Before chromosome expulsion into polar bodies during female meiosis, the APC inhibits CDK-1 to allow dynein-driven spindle rotation.
Abstract
In animals, the female meiotic spindle is positioned at the egg cortex in a perpendicular orientation to facilitate the disposal of half of the chromosomes into a polar body. In Caenorhabditis elegans, the metaphase spindle lies parallel to the cortex, dynein is dispersed on the spindle, and the dynein activators ASPM-1 and LIN-5 are concentrated at spindle poles. Anaphase-promoting complex (APC) activation results in dynein accumulation at spindle poles and dynein-dependent rotation of one spindle pole to the cortex, resulting in perpendicular orientation. To test whether the APC initiates spindle rotation through cyclin B–CDK-1 inactivation, separase activation, or degradation of an unknown dynein inhibitor, CDK-1 was inhibited with purvalanol A in metaphase-I–arrested, APC-depleted embryos. CDK-1 inhibition resulted in the accumulation of dynein at spindle poles and dynein-dependent spindle rotation without chromosome separation. These results suggest that CDK-1 blocks rotation by inhibiting dynein association with microtubules and with LIN-5–ASPM-1 at meiotic spindle poles and that the APC promotes spindle rotation by inhibiting CDK-1.
Introduction
Female meiotic spindles are attached to the egg cortex by one pole during anaphase (Fabritius et al., 2011) to allow efficient expulsion of chromosomes into polar bodies. In Xenopus laevis (Gard, 1992), Drosophila melanogaster (Endow and Komma, 1997), leech (Fernández et al., 1990), Caenorhabditis elegans (Yang et al., 2003), and mouse meiosis-II (Maro et al., 1984), the spindle is initially associated with the cortex in a parallel orientation and then rotates 90° to a perpendicular orientation. In C. elegans, spindle rotation is one of a series of precisely timed events that require the anaphase-promoting complex (APC; Yang et al., 2005; Ellefson and McNally, 2009). In wild-type embryos, the spindle is initially positioned parallel to the cortex in a kinesin-1–dependent but APC-independent manner (Yang et al., 2005). The metaphase spindle maintains an ∼8-µm-long steady-state length for 6 min before undergoing APC-dependent shortening in the pole-to-pole axis to a 4.8-µm spindle length. 1.7 min after initiating spindle shortening, one spindle pole moves toward the cortex in a cytoplasmic dynein–dependent manner, after which homologous chromosomes begin to separate (Yang et al., 2003; McNally and McNally, 2005). After rotation and during chromosome segregation, the spindle undergoes further changes that include further shortening, changes in shape, reduction in the density of microtubules (McNally et al., 2006), and finally, elongation of the spindle (Yang et al., 2003; Dumont et al., 2010) and polar body extrusion.
Spindle shortening and all subsequent changes require the APC, raising the question of which APC substrates must be degraded to drive these highly ordered events. During mitosis in budding yeast (Thornton and Toczyski, 2003) and Drosophila embryos (Oliveira et al., 2010), cyclin B and securin are the only essential substrates of the APC that must be degraded to allow accurate chromosome segregation. Ubiquitination and degradation of cyclin B leads to inactivation of CDK-1 (Murray, 2004), whereas degradation of securin activates separase, which mediates sister chromatid separation through proteolytic cleavage of one cohesin subunit (Nasmyth, 2002). However, additional essential APC substrates have been identified in other species. For example, anaphase chromosome separation in Xenopus requires APC-dependent degradation of the chromokinesin, xKid (Funabiki and Murray, 2000).
Because meiotic spindle rotation in C. elegans requires cytoplasmic dynein and cytoplasmic dynein concentrates on meiotic spindle poles just before rotation (Ellefson and McNally, 2009), we postulated that the APC targets an inhibitor of dynein activity. Cyclin B–CDK-1 might inhibit dynein activity through direct inhibitory phosphorylation of a dynein subunit or inhibitory phosphorylation of a regulator of dynein. Separase has poorly characterized activities in addition to its function as a cohesin-cleaving protease (Kudo et al., 2006; Bembenek et al., 2007); thus, the APC might induce rotation by activating separase. Finally, the APC might target a dynein-specific inhibitor protein analogous to She1 in budding yeast, which inhibits dynein during metaphase (Woodruff et al., 2009).
Results
Inhibition of CDK-1 in APC mutant embryos results in meiotic spindle shortening and rotation
To understand how the APC activates dynein-dependent meiotic spindle rotation, we chose to address the role of a known APC target, cyclin B, whose destruction leads to inactivation of CDK-1 (Murray, 2004; Pesin and Orr-Weaver, 2008). If cyclin B is the APC target that must be degraded to allow spindle rotation, depletion of cyclin B by RNAi should bypass the requirement for the APC. In C. elegans, there are four cyclin B homologues, which have overlapping and redundant roles in germinal vesicle breakdown and metaphase spindle assembly (van der Voet et al., 2009b; Deyter et al., 2010); therefore, it would be difficult to consistently knock down cyclin B activity and assay spindle rotation. The C. elegans CDK-1 homologue, NCC-1, is also required for normal germinal vesicle breakdown and metaphase spindle assembly (Boxem et al., 1999; Chase et al., 2000; van der Voet et al., 2009b). In cdk-1(RNAi) embryos, no discernible bipolar meiotic spindle formed (Fig. S1, A and B); therefore, spindle rotation could not be assayed after RNAi depletion of CDK-1. To circumvent this problem, we injected the CDK-1 inhibitor purvalanol A (PA; Villerbu et al., 2002; Goga et al., 2007) into the uterus of adult worms and tracked the progress of meiotic embryos in utero. In PA-injected wild-type worms, three out of seven embryos failed to undergo germinal vesicle breakdown. In the four out of seven PA-treated embryos that did undergo germinal vesicle breakdown, no discernable bipolar meiotic spindles formed, and the embryos failed to ovulate (Fig. S1 C); therefore, we were unable to assay meiotic spindle rotation.
To consistently test the effect of CDK-1 inhibition on meiotic spindle rotation, we modified our assay and injected PA into the uterus of worms depleted of the APC subunit MAT-1 (Golden et al., 2000) or MAT-2 (Davis et al., 2002), where metaphase-I–arrested embryos with fully formed bipolar meiotic spindles accumulate. In these embryos, the APC cannot target cyclin B or other substrates such as IFY-1/securin (Golden et al., 2000) for degradation. Embryos were imaged 5 min after injection of PA, and only the youngest embryo adjacent to the spermatheca was imaged and scored, as embryos arrested for longer periods of time exhibit structural defects (Sonneville and Gönczy, 2004). Because oocytes mature and ovulate every 23 min (McCarter et al., 1999), the embryos in this experiment were on average arrested in metaphase-I for 12 min before exposure to PA for 5 min.
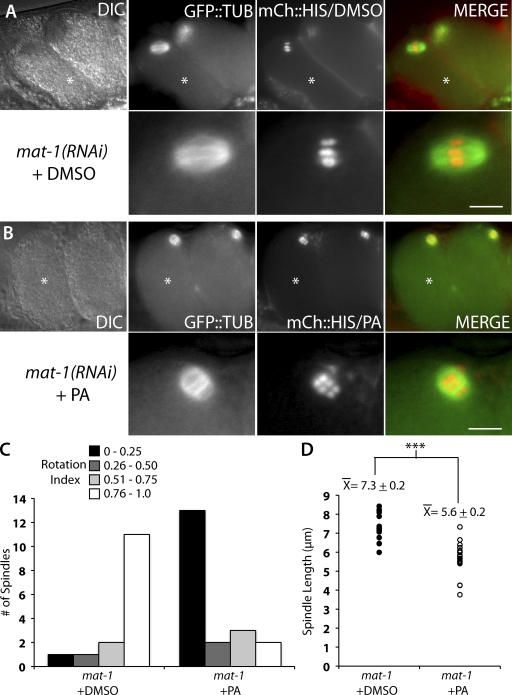
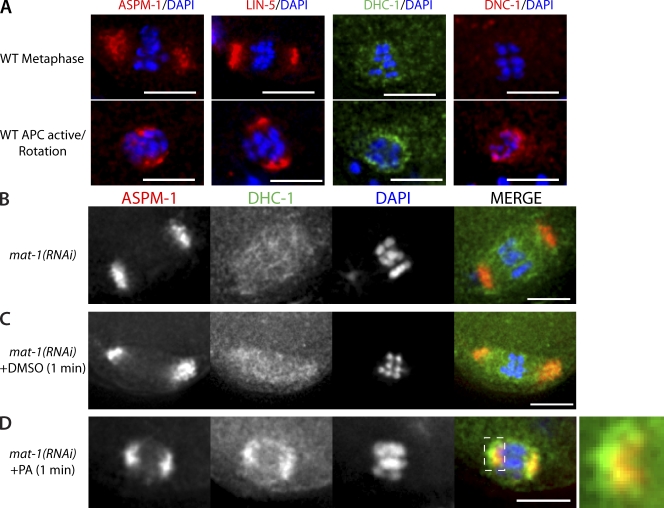
In meiotic embryos in which CDK-1 was inhibited by injection of PA, the metaphase-I–arrested meiotic spindles adopted a perpendicular position with respect to the embryo cortex (Fig. 1 B). This perpendicular position of the spindle resembled the rotated position that a wild-type meiotic spindle adopts after APC activation in meiosis-I and -II (Yang et al., 2003, 2005; Ellefson and McNally, 2009). In control mat-1(RNAi) meiotic embryos exposed to DMSO as a solvent control, the metaphase-I–arrested meiotic spindle was oriented with its pole-to-pole axis parallel to the embryo cortex (Fig. 1 A). This parallel position is similar to that observed for metaphase-I–arrested meiotic spindles in noninjected worms and wild-type metaphase-I and -II meiotic spindles (Yang et al., 2005). To quantify the position of the meiotic spindle, we devised a parameter termed rotation index (R). The rotation index is defined as the ratio of the distances measured between each spindle pole and the closest point of the embryo cortex (Fig. S1, D and E). A parallel spindle, where the distances between each pole and the cortex are similar, would result in a ratio with 0.76 ≤ R ≤ 1.0. Conversely, a rotated spindle that is perpendicular to the cortex would result in a ratio with 0.0 ≤ R ≤ 0.25. In control mat-1(RNAi) worms injected with DMSO, most meiotic spindles measured were parallel to the cortex (Fig. 1 C). Conversely, in embryos exposed to PA, most meiotic spindles were rotated (Fig. 1 C).
