for 2 h. Phage dilutions (100 μl) were prepared in PBS, transferred in duplicate to the coated blocked wells, and incubated for 1 h with shaking. The wells were washed five times with PBS containing 0.05% Tween 20, and 100 μl of anti-M13 antibody conjugated to horseradish peroxidase (HRP) (Pharmacia) at 1/5,000 dilution was added to each well. After a further 1-h incubation and washing as described above, bound phage was detected with o-phenylenediamine substrate (Sigma). A similar format was used for AMA1 and other antigen ELISAs, using 5 μg/ml for coating, and anti-human immunoglobulin G (IgG)-HRP or anti-rabbit IgG-HRP (both at 1/1,000; Silenus) conjugates were added for 1 h. For competition experiments with AMA1 or peptide, the same concentration of analyte (50 μl) was mixed with various concentrations of competitor (50 μl) for 30 min before being adding to the coated wells, and the same procedure was followed.
Peptide immobilizer plates (Exiqon, Vedbaek, Denmark) were used for synthetic peptide ELISAs. Briefly, the wells were coated with 10 μg of peptide/ml overnight at 4°C with gentle agitation; the wells were then washed three times with PBS-Tween, and dilutions of antibody or plasma in PBS-Tween were added for 1 h. Following a further five washes, the appropriate anti-human or anti-rabbit IgG-HRP conjugate was added for 1 h. The wells were washed five times, and peptide-reactive antibody was detected with 3,3′, 5,5′-tetramethylbenzidine substrate (Sigma).
All ELISAs were performed in duplicate, and the means ± individual values are shown in the figures.
Peptide synthesis and conjugations.
Peptides were synthesized to >70% purity by AusPep Pty Ltd. (Melbourne, Australia). For immunizations, the peptides were conjugated via the N terminus to keyhole limpet hemocyanin (KLH) with glutaraldehyde (19).
Immunizations and purification of peptide antisera.
New Zealand White rabbits were injected intramuscularly with 200 μg of peptide-KLH conjugates emulsified in 0.5 ml of Freund's complete adjuvant. Two biweekly booster doses followed by three additional booster doses with 0.5 mg of free peptide diluted 1:1 in Freund's incomplete adjuvant were performed.
An affinity column consisting of 5 mg of 3D7 AMA1 coupled to 5 ml of CNBr-activated Sepharose 4B (Pharmacia Biotech) was prepared according to the manufacturer's instructions. Antisera were diluted 1:5 in PBS and passed through a 0.2-μm-pore-size syringe filter prior to being applied to the affinity resin. AMA1-binding antibodies were eluted with 0.1 M glycine (pH 2.2), neutralized, and dialyzed against PBS.
Affinity purification of human IgG using mimotopes.
Affinity matrices were prepared by conjugating 2 mg of J1 or J7 peptide to 2 ml of N-hydroxysuccinimide-activated Sepharose 4B (Pharmacia Biotech) according to the manufacturer's instructions. Immobilized peptides were used to affinity purify antibodies from human plasma using a procedure similar to that described above. Plasma samples (outdated samples) were obtained from the PNG Red Cross Blood Transfusion Service (5).
Western blotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%) under nonreducing conditions and transferred to polyvinylidene difluoride membranes (Millipore). The membranes were blocked overnight with 10% BLOTTO and probed with antipeptide antibodies (10 μg/ml), followed by goat anti-rabbit (1/2,000) antibody conjugated to HRP (Amersham-Pharmacia). Protein binding was detected using enhanced-chemiluminescence reagent (Pierce).
Merozoite invasion-inhibition and immunofluorescence.
P. falciparum parasites (3D7 line) were cultured in vitro, and invasion-inhibition experiments were performed as described previously (17). Briefly, late trophozoite or early schizont stage 3D7 parasites were purified with Percoll following synchronization with sorbitol. Parasites were adjusted to 1% parasitemia and 2% hematocrit with uninfected erythrocytes. Antibodies in PBS (100 μl) were added to 100 μl of parasites in 96-well plates, and the cultures were incubated for 24 h at 37°C in 1% O2-5% CO2 in N2. After 24 h, the level of parasitemia was ∼6 to 10% for PBS controls, and thin smears were made, fixed in methanol, and stained with Giemsa stain. The number of ring stage parasites per 1,000 erythrocytes was determined, and the percentage of inhibition of invasion was calculated by comparison to the PBS control and expressed as the mean of duplicate wells. For a positive control, anti-AMA1 polyclonal antibodies (affinity purified on protein G) generated by immunizing rabbits with recombinant AMA1 were used (10). Rabbit and human antibodies, also protein G purified with very low binding to AMA1 by ELISA, were used as negative control antibodies.
For immunofluorescence experiments, synchronized schizont stage parasites were prepared as described previously (10). Affinity-purified antibodies were diluted in 0.5% bovine serum albumin (BSA)-PBS, layered onto the slides, and allowed to incubate for 1 h at room temperature in a humidified container. The slides were washed extensively with PBS-0.5% BSA, and anti-human-fluorescein isothiocyanate (FITC) (Silenus), anti-rat-rhodamine (Jackson Immunoresearch Laboratories), or anti-rabbit-FITC (Silenus) conjugate, all at 1:200 dilutions in 0.5% BSA-PBS, was allowed to incubate for 1 h. The slides were washed as described above, mounted with glycerol, and examined by fluorescence microscopy.
RESULTS
Screening for AMA1 mimotopes.
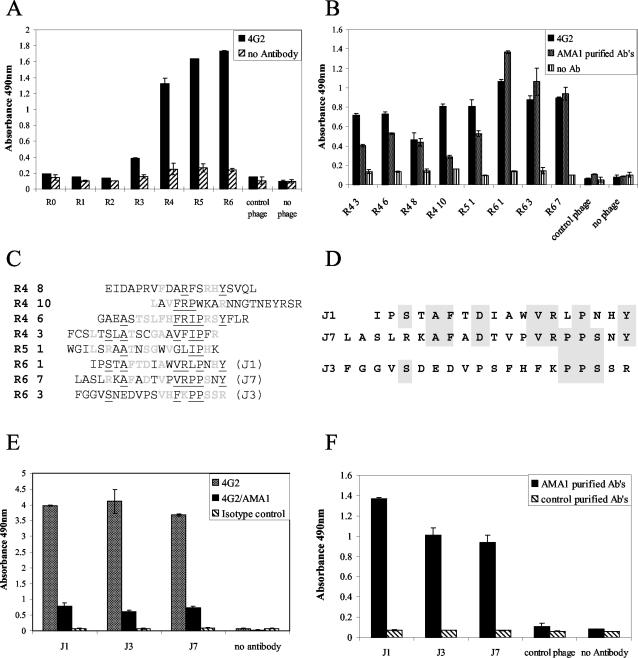
Peptides mimicking the 4G2dc1 epitope of AMA1 were selected from a 20-mer random linear peptide library by multiple rounds of panning on antibody-coated microtiter plates. To bias selection toward peptides binding with high affinity, the stringency of washing was increased with successive rounds of panning. An increased number of bound phage was detected after the third round of panning, with further increases in subsequent rounds (Fig. 1A). Eight phage clones shown by DNA sequencing to be displaying different peptides were selected in rounds 4 to 6, and their levels of binding to 4G2dc1 were compared by ELISA (Fig. 1B). The three clones with the highest binding signals in ELISA (J1, J3, and J7), all isolated from the sixth round of panning, were selected for further study.
FIG. 1.
Selection and characteristics of phage clones binding to MAb 4G2dc1. (A) Reactivities of selected phages from each round (R) of panning on 4G2dc1 detected by ELISA. (B) Binding of eight selected clones to 4G2dcl and human anti-AMA1 antibodies (Ab) affinity purified from a pool of plasma samples from PNG by ELISA. (C) Sequences of eight selected clones from rounds (R) 4 to 6. Amino acids that were identical in two sequences are in light gray, and those identical in three or more sequences are underlined. (D) Aligned amino acid sequences of J1, J3, and J7 selected phage peptides; the shaded areas indicate identical amino acids. (E) Selected phage clones bound to 4G2dc1, as shown by ELISA, but not to an antibody of the same isotype. Recombinant AMA1 (10 μg/ml) competed with the phage clones for binding to 4G2dc1. (F) Selected phage clones bound to human anti-AMA1 antibodies affinity purified from a pool of plasma samples from PNG but not to purified human IgG from individuals not exposed to malaria. Error bars indicate the ranges of individual values.
None of the eight peptide sequences had any obvious relationship to the primary sequence of AMA1. This was not unexpected, since the epitope of 4G2dc1 is known to be conformational. In addition, there was a lack of a linear motif common to all eight sequences (Fig. 1C). However, of the three peptides selected for further study, the J1 and J7 sequences were clearly related: both contained the motif -AFXDXXXVRXPXXY- (Fig. 1D). This region of the peptides consists of largely hydrophobic residues: A, F followed by an acidic residue, D, three or four hydrophobic residues, and then a V and a basic R residue, followed by a proline, which would provide a turn in the predicted structure of the peptides. The J3 sequence was unrelated to the J1 sequence, but the motif -PPS- was common to both J3 and J7.