Figure 1.
CDK-1 inhibition by PA results in short and rotated meiotic spindles in metaphase-I–arrested APC-depleted meiotic embryos. (A and B) Live images of mat-1(RNAi) meiotic embryos expressing mCherry::histone and GFP::tubulin in utero after treatment with DMSO (A) or PA (B). The bottom panels are magnified images of the top panels. Asterisks mark the +1 embryo. DIC, differential interference contrast. Bars, 5 µm. (C) Quantification of rotation index in mat-1(RNAi) meiotic embryos after treatment with DMSO (left; n = 15) or PA (right; n = 20). The y axis is the number of meiotic embryos. The data shown include measurements from over seven independent injections for each treatment. (D) Quantification of meiotic spindle length in mat-1(RNAi) meiotic embryos after treatment with DMSO (closed circles; n = 15) or PA (open circles; n = 24). Each data point represents an individual meiotic spindle length measurement. The ±SEM is rounded to the nearest tenth. ***, P < 0.0005.
Inhibition of CDK-1 not only resulted in a change in spindle orientation but also caused a change in spindle length. Control metaphase-I–arrested meiotic embryos exposed to DMSO only had a mean pole-to-pole spindle length of 7.3 ± 0.2 µm (Fig. 1 D). This was similar to the wild-type metaphase-I meiotic spindle length of 8.0 µm (Yang et al., 2003). In contrast, metaphase-I–arrested meiotic spindles exposed to PA had a mean pole-to-pole length of 5.6 ± 0.2 µm (Fig. 1 D). This shorter spindle length resembled the 4.8-µm length of wild-type meiotic spindles during rotation (Yang et al., 2003). The changes that occurred after CDK-1 inhibition by PA in otherwise metaphase-I–arrested meiotic embryos suggested that CDK-1 inhibits both the early phase of meiotic spindle shortening (McNally et al., 2006) and spindle rotation during metaphase.
PA-induced spindle rotation mimics wild-type rotation, requiring the cytoplasmic dynein heavy chain, DHC-1, and the dynein regulator, LIN-5/nuclear mitotic apparatus, but not UNC-116/Kinesin-1
Kinesin-1 is required for parallel meiotic spindle positioning before APC activation but not for meiotic spindle rotation (Yang et al., 2005). In the absence of the C. elegans kinesin-1 heavy chain, UNC-116, the meiotic spindle remains several micrometers away from the cortex during metaphase, but after activation of the APC, the spindle undergoes a dynein-dependent pole-leading late translocation that positions the spindle perpendicular to the cortex. In mat-2(ts);unc-116 meiotic embryos (Yang et al., 2005) and dhc-1(RNAi);unc-116 meiotic embryos (Ellefson and McNally, 2009), the spindles are positioned away from the cortex and do not undergo directed movement to the cortex. Thus, the late translocation event that occurs in the absence of UNC-116 is mechanistically identical to that of wild-type rotation.
If PA-induced meiotic spindle rotation is mechanistically identical to wild-type rotation, it should also be sufficient to position the meiotic spindle at the cortex in a perpendicular orientation in metaphase-I–arrested embryos depleted of kinesin-1. To test this hypothesis, PA was injected into the uterus of unc-116(f130);mat-1(RNAi) worms. Because the meiotic spindles were more centrally located within control unc-116(f130);mat-1(RNAi) embryos, comparing measurements of the rotation index was not informative. Therefore, we measured the distance between the spindle and cortex to determine whether inhibition of CDK-1 resulted in metaphase-I–arrested meiotic spindles juxtaposed to the cortex. In control unc-116(f130);mat-1(RNAi) embryos exposed to DMSO only, the metaphase-I–arrested meiotic spindle remained a mean of 4.7 ± 2.6 µm away from the cortex (Fig. S2, A and C). This distance is similar to the reported 5.3-µm distance between the metaphase meiotic spindle and the cortex in embryos depleted of kinesin-1 only (Yang et al., 2005). However, in unc-116(f130);mat-1(RNAi) embryos exposed to PA, the metaphase-I–arrested meiotic spindle was within 1.4 ± 1.1 µm of the cortex (Fig. S2, B and C). These results suggest that PA-induced meiotic spindle positioning in APC-depleted, metaphase-I–arrested embryos is mechanistically similar to wild-type meiotic spindle rotation and late translocation in kinesin-1–depleted embryos.
Wild-type meiotic spindle rotation in C. elegans requires a complex of proteins including cytoplasmic dynein (DHC-1), nuclear mitotic apparatus–related LIN-5, ASPM-1 (abnormal spindlelike and microcephaly associated), and CMD-1 (calmodulin; Ellefson and McNally, 2009; van der Voet et al., 2009a). RNAi of any of these proteins blocks meiotic spindle rotation in wild-type embryos. To determine whether PA-induced meiotic spindle rotation in metaphase-I–arrested embryos requires the same complex of proteins as wild-type meiotic spindle rotation, we used RNAi to deplete DHC-1 in APC mutant metaphase-I–arrested embryos and treated the embryos with PA via in utero injections. Because a complete loss of dynein results in disorganized meiotic spindles and leads to adult sterility (Gönczy et al., 1999a; Yang et al., 2005), a partial depletion of dynein was used in this experiment, as previously described (Ellefson and McNally, 2009), to circumvent the requirement for dynein in spindle assembly.
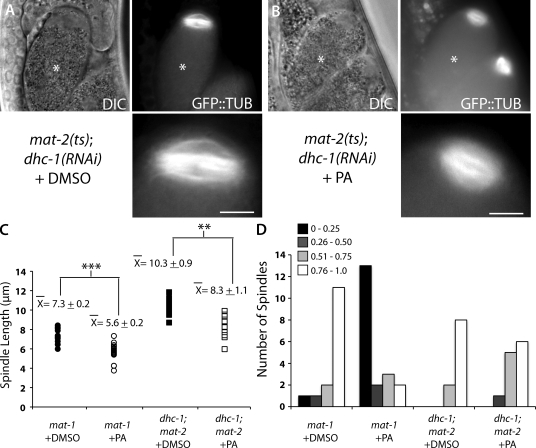
In control mat-2(ts);dhc-1(RNAi) meiotic embryos exposed to DMSO only, the meiotic spindles were parallel to the cortex with a mean pole-to-pole spindle length of 10.3 ± 0.9 µm (Fig. 2, A, C, and D). mat-2(ts);dhc-1(RNAi) meiotic spindles treated with PA failed to rotate (Fig. 2, B and D), but the spindles were shorter in the pole-to-pole axis than mat-2(ts);dhc-1(RNAi) control spindles (Fig. 2 C). These results demonstrate that dynein is required for PA-induced meiotic spindle rotation just as it is required for wild-type rotation.
Figure 2.
DHC-1 is required for purvalanol-induced meiotic spindle rotation but not spindle shortening. (A and B) Live images of dhc-1(RNAi);mat-2(ts) embryos expressing GFP::tubulin in utero after treatment with DMSO (A) or PA (B). The bottom panels are magnified images of the top panels. Asterisks mark the +1 embryo. DIC, differential interference contrast. Bars, 5 µm. (C) Quantification of meiotic spindle length in dhc-1(RNAi);mat-2(ts) meiotic embryos after treatment with DMSO (closed squares; n = 10) or PA (open squares; n = 12) compared with mat-1(RNAi) embryos. Data for mat-1(RNAi) are from Fig. 1. Each data point represents an individual meiotic spindle. The ±SEM is rounded to the nearest tenth. **, P < 0.005; ***, P < 0.0005. (D) Quantification of rotation index in dhc-1 (RNAi);mat-2(ts) meiotic embryos after treatment with DMSO (n = 10) or PA (n = 12) compared with mat-1(RNAi) embryos. The y axis is the number of meiotic embryos. The graph represents the rotation indices calculated for all of the spindles imaged in multiple experiments.
To further test whether the requirements for PA-induced meiotic spindle rotation are identical to wild-type spindle rotation, we repeated PA injections in metaphase-I–arrested embryos depleted of LIN-5 function using the lin-5(ev571ts) mutant (Lorson et al., 2000). Loss of LIN-5 reduced the extent of PA-induced meiotic spindle rotation in metaphase-I–arrested embryos (Fig. S3 D). This reduction is not as strong as that caused by dhc-1(RNAi), possibly as a result of residual LIN-5 function in the ts mutant. PA did cause spindle shortening in lin-5(ev571);mat-1(RNAi) embryos (Fig. S3 C). The requirement for LIN-5 in PA-induced meiotic spindle rotation provides further support that PA-induced meiotic spindle rotation is mechanistically identical to wild-type meiotic spindle rotation.