Importantly, the binding of all three phage clones to 4G2dcl was inhibited by AMA1, and none of the three bound to an isotype control MAb (Fig. 1E). Thus, the peptides appear to be true epitopic mimics, which share the antibody binding specificity of the 4G2dc1 epitope on AMA1.
Previous studies have shown the 4G2dc1 epitope to be a target of anti-AMA1 antibodies induced by malarial infections (12). Consistent with this observation, phage clones displaying J1, J3, and J7 peptide mimics of the 4G2dc1 epitope bound antibodies in the plasma of individuals living in regions of PNG where malaria is endemic. No such antibodies were detected in plasma from individuals not exposed to malaria (Fig. 1F).
Synthetic peptides corresponding to J1, J3, and J7 were also bound by 4G2dc1 (data not shown), confirming that the 4G2dc1 epitope was contained within the peptide sequence and was not dependent on the phage context.
Mimotopes induce AMA1-specific antibodies.
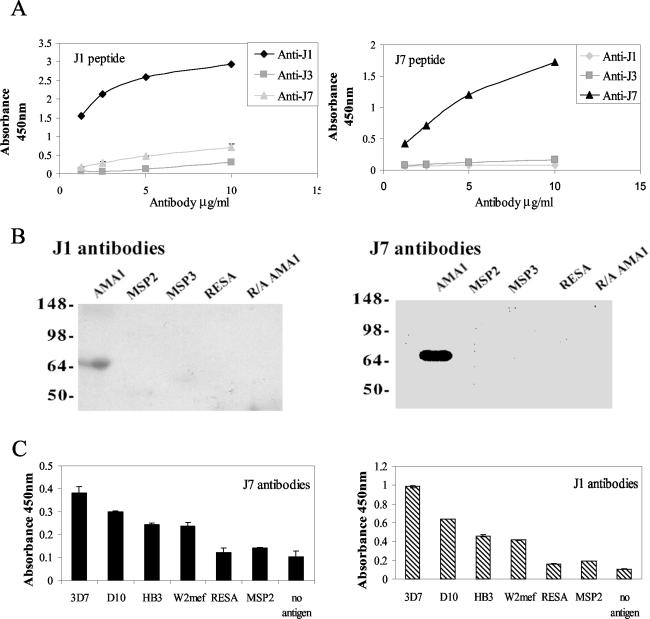
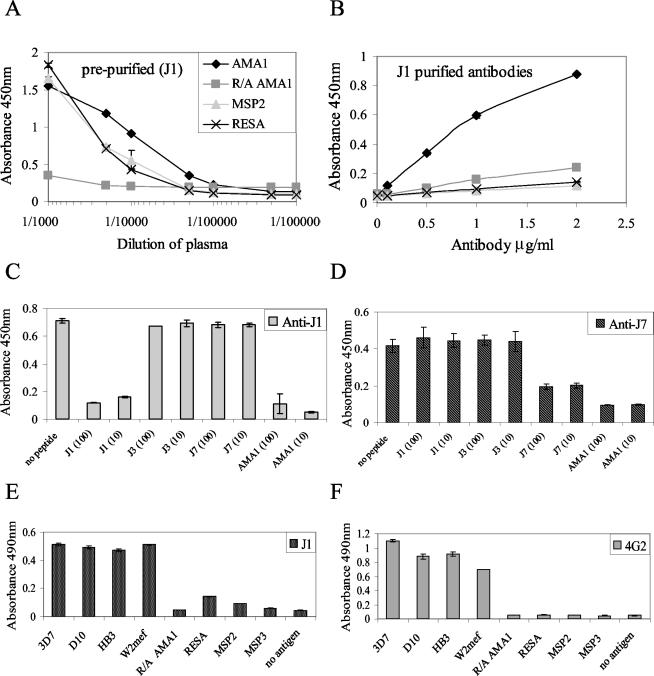
Immunization of rabbits with peptides conjugated to KLH resulted in high titers of peptide-specific antibodies (>1/1,000); further booster doses with free peptide increased the titer for J1 and J7 (>1/10,000), but not for J3 (>1/2,000). Because of the weak immune response generated by J3, the peptide was not characterized further. These rabbit antisera also reacted with recombinant AMA1, indicating that the peptides behave as immunogenic mimics of the natural antigen. Anti-AMA1 antibodies affinity purified from the antipeptide antisera were shown by ELISA to be specific for the immunizing peptide with no cross-reaction with the other mimotopes (Fig. 2A). This was surprising, since the peptides are mimicking the same epitope shape. Antibodies affinity purified on J1 and J7 peptides bound specifically to AMA1 and showed little binding to several other P. falciparum antigens (Fig. 2B and C). Significantly, antibodies generated by both J1 and J7 peptides did not react with R-A AMA1, indicating that these antibodies recognize epitopes that are dependent on intramolecular disulfide bonds, a characteristic of the 4G2dc1 epitope (12). Both J1 and J7 antibodies bound to AMA1 expressed by four different lines of P. falciparum (Fig. 2C), and the degree of reactivity reflected the sequence differences among the four forms of AMA1 studied.