Inhibition of CDK-1 does not cause anaphase chromosome segregation
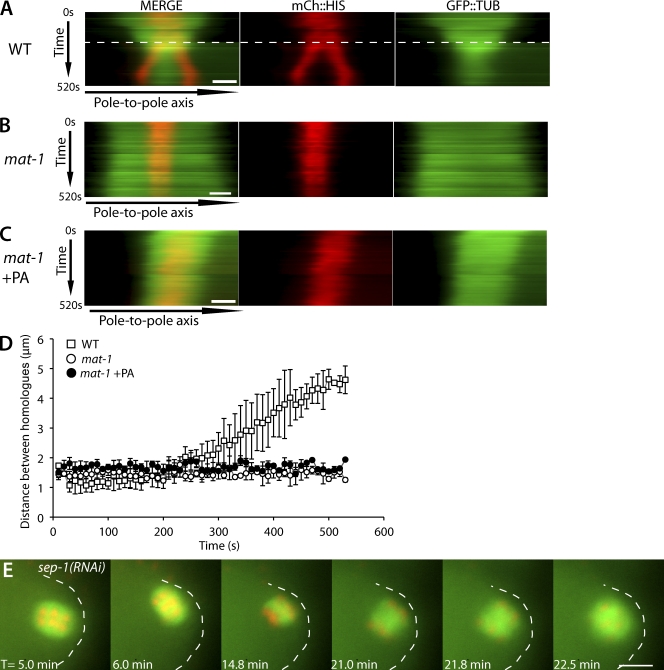
The conclusion that cyclin B is the specific APC target whose destruction leads to spindle rotation relies on the assumption that other APC substrates remain intact in APC mutant metaphase-I–arrested embryos treated with PA. To test whether IFY-1/securin was targeted for destruction in APC mutant PA-treated embryos, we assayed chromosome behavior in wild-type, mat-1(RNAi), and mat-1(RNAi) + PA embryos expressing mCherry::histone and GFP::tubulin. Degradation of securin would be expected to activate separase and drive homologue separation in APC mutant embryos (Nasmyth et al., 2000; Kitagawa et al., 2002; Oliveira et al., 2010). Kymographs of live time-lapse images show that in wild-type embryos, homologous chromosomes begin to separate after APC activation, which is indicated by spindle shortening (GFP::tubulin) and spindle rotation (Fig. 3 A). In contrast, in both mat-1(RNAi) and mat-1(RNAi) + PA embryos, homologous chromosomes failed to separate (Fig. 3, B and C). To further quantitate homologue separation, the change in distance between homologous chromosomes over time was measured. The measurements confirmed that homologue separation does not occur after treatment with PA (Fig. 3 D). These results demonstrate that inhibition of CDK-1 in APC mutant embryos does not result in activation of separase. The result that CDK-1 inhibition uncouples spindle rotation and separase activation suggests that separase is not required for meiotic spindle rotation. Indeed, sep-1(RNAi) meiotic embryos exhibited spindle rotation (Fig. 3 E) within 7.9 ± 1.8 min of exit from the spermatheca (n = 5), which is similar to the timing of wild-type meiotic spindle rotation (Ellefson and McNally, 2009). All sep-1(RNAi) embryos had incomplete chromosome segregation and failed polar body extrusion as previously reported (Siomos et al., 2001; Bembenek et al., 2007). These results strongly suggest that during wild-type metaphase, active CDK-1–cyclin B inhibits dynein-dependent meiotic spindle rotation and spindle shortening.
Figure 3.
Inhibition of CDK-1 does not result in homologous chromosome separation. (A–C) Kymographs from live time-lapse image sequences. For each kymograph, a line was drawn along the pole-to-pole axis (x axis), and each kymograph was compiled from 52 images taken at 10-s intervals, totaling 520 s (y axis). T = 0 started when chromosomes aligned at the metaphase plate and the distance between homologues could be measured. Images are from worms expressing mCherry::histone and GFP::tubulin comparing wild type (WT; A), mat-1(RNAi) (B), and mat-1(RNAi) + PA (C). The dashed line in wild type indicates the time of rotation in which the pole-to-pole axis line had to be repositioned because of the change in spindle position. Bars, 2 µm. (D) Quantification of distance between homologous chromosomes in wild type (n = 6), mat-1(RNAi) (n = 4), and mat-1(RNAi) + PA (n = 6). Each data point represents the mean distance measured between homologues for each time point. Error bars represent one standard deviation greater than and less than the mean. The y axis is the distance between homologous chromosomes. (E) Representative live time-lapse sequence of meiotic spindle rotation in sep-1(RNAi) embryos expressing GFP::tubulin and mCherry::histone (n = 5/5). T = 0 starts at the exit from the spermatheca. The embryo cortex is outlined for clarity. Bar, 5 µm.
Inhibition of CDK-1 causes redistribution of DHC-1 in metaphase-I–arrested embryos
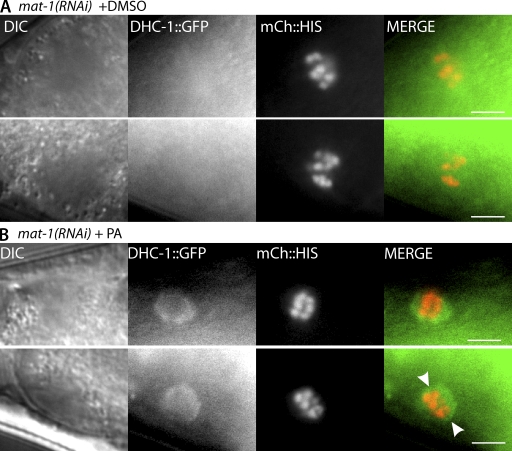
CDK-1 might inhibit spindle rotation by inhibiting dynein function. One way to examine the effect of CDK-1 on dynein is to determine whether inhibition of CDK-1 results in a change in localization of DHC-1 in metaphase-I–arrested embryos. In wild-type embryos, DHC-1 is dispersed throughout the metaphase meiotic spindle. Upon activation of the APC, DHC-1 concentrates on meiotic spindle poles just before rotation (Ellefson and McNally, 2009). To determine whether inhibition of CDK-1 alters DHC-1 localization, we repeated microinjections with either DMSO alone or PA in mat-1(RNAi) worms expressing GFP::DHC-1 and mCherry::histone (Gassmann et al., 2008). In live metaphase-I–arrested embryos exposed to DMSO, DHC-1::GFP was not readily detected on the meiotic spindle, and the spindles were in a parallel orientation with the cortex as judged by the orientation of the metaphase plate (n = 13/13; Fig. 4 A). However, in metaphase-I–arrested meiotic embryos treated with PA, DHC-1::GFP was concentrated on the meiotic spindle (Fig. 4 B). 24/33 embryos exposed to PA displayed DHC-1::GFP localization at meiotic spindles, and 21 of those 24 appeared to be rotated as judged by the orientation of the metaphase plate. Of the embryos exposed to PA where DHC-1::GFP was not detected at the meiotic spindle, eight out of nine appeared to be parallel to the cortex, suggesting that the PA treatment was not fully penetrant. These results suggest that CDK-1 inhibits meiotic spindle rotation through inhibition of dynein localization to meiotic spindles.
Figure 4.
Inhibition of CDK-1 causes redistribution of DHC-1 to the spindle poles in metaphase-I–arrested embryos. (A and B) Live images of mat-1(RNAi) embryos expressing DHC-1::GFP and mCherry::histone in utero after treatment with DMSO (A) or PA (B). The bottom and top panels are different examples of the DHC-1 localization observed. The arrowheads in B indicate a circular pattern of DHC-1::GFP indicative of nuclear envelope reassembly. DIC, differential interference contrast. Bars, 5 µm.
Recruitment of DHC-1 to meiotic spindles brings it into proximity with its activators LIN-5 and ASPM-1
A complex of LIN-5–ASPM-1–CMD-1 is required for localization of DHC-1 to anaphase spindle poles and is required for meiotic spindle rotation (van der Voet et al., 2009a). We were interested in determining when these complexes colocalize with DHC-1 and whether CDK-1 inhibits dynein-dependent spindle rotation by preventing the interactions between CMD-1, ASPM-1, LIN-5, and DHC-1. Immunofluorescence of fixed wild-type meiotic embryos revealed that both ASPM-1 and LIN-5 localize to meiotic spindle poles during metaphase-I (Fig. 5 A), in contrast with the diffuse localization of DHC-1. The p150 glued subunit of dynactin (DNC-1) was not detected on the metaphase meiotic spindle (Fig. 5 A; Ellefson and McNally, 2009). After APC activation, ASPM-1 and LIN-5 remained at spindle poles, whereas localization of DHC-1 and DNC-1 changed such that both concentrated at the spindle poles as well (Fig. 5 A; Ellefson and McNally, 2009). The observation that the wild-type metaphase-I localization of ASPM-1–LIN-5 differs from that of DHC-1–DNC-1 raised the possibility that CDK-1 may inhibit spindle rotation by blocking the interaction of CMD-1–ASPM-1–LIN-5 with DHC-1–DNC-1. If this were true, we would expect PA treatment to result in colocalization of DHC-1 with ASPM-1–LIN-5 at the spindle poles.
Figure 5.
CDK-1 inhibits recruitment of DHC-1 to meiotic spindle poles. Immunostaining of fixed meiotic embryos. (A) Examples of localization of ASPM-1, LIN-5, DHC-1::GFP, and DNC-1, all costained with DAPI, in wild-type (WT) metaphase (top: n = 5 for ASPM-1 and LIN-5, and n = 15 for DHC-1 and DNC-1) and after APC activation (bottom: n = 6 for ASPM-1 and LIN-5, and n = 17 for DHC-1 and DNC-1). (B–D) Localization of ASPM-1 and DHC-1::GFP on meiotic spindles costained with DAPI in mat-1(RNAi) embryos alone (B), mat-1(RNAi) embryos exposed to DMSO for 1 min (C), and mat-1(RNAi) embryos exposed to PA for 1 min (D). The farthest right panel is a magnification of the dashed area showing that DHC-1 does not completely overlap with ASPM-1 at the spindle poles. Bars, 5 µm.
We tested whether inhibition of CDK-1 in metaphase-I–arrested meiotic embryos results in colocalization of DHC-1 with ASPM-1 using immunohistochemistry. For these experiments, embryos were exposed to PA by dissecting worms in buffer containing PA. In all untreated metaphase-I–arrested embryos (n = 11/11) and the majority of DMSO-treated metaphase-I–arrested embryos (n = 9/11), ASPM-1 and DHC-1 did not colocalize. ASPM-1 was found at meiotic spindle poles, and DHC-1 was diffusely localized to the meiotic spindle region between spindle poles (Fig. 5, B and C). In contrast, ASPM-1 and DHC-1 colocalized at meiotic spindle poles in metaphase-I–arrested embryos exposed to PA (n = 18/19; Fig. 5 D). These results support the hypothesis that in wild-type metaphase, CDK-1 inhibits dynein-dependent meiotic spindle rotation by blocking the interaction of CMD-1–LIN-5–ASPM-1 with DHC-1.