FIG. 2.
Specificities of anti-J1 and anti-J7 rabbit antibodies affinity purified on AMA1. (A) ELISA showing antibodies binding to soluble peptide on microtiter plates. (B) Immunoblots showing specific reactivity with recombinant AMA1. (C) ELISA showing that antibodies bind to bacterially expressed recombinant AMA1 from four P. falciparum lines. Error bars indicate the ranges of individual values.
AMA1 antibodies from human plasma samples bind to mimotopes.
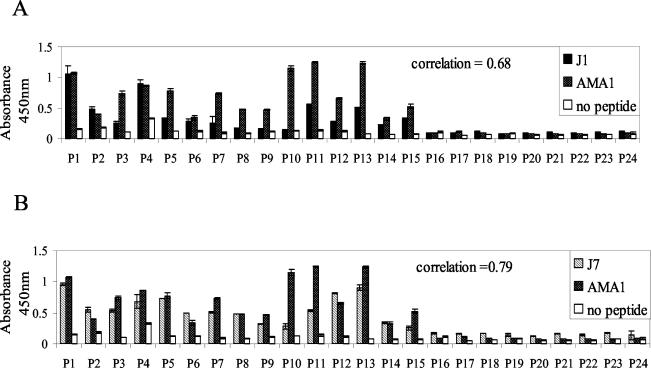
A panel of plasma samples from PNG blood donors was analyzed for reactivity to AMA1 and to the J1 and J7 peptides (Fig. 3). Plasma antibodies in most, but not all, samples recognized the peptides, and recognition correlated with the AMA1 antibody titer. The correlation coefficients (Microsoft Excel; a perfect correlation was equal to 1.0) of the two sets of data were analyzed; J7 showed a slightly higher correlation with AMA1 (0.78) than J1 (0.69). However, antibody titers to J1 and J7 were low, which was not unexpected, as they would encode a single or a very restricted number of AMA1 epitopes.
FIG. 3.
Human antibody titer to AMA1 correlates with the titer to J1 and J7 peptides. Plasma samples (P1 to P 24) from individuals living in an area of PNG where malaria is endemic were assayed by ELISA at a dilution of 1:10,000 on AMA1 and 1:100 on J1 (A) and J7 (B). The correlation coefficient was used to compare data sets. Error bars indicate the ranges of individual values.
Mimotopes as affinity ligands.
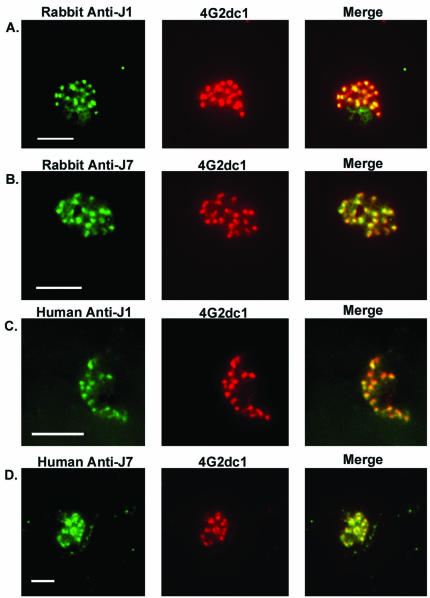
Peptides (J1 and J7) were used to affinity purify antibodies from the plasma of individuals who had been exposed to chronic malaria infections. Approximately 100 to 300 μg of antibody was purified from 5 ml of high-titer plasma. To assess the reactivities of antipeptide antibodies with the native antigen, we examined P. falciparum-infected erythrocytes by immunofluorescence (Fig. 4). A distinct punctate pattern typical of apical organelle location within the merozoite was observed for both purified human and rabbit antipeptide antibodies (Fig. 4, left). This observation agrees with previous reports of the location of AMA1 in rhoptries prior to its release onto the merozoite surface (10, 12). An identical staining pattern was observed for 4G2dc1 (Fig. 4, middle), and when the two images were merged, colocalization was apparent (Fig. 4, right). When free peptide was incubated with anti-J1 or anti-J7 antibodies in this assay, there was total elimination of fluorescence, indicating that the binding was peptide specific (data not shown), emphasizing again the capacity of the peptide to mimic the 4G2dc1 epitope of AMA1.