Dissection of worms in the presence of PA, followed by flash freezing for immunofluorescence, allowed us to observe the relocalization of DHC-1 to meiotic spindle poles within 1 min of exposure to PA (Fig. 5 D) as compared with the 5-min gap between microinjection of PA and live imaging (Fig. 4 B). This timing is critical, as dissected metaphase-I–arrested meiotic embryos exposed to PA for 5 min before freezing exhibited the same circular DHC-1 localization observed in utero after a 5-min exposure to PA (Fig. 4 B, bottom row) in addition to pole localization (Fig. S4). This result suggests that PA causes DHC-1 to concentrate at spindle poles in <1 min and then to concentrate in a circular pattern that is possibly the reforming nuclear envelope by 5 min.
PA induces accumulation of DHC-1 and DNC-1 on meiotic spindle microtubules, spindle poles, and the embryo cortex
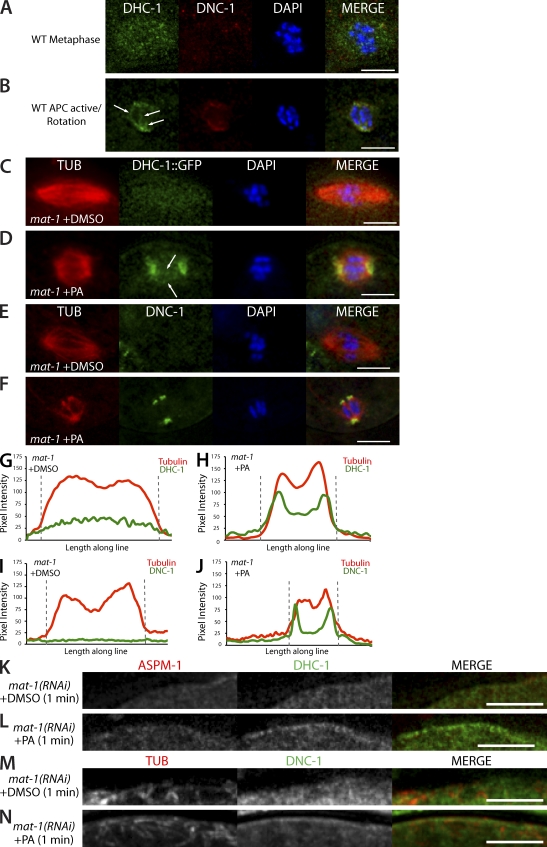
Because the localization of both DHC-1 and the dynactin subunit DNC-1 changes after APC activation (Ellefson and McNally, 2009), we looked closer at the colocalization of DHC-1 with DNC-1 in wild-type meiotic embryos. In wild-type metaphase-I embryos, DHC-1 was detected in a diffuse pattern within the meiotic spindle area, whereas DNC-1 was not detected (Fig. 6 A). In contrast, DHC-1 and DNC-1 colocalized at meiotic spindle poles after APC activation (Fig. 6 B). In addition, DHC-1, but not DNC-1, was detected in a linear pattern between the poles (Fig. 6 B, arrows), presumably on interpolar microtubules. These observations suggest that APC activation increases the colocalization of DHC-1 with DNC-1 at meiotic spindle poles.
Figure 6.
Inhibition of CDK-1 results in accumulation of DHC-1 and DNC-1 on meiotic spindle microtubules, spindle poles, and the embryo cortex. Immunostaining of fixed meiotic embryos. (A and B) Representative example of DHC-1::GFP and anti–DNC-1 localization with DAPI in wild-type (WT) metaphase-I (A; n = 15/15 DHC-1 and 13/15 DNC-1) and wild-type rotation (B; n = 17/17). (C) Representative example of DHC-1::GFP and tubulin (DM1α) localization in control mat-1(RNAi) embryos treated with DMSO (n = 21/22). (D) Localization of DHC-1::GFP and tubulin in mat-1(RNAi) embryos treated with PA (n = 14/21). (B and D) The arrows indicate DHC-1 localization on interpolar microtubules. (E) mat-1(RNAi) embryo treated with DMSO and stained for DNC-1 and tubulin (n = 6/7). (F) mat-1(RNAi) embryo treated with PA and stained for DNC-1 and tubulin (n = 7/12). (G–J) Graphs of pixel intensity along a line (4-µm thick and 12-µm long) drawn down the pole-to-pole axis of the spindle in representative background-subtracted images in C–F of mat-1(RNAi) embryos treated with DMSO (G and I) and PA (H and J). Red, tubulin pixel intensity; green, DHC-1 (G and H) and DNC-1 (I and J). The dashed gray lines indicate the spindle boundaries. (K and L) Cortical localization of ASPM-1 and DHC-1::GFP in mat-1(RNAi) embryos exposed to DMSO for 1 min (K; n = 18/22) and mat-1(RNAi) embryos exposed to PA for 1 min (L; n = 18/21). (M and N) Cortical localization of tubulin and DNC-1 in mat-1(RNAi) embryos exposed to DMSO for 1 min (M; n = 7/7) and mat-1(RNAi) embryos exposed to PA for 1 min (N; n = 12/12). Bars, 5 µm.
To determine whether inhibition of CDK-1 leads to accumulation of DHC-1 and DNC-1 specifically on meiotic spindle poles or on all meiotic spindle microtubules, we investigated the colocalization patterns of DHC-1 and DNC-1 with tubulin in metaphase-I–arrested embryos treated with and without PA. Similar to wild-type metaphase-I, DHC-1 localized in a faint pattern on the spindle microtubules in metaphase-I–arrested embryos exposed to DMSO (Fig. 6 C). The increase in the fluorescence intensity of GFP::DHC-1 on the metaphase spindle relative to the adjacent cytoplasm was readily apparent in pixel intensity plots (Fig. 6 G). Upon treatment with PA, the fluorescence intensity of GFP::DHC-1 increased on both meiotic spindle poles and on interpolar microtubules in 14/21 metaphase-I–arrested embryos (Fig. 6, D and H). In 4/21 PA-treated, metaphase-I–arrested embryos, DHC-1 remained in a diffuse pattern on the spindle microtubules, and in 3/21 PA-treated, metaphase-I–arrested embryos, DHC-1 accumulated on only one of the meiotic spindle poles. The localization of DNC-1 differed from that of DHC-1 in two ways. First, in metaphase-I–arrested embryos, no increase in DNC-1 on the spindle relative to the adjacent cytoplasm was detected (Fig. 6, E and I). Second, in 7/12 metaphase-I–arrested meiotic embryos treated with PA, DNC-1 accumulated much more specifically on spindle poles (Fig. 6 F) than DHC-1, as indicated by the pixel intensity plot in Fig. 6 J. In 2/12 metaphase-arrested meiotic embryos treated with PA, DNC-1 accumulated on only one spindle pole. In 2/12 embryos, DNC-1 localization increased on the entire spindle, and in 1/12 embryos, DNC-1 was not detected. These results suggest that CDK-1 prevents any association of DNC-1 with the metaphase-I spindle and that inactivation of CDK-1 results in specific accumulation of DNC-1 on spindle poles. In contrast, a population of CDK-1–resistant DHC-1 resides along the entire length of the metaphase-I spindle, and inhibition of CDK-1 results in a dramatic increase in DHC-1 on both the poles and the interpolar microtubules.
In addition to the dramatic accumulation of DHC-1 and DNC-1 on meiotic spindles induced by the inhibition of CDK-1, treatment with PA also resulted in the localization of both DHC-1 and DNC-1 to the embryo cortex. In metaphase-I–arrested meiotic embryos exposed to DMSO, DHC-1 did not appear enriched at the cortex (Fig. 6 K). However, inhibition of CDK-1 in metaphase-I–arrested meiotic embryos resulted in cortical enrichment of DHC-1 around the entire embryo (Fig. 6 L). Cortical localization of DNC-1 was also observed in metaphase-I–arrested meiotic embryos treated with PA (Fig. 6 N) but not in metaphase-I–arrested embryos treated with DMSO (Fig. 6 M). Because inhibition of CDK-1 did not result in noticeable cortical enrichment of ASPM-1 (Fig. 6, K and L), it is not clear whether PA-induced cortical enrichment of DHC-1 and DNC-1 is a result of DHC-1–DNC-1 binding to the cortex through some unknown cortical anchor or a result of dynein/dynactin motoring to cytoplasmic microtubule minus ends anchored at the cortex.
Inhibition of CDK results in increased microtubule association by the N-terminal microtubule-binding domain of DNC-1
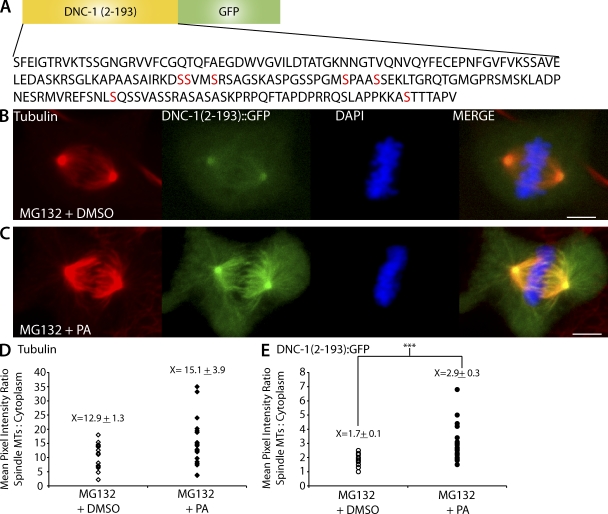
From the observation that DNC-1 is not detected on metaphase-I meiotic spindles but localizes to meiotic spindle poles after APC activation and because mitotic phosphorylation of the p150 N-terminal domain inhibits its association with spindle microtubules in Drosophila cells (Romé et al., 2010), we hypothesized that CDK-1 might inhibit spindle association of DHC-1 by inhibiting the microtubule binding activity of the N-terminal domain of the p150 dynactin subunit. To test this hypothesis, we fused the N-terminal 193 aa of C. elegans DNC-1 to GFP (Fig. 7 A) and expressed this construct in Xenopus A6 cells. This construct contains the cytoskeleton-associated protein (CAP)–Gly/microtubule-binding domain (Waterman-Storer et al., 1995; Weisbrich et al., 2007), the basic skating domain (Culver-Hanlon et al., 2006), and seven out of eight in vivo phosphorylation sites (Bodenmiller et al., 2008; Zielinska et al., 2009) but lacks any coiled coil and therefore should not dimerize with endogenous p150, and it should not interact with the dynein intermediate chain. DNC-1(2–193)::GFP was observed on centrosomes and faintly on the mitotic spindles of A6 cells that were arrested at metaphase with the proteasome inhibitor MG132 and treated with DMSO (n = 15; Fig. 7 B). In contrast, when MG132-arrested A6 cells were treated with PA for 5 min, a strong DNC-1(2–193)::GFP signal was detected on centrosomes, spindle microtubules, and astral microtubules (n = 20; Fig. 7 C). To quantify DNC-1(2–193)::GFP levels on microtubules in MG132-arrested cells with and without CDK-1 inhibition, we measured the mean pixel intensity of DNC-1(2–193)::GFP. To control for difference in protein expression, we compared the ratios of the mean pixel intensity of DNC-1(2–193)::GFP on the spindle/centrosomes with the cytoplasmic background. PA induced nearly a twofold increase in this ratio (Fig. 7 E), whereas similar measurements of antitubulin staining intensity revealed no significant difference (Fig. 7 D). A previous study revealed that the inhibition of CDK-1 with another small molecule inhibitor, Flavopiridol, in MG132-arrested Xenopus cells induces reformation of the nucleus and extension of interphase-like microtubules (Potapova et al., 2006). Indeed, a longer 10-min PA treatment in MG132-arrested A6 cells resulted in reformation of the nucleus and extension of interphase-like microtubules (Fig. S5), indicating that treatment with PA is effectively inhibiting CDK-1 function in MG132-arrested A6 cells. These results suggest that inhibition of CDK-1 increases the association of the GFP-tagged C. elegans DNC-1 microtubule-binding domain with microtubules in A6 cells. This could result from increased DNC-1 microtubule binding affinity in the absence of CDK-1, or, alternatively, CDK-1 inhibition could result in the removal of an unknown protein from the spindle that allows DNC-1 to access spindle microtubules.
Figure 7.
Inhibition of CDK-1 leads to association of the DNC-1 CAP-Gly domain with microtubules. (A) A schematic of the C. elegans DNC-1 CAP-Gly construct transfected into Xenopus A6 cells where DNC-1(2–193) was fused to GFP. The construct contains the CAP-Gly domain of DNC-1, the basic skating domain, as well as seven out of eight phosphorylation sites (red). (B and C) Representative images of Xenopus A6 cells expressing DNC-1(2–193)::GFP stained for tubulin and DAPI. (B) MG132 metaphase-arrested control cell treated with DMSO. (C) MG132 metaphase-arrested cell treated with PA. Bars, 5 µm. (D) Quantification of tubulin ratio of the mean pixel intensity of spindle/cytoplasm in MG132 metaphase-arrested cells treated with DMSO (open diamonds; n = 15) and PA (closed diamonds; n = 20). (E) Quantification of DNC-1(2–193)::GFP ratio of the mean pixel intensity of spindle/cytoplasm in MG132 metaphase-arrested cells treated with DMSO (open circles; n = 15) and PA (closed circles; n = 20). Each data point represents an individual cell. The ±SEM is rounded to the nearest tenth. ***, P < 0.0005. MTs, microtubules.
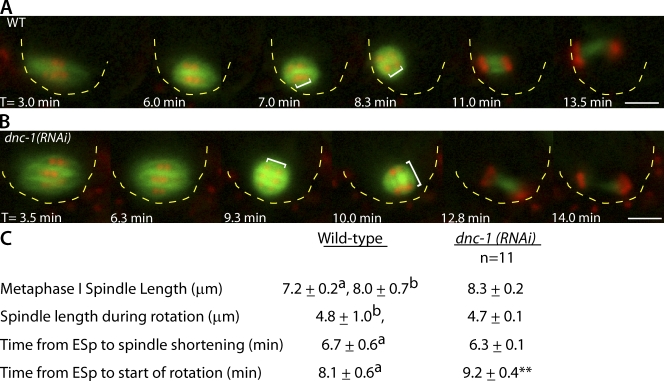
If the regulation of DNC-1 microtubule binding by CDK-1 is relevant to meiotic spindle rotation, DNC-1 should be required for meiotic spindle rotation. Knockdown of DNC-1 through RNAi by feeding resulted in a significant delay in meiotic spindle rotation (Fig. 8). In addition to a timing delay, in 8/11 dnc-1(RNAi) meiotic embryos, homologous chromosomes began to separate before rotation, and anaphase chromosome segregation was already in progress at the time when the spindle completed rotation (Fig. 8 B). This differs from wild-type meiosis, in which anaphase chromosome segregation does not occur until after the meiotic spindle completes rotation (Fig. 8 A; Yang et al., 2003; McNally and McNally, 2005). In dnc-1(RNAi) embryos with delayed rotation, the anaphase spindles did not remain perpendicular to the cortex in 7/11 embryos, leading to defects in polar body extrusion (Fig. 8 B). Spindle length and timing of APC activation in dnc-1(RNAi) embryos were similar to wild type (Fig. 8 C).
Figure 8.
DNC-1 is partially required for wild-type meiotic spindle rotation. (A and B) Representative images from live in utero time-lapse sequences of meiotic embryos expressing GFP::tubulin and mCherry::histone. (A) Wild-type (WT) meiotic spindle rotation. (B) Meiotic spindle movements in a dnc-1(RNAi) embryo (n = 8/11). The white brackets indicate the distance between homologues at the start and end of rotation. Yellow dashed lines outline the cell cortex for clarity. T = 0 is the exit from the spermatheca. Bars, 5 mm. (C) Quantification of meiotic spindle movements and length in dnc-1(RNAi) embryos compared with wild type. a, Ellefson and McNally, 2009; b, Yang et al., 2003; ESp, exit from the spermatheca; **, P < 0.05. The rotation defect observed in dnc-1(RNAi) was weaker than that observed in dhc-1(RNAi) embryos (Ellefson and McNally, 2009). This could be caused by incomplete knockdown of DNC-1. In C. elegans, dynactin knockdown has been previously found to result in two female pronuclei (Gönczy et al., 1999b; Terasawa et al., 2010), which might be a result of rotation failure.
Discussion
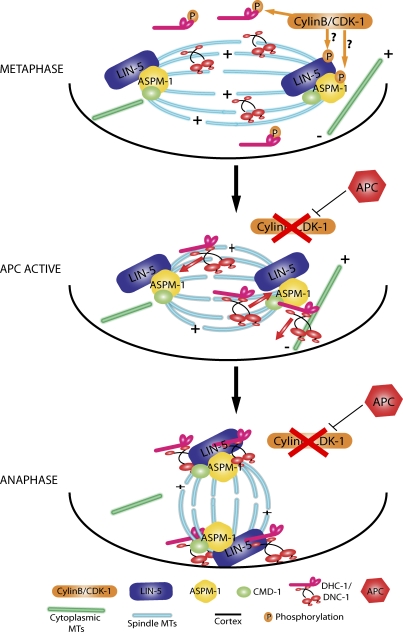
Based on these findings, we propose a model in which active cyclin B–CDK-1 inhibits microtubule binding by the CAP-Gly domain of p150 dynactin from germinal vesicle breakdown through metaphase-I (Fig. 9). Cytoplasmic dynein participates in assembly of the metaphase-I meiotic spindle in a manner that does not require microtubule binding by the CAP-Gly domain of p150 dynactin. Both C. elegans and mammals express isoforms of p150 that lack this domain, and these isoforms are sufficient to drive dynein-dependent membrane motility (Kim et al., 2007; Dixit et al., 2008) and processive movement (Kardon et al., 2009). Upon activation of the APC, cyclin B is degraded, and CDK-1–dependent phosphorylations are reversed, leading to microtubule binding by the CAP-Gly domain of p150 dynactin. This binding leads to a dramatic increase in the amount of dynein and dynactin associated with the spindle, where they can associate with the dynein activators ASPM-1 and LIN-5 to generate a complete rotation complex. A study in budding yeast indicates that the CAP-Gly domain of p150 dynactin is specifically required for the movement against a high load (Moore et al., 2009). In our model, spindle rotation might involve movement against a high load, or rotation may simply require a higher local concentration of dynein than is required for spindle assembly. During metaphase, cyclin B–CDK-1 also inhibits spindle shortening by targeting different proteins than those that are phosphorylated to prevent dynein-dependent rotation.
Figure 9.
Model for the mechanism of CDK-1 inhibition of dynein-dependent meiotic spindle rotation. During wild-type metaphase, cyclin B–CDK-1 reduces the association of DNC-1 with microtubules via inhibitory phosphorylations, which limit the accumulation of DHC-1–DNC-1 on the spindle microtubules (MTs). Cyclin B–CDK-1 may also block the protein–protein interactions between DHC-1–DNC-1 with CMD-1–ASPM-1–LIN-1 at meiotic spindle poles via inhibitory phosphorylations. After APC activation, cyclin B is targeted for degradation, and CDK-1 kinase activity ceases. Inhibitory phosphorylations of DNC-1 are removed, and DHC-1–DNC-1 associates with spindle and cytoplasmic microtubules, motors toward microtubule minus ends, and accumulates on spindle poles and the embryo cortex. DHC-1–DNC-1 at the meiotic spindle poles interacts with CMD-1–ASPM-1–LIN-5, thus forming a functional rotation complex. DHC-1, with its cargo-binding domain bound to spindle poles, engages cytoplasmic microtubules with minus ends anchored at the cortex and motors toward the cortex, resulting in meiotic spindle rotation.