FIG. 4.
Colocalization of MAb 4G2dc1 and antipeptide antibodies. Immunofluorescence analysis of parasite smears with rabbit anti-J1 (A), rabbit anti-J7 (B), human anti-J1 (C), and human anti-J7 (D) purified antibodies probed with FITC (left), 4G2dc1 probed with anti-rat rhodamine (middle), and merged images (right). Scale bars, 5 μm.
The human plasma samples used in the immunofluorescence experiments were characterized prior to purification and demonstrated a high level of binding to AMA1 and other malaria antigens (MSP2 and RESA), indicating prior exposure to these antigens. There was a relatively low level of binding to R-A AMA1, implying that these individuals produced antibodies mainly to conformational epitopes and not to linear epitopes of AMA1 (Fig. 5A). After purification, the eluted antibodies showed higher relative binding to AMA1 than to RESA and MSP2, indicating enrichment for AMA1 antibodies that bind the mimotope sequences (Fig. 5B). The purified antibodies were found to be highly specific for the peptide they were purified against, as the J1 peptide, but not the J7 and J3 peptides, inhibited binding of J1 antibodies to AMA1 (Fig. 5C); similar specificity was observed for J7 antibodies (Fig. 5D). Again, as for the rabbit anti-J1 and anti-J7 antibodies, this was surprising, since the peptides are mimicking the same epitope and it might be predicted that there would be some degree of cross-reactivity of the antibodies. The J1 and J7 peptides could form a different structure in solution when anchored to the carrier molecule KLH for immunization studies or when bound to microtiter plates, which could account for the antisera not cross-competing.
FIG. 5.
Specificities of J1 and J7 affinity-purified human antibodies. ELISAs show human plasma antibodies binding to antigens before (A) and after (B) purification on immobilized J1. AMA1 and J1 (C) or J7 (D) peptide at 10 and 100 μg/ml competed with J1 (C) or J7 (D) affinity-purified human antibodies for binding to AMA1. J1 antibodies (E) and 4G2dc1 (F) bound to bacterially expressed recombinant AMA1 from four P. falciparum lines. Error bars indicate the ranges of individual values.
Recombinant AMA1 was also found to inhibit the binding of both J1 and J7 purified antibodies to immobilized peptides. Purified J1 antibodies showed a binding profile remarkably similar to that of 4G2dc1 (Fig. 5E and F) in recognizing AMA1 from four different strains of P. falciparum and in not binding to other malaria antigens, such as RESA and MSP2. Importantly, the peptide-purified antibodies did not recognize R-A AMA1, reflecting the specificity of the 4G2dc1 antibody used in the original panning. J7 antibodies showed an identical pattern of binding (data not shown). These results clearly illustrate an enrichment of AMA1 antibodies with binding properties similar to those of 4G2dc1.
Antipeptide antibodies inhibit P. falciparum invasion of erythrocytes.
The MAb 4G2dc1 has previously been reported to inhibit P. falciparum invasion of erythrocytes by 60 to 70% at 1 mg/ml (12). It is generally recognized that the ultimate test of a mimotope is the capacity to induce an immune response that has the biological activity of the original antibody used to select the peptide. Anti-J1 and anti-J7 antibodies were found to inhibit merozoite invasion by 53 and 47% at 1 mg/ml compared with polyclonal rabbit anti-AMA1 antibodies as a positive control. Rabbit IgG control antibodies did not inhibit invasion (Table 1). For comparison, antibodies raised to the J3 peptide did not have a significant effect on invasion (17%), presumably reflecting differences in the antibody titer and the specificity of the antibody response to this peptide. In addition, human antibodies purified using immobilized J1 and J7 peptides were found to inhibit merozoite invasion by 41 and 43%, respectively, at 0.5 mg/ml compared with a control human IgG.
TABLE 1.