Our model predicts that the CAP-Gly domain of p150 dynactin should not be associated with the metaphase-I spindle, and indeed immunofluorescence with an antibody specific for this domain (Dinkelmann et al., 2007) showed no labeling of metaphase spindles (Fig. 6). Our model also predicts that p150 dynactin should be required for spindle rotation, and we indeed observed a strong delay in spindle rotation in dnc-1(RNAi) embryos and instability in the perpendicular orientation of anaphase spindles. It is possible that p150 was not completely depleted in our RNAi experiments or that additional mechanisms participate in inhibiting spindle rotation during metaphase. For example, phosphorylation of the dynein intermediate chain might inhibit binding of dynein to p150 dynactin (Vaughan et al., 2001; King et al., 2003; Whyte et al., 2008). Three in vivo phosphorylation sites (Zielinska et al., 2009) have been identified in the p150-binding domain (Vaughan et al., 1995) of C. elegans DYCI-1. One of these (S77) matches the minimal CDK consensus, and another one of these (S84) inhibits interaction with p150 dynactin when phosphorylated in mammalian cells (Vaughan et al., 2001). Alternatively, cyclin B–CDK-1–dependent phosphorylation might inhibit binding of dynein/dynactin to LIN-5 or ASPM-1. We do not favor this possibility as the sole mechanism of regulation because DHC-1 only partially colocalizes with ASPM-1–LIN-5 in purvalanol-treated embryos (Fig. 5 D, inset). In addition, purvalanol induces a strong relocalization of dynein and dynactin to the embryo cortex, where there is no obvious enrichment for ASPM-1.
Although the inhibition of CDK-1 resulted in increased in vivo microtubule association of the DNC-1 microtubule-binding domain in metaphase-arrested cells, it is not clear whether the limited spindle association during normal metaphase is a result of direct phosphorylation of the DNC-1 microtubule-binding domain by cyclin B–CDK-1. A global mass spectrometry–based analysis of phosphorylations that decrease upon inhibition of CDK-1 in yeast revealed that over half of the sites did not match a minimal CDK consensus sequence (Holt et al., 2009). A more thorough analysis of 1,940 phosphopeptides enriched in HeLa cell mitotic spindles revealed that 39% matched a minimal CDK consensus, 9% matched a polo consensus, 11% matched an aurora consensus, and 30% were distributed among kinases that are not mitosis specific (Malik et al., 2009). Thus, CDK-1 drives global phosphorylation of proteins by other kinases. In Drosophila, aurora A kinase is responsible for the inhibitory phosphorylation of eight serines within the basic skating domain adjacent to the CAP-Gly domain of p150 (Romé et al., 2010); however, none of the Drosophila phosphorylation sites are conserved across phyla. Eight in vivo phosphorylation sites have been identified in the same basic skating domain of C. elegans DNC-1 by mass spectrometry (Bodenmiller et al., 2008; Zielinska et al., 2009), but none of the C. elegans sites match an aurora consensus, and only two of these sites match a minimal CDK consensus. The only things about the basic skating domain that are conserved across animal phyla are a similar length (∼200 aa), a net-positive charge in the dephosphorylated state (+13 C. elegans, +7 Drosophila, and +8 mouse) that might promote microtubule binding by the adjacent CAP-Gly domain, and an abundance of serines that, if phosphorylated, would neutralize the positive charges and might reduce the microtubule binding affinity of the adjacent CAP-Gly domain. If this charge neutralization model is correct, charge neutralization could be mediated by different kinases and different phosphorylation sites in different species.
An important unanswered question is whether dynein is activated at one spindle pole first to allow spindle rotation rather than isometric pulling on both spindle poles. In Drosophila, destruction of cyclin B is spatially regulated on the mitotic spindle (Huang and Raff, 1999). In human cells (Tugendreich et al., 1995) and C. elegans embryos (Kitagawa et al., 2002), components of the APC localize to the mitotic spindle, but the significance of this localization has not yet been determined.
Asymmetric positioning of the spindle occurs after the metaphase/anaphase transition in the asymmetric mitotic divisions of Saccharomyces cerevisiae (Sheeman et al., 2003; Grava et al., 2006; Woodruff et al., 2009), C. elegans mitotic embryos (McCarthy Campbell et al., 2009), and Drosophila neuroblasts (Kaltschmidt et al., 2000), in meiosis of C. elegans (Yang et al., 2003; Ellefson and McNally, 2009) and Drosophila (Endow and Komma, 1997), and in meiosis-II in the mouse (Zhu et al., 2003; Matson et al., 2006). The observation that asymmetric spindle positioning occurs after APC activation in both mitotic and meiotic divisions of different species suggests that it may confer some selective advantage. Thus, the mechanisms elucidated here for APC-dependent activation of dynein will likely be relevant to cell divisions in a wide range of species and cell types.
Materials and methods
C. elegans strains
Strains were cultured according to standard procedures (Brenner, 1974). In this study, wild type indicates the integrated GFP::tubulin and mCherry::histone strain created in our laboratory and designated FM125. FM125 (unc-119(ed3);ruIs57[pAZ147:pie-1/β-tubulin::GFP;unc-119(+)];itIs37[unc-119(+) pie-1::mCherry::H2B]) was created by crossing OD57 (unc-119(ed3);ruIs57[pAZ147:pie-1/β-tubulin::GFP;unc-119(+)];itIs37[unc-119(+) pie-1::mCherry::H2B]) (provided by K. McNally, University of California, Davis, Davis, CA; McNally et al., 2006) with AZ244 (unc-119(ed3);ruIs57[pAZ147:pie-1/β-tubulin::GFP;unc-119(+)]), which is derived from N2 (Praitis et al., 2001). FM55 (unc-36(e251); unc-116(f130);unc-119(ed3);ruIs57[pAZ147:pie-1/β-tubulin::GFP;unc-119(+)];itIs37[unc-119(+) pie-1::mCherry::H2B]) carries the unc-116(f130) temperature-sensitive maternal effect lethal mutation (McNally et al., 2010). FM102 (lin-5(ev571ts) unc-119(ed3);ruIs57 [pAZ147:pie-1/β-tubulin::GFP;unc-119(+)]) (a gift from L. Rose, University of California, Davis, Davis, CA; Park and Rose, 2008) carries the lin-5(ev571) allele, which is a recessive loss-of-function temperature-sensitive mutation (Lorson et al., 2000). FM157 (mat-2(ax102)II; ltIs24 [pAZ132; pie-1::GFP::tba-2 + unc-119(+)]) carries the mat-2(ax102)II temperature-sensitive mutation (Davis et al., 2002) and was obtained from crossing DS98 (mat-2(ax102)II) with OD73 (tIs38 [pAA1; pie-1::GFP::PH(PLC1d1) + unc-119(+)]; itIs24 (pAZ132; pie-1::GFP::tba-2 + unc-119(+)) was provided by A. Fabritius (University of California, Davis, Davis, CA). Both DS98 and OD73 were obtained from the Caenorhabditis Genetics Center at the University of Minnesota. EU1561 was the strain used for visualization of DHC-1. EU1561 (orIs[dhc-1::GFP::DHC-1, unc-119(+)];itIs37[unc-119(+) pie-1::mCherry::H2B]) has integrated GFP::dynein (DHC-1) and mCherry::histone H2B transgenes (a gift from B. Bowerman, University of Oregon, Eugene, OR; Gassmann et al., 2008). FM125 and EU1561 were maintained at 20°C. All temperature-sensitive mutants (FM55, FM102, and FM157) were maintained at 16°C. All strains were shifted to 25°C for 24 h before imaging.
RNAi
All of the RNAi experiments were performed by feeding bacteria (HT115) induced to express double-stranded RNA corresponding to each gene as described by Timmons et al. (2001) and by Kamath et al. (2001). L4 hermaphrodites were transferred to RNAi plates and were allowed to feed on the RNAi bacterial lawn for 16–24 h. The following clones from the genomic RNAi feeding library (Medical Research Council Gene Services, Source BioScience; Kamath et al., 2001) were used: mat-1 clone I-2C18, dhc-1 clone I-1PO4, and dnc-1 clone IV-5D24. For live imaging and injections, a 24-h feeding was used for mat-1(RNAi) and dnc-1(RNAi) at 25°C in the FM125, unc-116(f130), lin-5(ev571), and EU1561 strains, and a 16-h feeding at 25°C was used for dhc-1(RNAi) in the mat-2(ax102) strain.
In utero microinjections and live imaging
Adult hermaphrodite worms were transferred to unseeded Modified Youngren’s, Only Bacto-peptone plates 10 min before injection to remove bacteria. In sets of two, worms were immobilized by placing them into halocarbon oil on a coverslip with a desiccated 2% agarose pad and were immediately transferred to the injection scope. For injections, 1 µl of 100 mM PA (a gift from A. Goga, University of California, San Francisco, San Francisco, CA; Tocris Bioscience) in 100% DMSO was loaded into the injection needle, or, for controls, the needle was loaded with 100% DMSO. This high concentration of PA was used to account for rapid dilution after injection of a small volume into the uterus of the worm. We estimate that this mimics treating the embryos with 200 µM PA directly. Injection mixtures also contained 0.1 µl Alexa Fluor 594–labeled secondary antibody, which revealed the extent of diffusion of the injected mixture within the uterus. To inject, the glass microinjection needle was directly inserted into the vulva of the adult worm, and then PA or DMSO was released directly into the uterus without puncturing the worm. The worms were immediately recovered for 15 s in M9 buffer, transferred to anesthesia with tricaine/tetramisole (Kirby et al., 1990; McCarter et al., 1999) for 30 s, and gently mounted between a coverslip and a thin 3% agarose pad on a glass slide. The slides were quickly transferred to the imaging microscope and imaged. The time from injection to the start of imaging was 5 min. Live imaging was performed on an inverted microscope (IX71; Olympus) equipped with a 60× PlanApo NA 1.42 objective and a deep-cooled C10600-10B digital mono charge-coupled device camera (Orca R2; Hamamatsu Photonics). Excitation light from a mercury burner power supply (U-RFL-T 100W; Olympus) was shuttered with a shutter (Sutter Instrument) controlled by a Lambda 10–3 controller (Sutter Instrument) and MetaMorph Imaging software (Universal Imaging) through a 32-mm excitation filter wheel (Sutter Instrument). The stage temperature was 22–24°C. Single exposures of 0.15–0.3 s were captured. For time-lapse imaging (Fig. 4), images were captured at 10-s intervals for 10 min, and T = 0 was set at the time at which homologous chromosomes were clearly in focus and measureable. All quantitative analysis was performed with MetaMorph software. Only the +1 embryo was imaged and scored for all experiments.