Inhibition of merozoite invasion of AMA1-purified rabbit antipeptide antibodies and peptide-purified human antibodies
| Antibody | Inhibition (%)a
|
|
|---|---|---|
| Rabbit (final concn, 1 mg/ml) | Human (final concn, 0.5 mg/ml) | |
| J1 | 53 | 41 |
| J3 | 17 | ND |
| J7 | 47 | 43 |
| IgG (control) | 0 | 0 |
| Anti-AMA1 | 95 | 82 |
Values represent the averages of duplicate assays. ND, not done.
DISCUSSION
The major antigenic determinant regions of AMA1 have not yet been characterized in any detail, although the disulfide bond-dependent conformation of AMA1 is essential for the induction of antibodies that block erythrocyte invasion by merozoites (2). The epitopes of two MAbs have been mapped using phage-displayed fragments of AMA1 (4), but this approach failed to identify the epitope of the inhibitory MAb 4G2dc1. 4G2dc1 binds to a reduction-sensitive epitope on AMA1 and is of particular interest because the epitope is conserved in all lines of P. falciparum that have been examined.
As an alternate approach to characterizing the 4G2dc1 epitope, we selected peptide mimotopes of the epitope from a phage-displayed random peptide library. We chose to construct a 20-residue library, which should provide almost complete coverage of short (5-, 6-, and 7-amino-acid) linear peptides, with peptides long enough to permit short turns and other structural features that could enhance the chance of selecting peptides with a shape complementary to the antibody paratope. Eight independent clones selected from the library displayed different peptide sequences, with no consensus sequence and no apparent sequence identity with AMA1. Reaction with these diverse peptides probably reflects the conformational nature of the 4G2dc1 epitope. It is not uncommon to find a number of peptides with different sequences binding to a single MAb that recognizes a discontinuous or conformational epitope (15). A single antibody may include several nonoverlapping and partially overlapping paratopes within the antigen-binding site defined by the complementarity-determining regions (15). Furthermore, it has recently been discovered that MAbs can adopt different binding site conformations and exist naturally as isomers (11). Consistent with this observation, the J1 and J7 peptides, both selected in round 6 of the panning process, bound more strongly to one isomer of 4G2dc1, perhaps providing a superior fit to the antigen-binding groove, whereas other, weaker binders in earlier rounds might only partially mimic the shape of the epitope and therefore could bind with lower affinity to another isomer of 4G2dc1.
Previous studies have identified peptides that mimic epitopes of other malarial antigens (1, 7, 21). Indeed, one of the first studies to show that peptide mimics could be selected on antibodies was done with a MAb to the sexual-stage malaria antigen Pfs25 (24). However, our study is the first to prove that a peptide mimic of an epitope in a malaria antigen can elicit a humoral response similar to that evoked by the original antigen.
The antipeptide antibodies raised by immunizing rabbits with the J1 or J7 peptide recognized both native merozoite AMA1 and recombinant, refolded AMA1 from four different strains of P. falciparum, but not R-A AMA1 or irrelevant plasmodial proteins. Additionally, antibodies affinity purified on the peptides from plasma samples from individuals with a history of malaria infection bound AMA1 irrespective of the strain from which the antigen was derived, again reflecting the binding characteristics of the parent antibody, 4G2dc1. The induction of an inhibitory immune response and the ability to selectively purify human antibodies that recognize AMA1 from a variety of P. falciparum strains suggests that the peptides have a three-dimensional structure that closely resembles that of the 4G2dc1 epitope on the folded surface of AMA1 and are true mimics of 4G2dc1.
Interestingly, the immune response generated by each 4G2dc1 peptide mimic was highly specific for the individual peptide. Similarly, antibodies purified from human plasma were also highly specific for the peptide used for their purification. This was surprising, since J1 and J7 share a common sequence that may be important for binding. Furthermore, soluble J1 and J7 peptides were found to cross-compete (albeit at concentrations of soluble peptide of ≥1 mg/ml) with the phage clones for binding to 4G2dc1 (data not shown). The behavior of the peptides when bound to microtiter plates could differ when in solution, on phage, or attached to KLH for immunizing, and this needs to be investigated further. The contributions of particular residues to the antigenicity, immunogenicity, and structural integrity of the mimotopes are not known, and we are analyzing the nuclear magnetic resonance structures of J1, J3, and J7 and mutating common residues to determine their importance in binding, with a view to potentially improving the binding affinity. This may result in an optimized consensus peptide that has been designed to more accurately resemble the atomic structure of the 4G2dc1 epitope.