Immunofluorescence
Adult hermaphrodite worms were washed in a watch glass in 0.8× egg buffer, and the worms were transferred in sets of 10 to a polylysine-coated glass slide (Superfrost Plus; Thermo Fisher Scientific). Excess liquid was wicked away, 200 µM PA or 0.2% DMSO in 0.8× egg buffer was added, and embryos were quickly dissected out of the worms using a 27G1/2 needle. Slides with dissected embryos in PA or DMSO were incubated for 1 or 5 min in a moist chamber, excess liquid was wicked away, a coverslip was gently applied, and the slides were submerged into liquid nitrogen for 10 min. After removal from liquid nitrogen, the coverslips were quickly flicked off, and the slides were submerged in −20°C MeOH for 25 min. Slides were washed twice for 10 min in 1× PBS and once for 15 min in PBS containing 0.05% Tween 20 (PBST). Slides were preblocked in 4% BSA in PBST for 45 min at RT followed by overnight incubation with primary antibodies at 4°C. Slides were washed twice for 10 min in 1× PBS and once for 15 min in PBST and then incubated with secondary antibodies for 1 h at RT. Slides were washed twice for 10 min in 1× PBS and once for 15 min in PBST and then mounted with a coverslip and 1,4-diazabicyclo[2.2.2]octane/Mowiol. Primary antibodies were diluted in PBST and 4% BSA in the following ratios: 1:50 anti–ASPM-1 (a gift from S. van den Huevel, Utrecht University, Utrecht, Netherlands; van der Voet et al., 2009a), 1:50 anti–LIN-5 (a gift from L. Rose; Park and Rose, 2008), 1:200 anti–DNC-1 (a gift from A. Skop and J. White, University of Wisconsin Madison, Madison, WI; Dinkelmann et al., 2007), and 1:200 anti–DM1α (Sigma-Aldrich). All secondary antibodies were diluted 1:200 in PBST. The secondary antibodies used were Alexa Flour 594 goat anti–rabbit IgG (Invitrogen), anti–rat (Jackson ImmunoResearch Laboratories, Inc.), and anti–mouse IgG (Invitrogen). DAPI staining was used to visualize DNA. Stained embryos were imaged using a 60× NA 1.4 objective on a real-time deconvolution microscope (Delta Vision; Applied Precision) equipped with a charge-coupled device camera (Coolsnap HQ; Roper Scientific). Images were deconvolved using SoftWorx Explorer Suite software (Applied Precision). Z stacks of images were taken in 0.20-µm step size. Metaphase-arrested meiotic embryos with highly abnormal spindle or chromosome morphology were excluded from this analysis, as our in utero analysis (Fig. 1) as well as that of Sonneville and Gönczy (2004) indicate that these represent embryos that have been arrested for >1 h.
Xenopus A6 cells
DNA encoding amino acids 2–193 of DNC-1 was synthesized by GenScript with a HindIII site and Kozak consensus at the 5′ end and an HA tag, six–amino acid flexible linker, and AgeI site at the 3′ end and was first cloned into pUC57. The HindIII–AgeI fragment was cloned into the pEGFP-N1 vector (Takara Bio Inc.) and was confirmed by sequencing.
Xenopus A6 cells, which were obtained from D. Furlow (University of California, Davis, Davis, CA), were grown at 27°C on coverslips in 70% Leibovits’s L15 medium supplemented with10% FBS and penicillin/streptomycin. Cells were transfected with DNC-1(2–193)::GFP using Lipofectamine 2000 (Invitrogen). 24 h after transfection, the cells were treated with 25 µM MG132 (EMD). After 60–90 min in MG132, cells were treated for 5–10 min with 200 µM PA or treated with DMSO as a control solvent. The coverslips were then fixed in −20°C MeOH and stained with antitubulin antibody (DM1α) and DAPI. Cells were imaged on an inverted microscope. All images of Xenopus A6 cells in Fig. 7 were taken with the same exposure times: 500 ms (mCherry), 2 s (GFP), and 500 ms (DAPI).
Quantification of pixel intensity was performed with iVision software (BioVision Technologies). A projection of five planes of a Z stack for each image was background subtracted. The mean pixel intensity was measured by drawing a segment over the spindle microtubules and centrosomes or over the cytoplasm. The ratio of mean pixel intensity of (spindle + centrosomes)/(cytoplasm) was calculated for each cell.
Measurements
All measurements of spindle length, distance from cortex, and rotation index were made from single focal plane live images of meiotic spindles in utero using MetaMorph software. Measurements were taken only for images in which the spindle orientation and pole-to-pole axis could be unambiguously determined from the direction of microtubule bundles within the spindle and in which both spindle poles were in focus. Variation between different embryos is indicated by the spread of values presented in histograms (Figs. 1 C and 2 D) or the SEM (Figs. 1 D, 2 C, 7 [D and E], and 8 C). All histograms and mean values include measurements from multiple experiments. The accuracy of individual length measurements was determined from the standard deviation between 10 separate measurements for the same spindle. This standard deviation was 0.2 µm and justifies reporting length measurements rounded to the nearest tenth of a micrometer.
Spindle length was measured as the distance along the pole-to-pole axis, which was determined from thick fluorescent bars of GFP::tubulin that represent dense bundles of microtubules extending along the pole-to-pole axis. Spindle length was measured as the longest length of the longest bar of GFP::tubulin fluorescence. The distance between the spindle and the cortex (Fig. S2 C) was measured as the shortest distance between the edge of the spindle and the cortex. Distance between the spindle pole and the cortex, which was used to determine the rotation index, was measured as the shortest distance between the spindle pole and the cortex.
Online supplemental material
Fig. S1 shows that cdk-1(RNAi) prevents meiotic spindle assembly. Fig. S2 shows that UNC-116 is not required for purvalanol-induced spindle shortening or rotation. Fig. S3 shows that LIN-5 is required for PA-induced meiotic spindle rotation. Fig. S4 shows that DHC-1 relocalizes to meiotic spindle poles after 5 min of PA treatment. Fig. S5 shows that inhibition of CDK-1 leads to mitotic exit in MG132-arrested cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201104008/DC1.
Acknowledgments
We thank Andrei Goga and Noelle L’Etoile for the generous gift of PA, Lesilee Rose, John White, Ahna Skop, and Sander van den Heuvel for antibodies, Karen McNally, Amy Fabritius, Bruce Bowerman, John White, and Lesilee Rose for strains, and Karen McNally and Amy Fabritius for critical reading of the manuscript.
This work was supported by the National Institute of General Medical Sciences grant 1R01GM-079421 to F.J. McNally.
Footnotes
Abbreviations used in this paper:
- APC
- anaphase-promoting complex
- CAP
- cytoskeleton-associated protein
- PA
- purvalanol A
References
- Bembenek J.N., Richie C.T., Squirrell J.M., Campbell J.M., Eliceiri K.W., Poteryaev D., Spang A., Golden A., White J.G. 2007. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development. 134:3837–3848 10.1242/dev.011361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., Picotti P., Schlapbach R., Aebersold R. 2008. PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 26:1339–1340 10.1038/nbt1208-1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem M., Srinivasan D.G., van den Heuvel S. 1999. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development. 126:2227–2239 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D., Serafinas C., Ashcroft N., Kosinski M., Longo D., Ferris D.K., Golden A. 2000. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis. 26:26–41 [DOI] [PubMed] [Google Scholar]
- Culver-Hanlon T.L., Lex S.A., Stephens A.D., Quintyne N.J., King S.J. 2006. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat. Cell Biol. 8:264–270 10.1038/ncb1370 [DOI] [PubMed] [Google Scholar]
- Davis E.S., Wille L., Chestnut B.A., Sadler P.L., Shakes D.C., Golden A. 2002. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics. 160:805–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter G.M., Furuta T., Kurasawa Y., Schumacher J.M. 2010. Caenorhabditis elegans cyclin B3 is required for multiple mitotic processes including alleviation of a spindle checkpoint-dependent block in anaphase chromosome segregation. PLoS Genet. 6:e1001218 10.1371/journal.pgen.1001218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M.V., Zhang H., Skop A.R., White J.G. 2007. SPD-3 is required for spindle alignment in Caenorhabditis elegans embryos and localizes to mitochondria. Genetics. 177:1609–1620 10.1534/genetics.107.078386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Levy J.R., Tokito M., Ligon L.A., Holzbaur E.L. 2008. Regulation of dynactin through the differential expression of p150Glued isoforms. J. Biol. Chem. 283:33611–33619 10.1074/jbc.M804840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., Desai A. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901 10.1038/ncb2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson M.L., McNally F.J. 2009. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Mol. Biol. Cell. 20:2722–2730 10.1091/mbc.E08-12-1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A., Komma D.J. 1997. Spindle dynamics during meiosis in Drosophila oocytes. J. Cell Biol. 137:1321–1336 10.1083/jcb.137.6.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabritius A.S., Ellefson M.L., McNally F.J. 2011. Nuclear and spindle positioning during oocyte meiosis. Curr. Opin. Cell Biol. 23:78–84 10.1016/j.ceb.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández J., Olea N., Téllez V., Matte C. 1990. Structure and development of the egg of the glossiphoniid leech Theromyzon rude: reorganization of the fertilized egg during completion of the first meiotic division. Dev. Biol. 137:142–154 10.1016/0012-1606(90)90015-B [DOI] [PubMed] [Google Scholar]
- Funabiki H., Murray A.W. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424 10.1016/S0092-8674(00)00047-7 [DOI] [PubMed] [Google Scholar]
- Gard D.L. 1992. Microtubule organization during maturation of Xenopus oocytes: assembly and rotation of the meiotic spindles. Dev. Biol. 151:516–530 10.1016/0012-1606(92)90190-R [DOI] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R., III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goga A., Yang D., Tward A.D., Morgan D.O., Bishop J.M. 2007. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat. Med. 13:820–827 10.1038/nm1606 [DOI] [PubMed] [Google Scholar]
- Golden A., Sadler P.L., Wallenfang M.R., Schumacher J.M., Hamill D.R., Bates G., Bowerman B., Seydoux G., Shakes D.C. 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151:1469–1482 10.1083/jcb.151.7.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Pichler S., Kirkham M., Hyman A.A. 1999a. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147:135–150 10.1083/jcb.147.1.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Schnabel H., Kaletta T., Amores A.D., Hyman T., Schnabel R. 1999b. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144:927–946 10.1083/jcb.144.5.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grava S., Schaerer F., Faty M., Philippsen P., Barral Y. 2006. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev. Cell. 10:425–439 10.1016/j.devcel.2006.02.018 [DOI] [PubMed] [Google Scholar]
- Holt L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., Morgan D.O. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 325:1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Raff J.W. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18:2184–2195 10.1093/emboj/18.8.2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt J.A., Davidson C.M., Brown N.H., Brand A.H. 2000. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat. Cell Biol. 2:7–12 10.1038/71323 [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2:RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon J.R., Reck-Peterson S.L., Vale R.D. 2009. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc. Natl. Acad. Sci. USA. 106:5669–5674 10.1073/pnas.0900976106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ling S.C., Rogers G.C., Kural C., Selvin P.R., Rogers S.L., Gelfand V.I. 2007. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176:641–651 10.1083/jcb.200608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.J., Brown C.L., Maier K.C., Quintyne N.J., Schroer T.A. 2003. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol. Biol. Cell. 14:5089–5097 10.1091/mbc.E03-01-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby C., Kusch M., Kemphues K. 1990. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev. Biol. 142:203–215 10.1016/0012-1606(90)90164-E [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Law E., Tang L., Rose A.M. 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12:2118–2123 10.1016/S0960-9822(02)01392-1 [DOI] [PubMed] [Google Scholar]
- Kudo N.R., Wassmann K., Anger M., Schuh M., Wirth K.G., Xu H., Helmhart W., Kudo H., McKay M., Maro B., et al. 2006. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 126:135–146 10.1016/j.cell.2006.05.033 [DOI] [PubMed] [Google Scholar]
- Lorson M.A., Horvitz H.R., van den Heuvel S. 2000. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J. Cell Biol. 148:73–86 10.1083/jcb.148.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R., Lenobel R., Santamaria A., Ries A., Nigg E.A., Körner R. 2009. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J. Proteome Res. 8:4553–4563 10.1021/pr9003773 [DOI] [PubMed] [Google Scholar]
- Maro B., Johnson M.H., Pickering S.J., Flach G. 1984. Changes in actin distribution during fertilization of the mouse egg. J. Embryol. Exp. Morphol. 81:211–237 [PubMed] [Google Scholar]
- Matson S., Markoulaki S., Ducibella T. 2006. Antagonists of myosin light chain kinase and of myosin II inhibit specific events of egg activation in fertilized mouse eggs. Biol. Reprod. 74:169–176 10.1095/biolreprod.105.046409 [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T. 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205:111–128 10.1006/dbio.1998.9109 [DOI] [PubMed] [Google Scholar]
- McCarthy Campbell E.K., Werts A.D., Goldstein B. 2009. A cell cycle timer for asymmetric spindle positioning. PLoS Biol. 7:e1000088 10.1371/journal.pbio.1000088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K.L., McNally F.J. 2005. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev. Biol. 282:218–230 10.1016/j.ydbio.2005.03.009 [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F.J. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K.L., Martin J.L., Ellefson M., McNally F.J. 2010. Kinesin-dependent transport results in polarized migration of the nucleus in oocytes and inward movement of yolk granules in meiotic embryos. Dev. Biol. 339:126–140 10.1016/j.ydbio.2009.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Sept D., Cooper J.A. 2009. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc. Natl. Acad. Sci. USA. 106:5147–5152 10.1073/pnas.0810828106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W. 2004. Recycling the cell cycle: cyclins revisited. Cell. 116:221–234 10.1016/S0092-8674(03)01080-8 [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science. 297:559–565 10.1126/science.1074757 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Peters J.M., Uhlmann F. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 288:1379–1385 10.1126/science.288.5470.1379 [DOI] [PubMed] [Google Scholar]
- Oliveira R.A., Hamilton R.S., Pauli A., Davis I., Nasmyth K. 2010. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 12:185–192 10.1038/ncb2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.H., Rose L.S. 2008. Dynamic localization of LIN-5 and GPR-1/2 to cortical force generation domains during spindle positioning. Dev. Biol. 315:42–54 10.1016/j.ydbio.2007.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesin J.A., Orr-Weaver T.L. 2008. Regulation of APC/C activators in mitosis and meiosis. Annu. Rev. Cell Dev. Biol. 24:475–499 10.1146/annurev.cellbio.041408.115949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova T.A., Daum J.R., Pittman B.D., Hudson J.R., Jones T.N., Satinover D.L., Stukenberg P.T., Gorbsky G.J. 2006. The reversibility of mitotic exit in vertebrate cells. Nature. 440:954–958 10.1038/nature04652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romé P., Montembault E., Franck N., Pascal A., Glover D.M., Giet R. 2010. Aurora A contributes to p150(glued) phosphorylation and function during mitosis. J. Cell Biol. 189:651–659 10.1083/jcb.201001144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M.A., Pellman D. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13:364–372 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Siomos M.F., Badrinath A., Pasierbek P., Livingstone D., White J., Glotzer M., Nasmyth K. 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11:1825–1835 10.1016/S0960-9822(01)00588-7 [DOI] [PubMed] [Google Scholar]
- Sonneville R., Gönczy P. 2004. Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development. 131:3527–3543 10.1242/dev.01244 [DOI] [PubMed] [Google Scholar]
- Terasawa M., Toya M., Motegi F., Mana M., Nakamura K., Sugimoto A. 2010. Caenorhabditis elegans ortholog of the p24/p22 subunit, DNC-3, is essential for the formation of the dynactin complex by bridging DNC-1/p150(Glued) and DNC-2/dynamitin. Genes Cells. 15:1145–1157 10.1111/j.1365-2443.2010.01451.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B.R., Toczyski D.P. 2003. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 5:1090–1094 10.1038/ncb1066 [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D.L., Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263:103–112 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Tugendreich S., Tomkiel J., Earnshaw W., Hieter P. 1995. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 81:261–268 10.1016/0092-8674(95)90336-4 [DOI] [PubMed] [Google Scholar]
- van der Voet M., Berends C.W., Perreault A., Nguyen-Ngoc T., Gönczy P., Vidal M., Boxem M., van den Heuvel S. 2009a. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat. Cell Biol. 11:269–277 10.1038/ncb1834 [DOI] [PubMed] [Google Scholar]
- van der Voet M., Lorson M.A., Srinivasan D.G., Bennett K.L., van den Heuvel S. 2009b. C. elegans mitotic cyclins have distinct as well as overlapping functions in chromosome segregation. Cell Cycle. 8:4091–4102 10.4161/cc.8.24.10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K.T., Holzbaur E.L., Vallee R.B. 1995. Subcellular targeting of the retrograde motor cytoplasmic dynein. Biochem. Soc. Trans. 23:50–54 [DOI] [PubMed] [Google Scholar]
- Vaughan P.S., Leszyk J.D., Vaughan K.T. 2001. Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J. Biol. Chem. 276:26171–26179 10.1074/jbc.M102649200 [DOI] [PubMed] [Google Scholar]
- Villerbu N., Gaben A.M., Redeuilh G., Mester J. 2002. Cellular effects of purvalanol A: a specific inhibitor of cyclin-dependent kinase activities. Int. J. Cancer. 97:761–769 10.1002/ijc.10125 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Karki S., Holzbaur E.L. 1995. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1). Proc. Natl. Acad. Sci. USA. 92:1634–1638 10.1073/pnas.92.5.1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrich A., Honnappa S., Jaussi R., Okhrimenko O., Frey D., Jelesarov I., Akhmanova A., Steinmetz M.O. 2007. Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 14:959–967 10.1038/nsmb1291 [DOI] [PubMed] [Google Scholar]
- Whyte J., Bader J.R., Tauhata S.B., Raycroft M., Hornick J., Pfister K.K., Lane W.S., Chan G.K., Hinchcliffe E.H., Vaughan P.S., Vaughan K.T. 2008. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J. Cell Biol. 183:819–834 10.1083/jcb.200804114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., Barnes G. 2009. Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol. Biol. Cell. 20:3003–3011 10.1091/mbc.E09-03-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Y., McNally K., McNally F.J. 2003. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev. Biol. 260:245–259 10.1016/S0012-1606(03)00216-1 [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Mains P.E., McNally F.J. 2005. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J. Cell Biol. 169:447–457 10.1083/jcb.200411132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.Y., Chen D.Y., Li J.S., Lian L., Lei L., Han Z.M., Sun Q.Y. 2003. Rotation of meiotic spindle is controlled by microfilaments in mouse oocytes. Biol. Reprod. 68:943–946 10.1095/biolreprod.102.009910 [DOI] [PubMed] [Google Scholar]
- Zielinska D.F., Gnad F., Jedrusik-Bode M., Wiśniewski J.R., Mann M. 2009. Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J. Proteome Res. 8:4039–4049 10.1021/pr900384k [DOI] [PubMed] [Google Scholar]