Several groups have proposed the use of peptide mimics that can induce protective antibodies as potential vaccine candidates (15, 7). However, it is unlikely that J1 and J7 peptides by themselves will be useful as vaccines, since they are specific for only one epitope and relatively high concentrations of peptide antibodies (0.5 to 1 mg/ml) were required to inhibit merozoite invasion. It should be noted, however, that other groups have found that equally high concentrations of MAbs to AMA1 and other merozoite surface antigens were required for inhibition (3, 12). Immunization with both J1 and J7 peptides, or perhaps a combination of all of the selected 4G2dc1 epitope mimics, may improve the potency of the immune response. Peptide vaccines can also be designed to stimulate specific B- and T-cell responses. In a recent study, chimeric peptides consisting of both T helper cell epitope and mimotope entities produced B- and T-cell responses that induced up to 100% protection of mice from measles virus-induced encephalitis (18). Therefore, future studies could also be directed toward tailoring the peptides to generation of both B- and T-cell responses.
A more immediate use for J1, J7, and other peptide mimotopes might be as surrogates of AMA1 in order to establish in vitro correlates of immunity. Immunization with AMA1, or infection, generates antibodies to many linear and conformational epitopes (10); however, it is assumed that only a small proportion of these will be important in protective immunity. A set of peptide mimotopes with the ability to distinguish between antibodies that inhibit merozoite invasion and irrelevant antibodies would be very useful for monitoring the generation of a protective immune response in human vaccine trials with AMA1.
Here, we have demonstrated that J1 and J7 mimotopes specific for a single epitope of AMA1 can recognize antibodies in human plasma samples from individuals exposed to malaria. The fact that individuals possess antibodies against short peptides selected from random libraries that they would not have been previously exposed to again emphasizes the mimetic nature of the peptides.
The detection of human antibodies to the J1 and J7 mimotopes is consistent with the observation that there are human antibodies that recognize the MAb 4G2dc1 epitope (13). However, because a single MAb represents a small proportion of the polyclonal antibody response, we are screening peptide libraries on plasma samples from individuals who have generated a high titer of protective polyclonal antibodies specific for AMA1 and other malarial antigens. Extensive screening of individual plasma samples using these peptides could reveal a correlation between mimotope binding and clinical outcome, such as severe malaria. Furthermore, this strategy may also be useful for identifying many new protective epitopes, since the methodology does not necessarily require knowledge of the original ligand (8). Recently, an antigen thought to be involved in metastatic disease progression was identified by using peptide mimics against circulating antibodies in the sera of cancer patients (16).
Recombinant AMA1 has been extensively studied as a potential malarial vaccine component, and clinical trials have commenced. At present, however, there are no cheap and reliable in vitro correlates of immunity for AMA1 or any other malaria vaccine candidates under clinical development. The technology used in this study could be applied to many other protective malaria antibodies and may provide a means of overcoming what has been viewed as a major hurdle in the development of an effective malaria vaccine.
Acknowledgments
This work was supported by the Cooperative Research Center for Diagnostics.
We thank George Smith (University of Missouri, Columbia) and Daniel Rajotte (Burnham Institute, La Jolla, Calif.) for providing the Fuse 5 vector and MC1061 cell line, Rosella Masciantonio (La Trobe University) for providing AMA1, and Andrew Pierce (WEHI) for providing MSP3.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Adda, C., L. M. Tilley, R. F. Anders, and M. Foley. 1999. Isolation of peptides that mimic epitopes on a malarial antigen from random peptide libraries displayed on phage. Infect. Immun. 67:4679-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunization with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 3.Blackman, M. J., H. G. Heidrich, S. Donachie, J. S. McBride, and A. A. Holder. 1990. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J. Exp. Med. 172:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coley, A. M., N. V. Campanle, J. L. Casey, A. N. Hodder, P. E. Crewther, R. F. Anders, L. M. Tilley, and M. Foley. 2001. Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides. Protein Eng. 14:691-698. [DOI] [PubMed] [Google Scholar]
- 5.Crewther, P. E., E. Bianco, G. V. Brown, R. L. Coppel, H. D. Stahl, D. J. Kemp, and R. F. Anders. 1986. Affinity-purification of human antibodies directed against cloned antigens of Plasmodium falciparum. J. Immunol. Methods 86:257-264. [DOI] [PubMed] [Google Scholar]
- 6.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune response to apical membrane antigen 1 of Plasmodium chabaudi involves recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demagel, C., S. Rouyre, F. N. Alzari, S. Longacre, P. Lafaye, and J. C. Mazie. 1998. Phage-displayed mimotopes elicit monoclonal antibodies specific for a malaria vaccine candidate. Biol. Chem. 379:65-70. [DOI] [PubMed] [Google Scholar]
- 8.Folgori, A., R. Tafi, A. Meola, F. Felici, G. Galfre, R. Cortese, P. Monaci, and A. Nicosa. 1994. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 13:2236-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen 1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 10.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James, L. C., P. Roversi, and D. S. Tawfik. 2003. Antibody multispecificity mediated by conformational diversity. Science 299:1362-1367. [DOI] [PubMed] [Google Scholar]
- 12.Kocken, C. H. M., A. M. van der Wel, M. A. Dubbeld, D. L. Narum, F. M. van de Rijke, G. J. V. Gemert, X. V. Linde, L. H. Bannister, A. P. Waters, and A. W. Thomas. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 273:15119-15124. [DOI] [PubMed] [Google Scholar]
- 13.Kocken, C. H. M., D. L. Narum, A. Massougbodiji, B. Ayivi, M. A. Dubbeld, A. van der Wel, D. J. Conway, A. Sanni, and A. W. Thomas. 2000. Molecular characterization of Plasmodium reichenowi apical membrane antigen 1 (AMA1), comparison with P. falciparum AMA1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 109:147-156. [DOI] [PubMed] [Google Scholar]
- 14.Kocken, C. H. M., C. Withers-Martinez, M. A. Dubbeld, A. van der Wel, F. Hackett, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 70:4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meloen, R., W. Puijk, and J. Slootstra. 2000. Mimotopes: realization of an unlikely concept. J. Mol. Recognit. 13:352-359. [DOI] [PubMed] [Google Scholar]
- 16.Mintz, P. J., J. Kim, K. A. Do, X. Wang, R. G. Zinner, M. Cristofanilli, M. A. Arap, W. K. Hong, P. Troncoso, C. J. Logothetis, R. Pasqualini, and W. Arap. 2003. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat. Biotechnol. 21:57-62. [DOI] [PubMed] [Google Scholar]
- 17.Nair, M., M. G. Hinds, A. M. Coley, A. N. Hodder, M. Foley, R. F. Anders, and R. S. Norton. 2002. Structure of domain III of the blood-stage malaria vaccine candidate Plasmodium falciparum apical membrane antigen 1. J. Mol. Biol. 322:741-753. [DOI] [PubMed] [Google Scholar]
- 18.Olszewska, W., C. D. Partidos, and M. W. Steward. 2000. Antipeptide antibody responses following intranasal immunization: effectiveness of mucosal adjuvants. Virology 272:98-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reichlin, M. 1980. Use of glutaraldehyde as a coupling agent for proteins and peptides. Methods Enzymol. 70:159-165. [DOI] [PubMed] [Google Scholar]
- 20.Smith, G. P., and J. K. Scott. 1993. Libraries of peptides and proteins displayed on phage. Methods Enzymol. 217:228-257. [DOI] [PubMed] [Google Scholar]
- 21.Stoute, J., W. Ballou, N. Kolodny, C. Deal, R. L. Wirtz, and L. Lindler. 1995. Induction of humoral immune response against Plasmodium falciparum sporozoites by immunization with a synthetic peptide mimotope whose sequence was derived from screening a filamentous phage epitope library. Infect. Immun. 63:934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stowers, A. W., M. C. Kennedy, B. P. Keegan, A. Saul, C. Long, and L. H. Miller. 2002. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect. Immun. 70:6961-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas, A. W., J. A. Deans, G. H. Mitchell, T. Alderson, and S. Cohen. 1984. The Fab fragments of monoclonal IgG to a merozoite surface antigen inhibit Plasmodium knowlesi invasion of erythrocytes. Mol. Biochem. Parasitol. 13:187-199. [DOI] [PubMed] [Google Scholar]
- 24.van Amerongen, A., P. A. Beckers, H. Plasman, R. Sauerwein, and R. Meloen. 1992. Peptides reactive with a transmission-blocking MAb against Plasmodium falciparum Pfs25: 2000-fold affinity increase by PEPSCAN-based amino acid substitutions. Peptide Res. 5:269-274. [PubMed] [Google Scholar]
- 25.Yaman, F. A., B. Genton, R. Anders, J. Taraika, M. Ginny, S. Mellor, and M. P. Alpers. 1995. Assessment of the role of the humoral response to Plasmodium falciparum MSP2 compared to RESA and SPf66 in protecting Papua New Guinean children from clinical malaria. Parasite Immunol. 17:493-501. [DOI] [PubMed] [Google Scholar]