Cdc42 regulates cardiac function in mice and flies downstream of a conserved Tinman/Nkx2-5–miR-1 signaling network.
Abstract
Unraveling the gene regulatory networks that govern development and function of the mammalian heart is critical for the rational design of therapeutic interventions in human heart disease. Using the Drosophila heart as a platform for identifying novel gene interactions leading to heart disease, we found that the Rho-GTPase Cdc42 cooperates with the cardiac transcription factor Tinman/Nkx2-5. Compound Cdc42, tinman heterozygous mutant flies exhibited impaired cardiac output and altered myofibrillar architecture, and adult heart–specific interference with Cdc42 function is sufficient to cause these same defects. We also identified K+ channels, encoded by dSUR and slowpoke, as potential effectors of the Cdc42–Tinman interaction. To determine whether a Cdc42–Nkx2-5 interaction is conserved in the mammalian heart, we examined compound heterozygous mutant mice and found conduction system and cardiac output defects. In exploring the mechanism of Nkx2-5 interaction with Cdc42, we demonstrated that mouse Cdc42 was a target of, and negatively regulated by miR-1, which itself was negatively regulated by Nkx2-5 in the mouse heart and by Tinman in the fly heart. We conclude that Cdc42 plays a conserved role in regulating heart function and is an indirect target of Tinman/Nkx2-5 via miR-1.
Introduction
Cardiovascular disease is the leading cause of mortality and morbidity in industrialized nations. Since the discovery of the first key determinant of heart development—tinman—in Drosophila (Azpiazu and Frasch, 1993; Bodmer, 1993), numerous regulators, including transcription factors, cell signaling molecules, extracellular matrix proteins, and microRNAs have provided an understanding of an increasingly complex network that guides cardiac specification, differentiation, function, and maturation in all organisms that possess a heart (Bier and Bodmer, 2004; Olson, 2006; Srivastava, 2006; Qian et al., 2008a).
The fruit fly Drosophila is an attractive model system to study various human diseases, including cardiac disease (St Johnston, 2002; Bier, 2005). The Drosophila heart is a simple linear tube comprised of myocardial and pericardial cells, which is reminiscent of the primitive vertebrate embryonic heart. Despite the simplicity of the structure, the genetic and molecular mechanisms that orchestrate heart formation are remarkably conserved between flies and vertebrates (Bodmer, 1995; Cripps and Olson, 2002). Moreover, recent studies using multiple functional assays revealed extensive conservation of gene functions required for maintaining normal heart physiology in the adult (Bier and Bodmer, 2004; Ocorr et al., 2007; Qian et al., 2008a,b). Combined with the powerful genetic tools available in the fly, screens to identify molecular–genetic pathways involved in cardiac contractility and rhythm can now be executed, leading to the identification of novel cardiac regulators that can then be examined in mammals (Adams and Sekelsky, 2002; St Johnston, 2002; Wessells and Bodmer, 2004; Neely et al., 2010).
Tinman and its mammalian homologue Nkx2-5 are homeobox transcription factors that play crucial roles in heart development and function (Prall et al., 2002). Whereas ablation of tinman in the Drosophila embryo abolishes heart formation, targeted deletion of tinman at later stages reveals additional requirements for tinman in maintaining normal adult heart function and physiology (Zaffran et al., 2006). This is reminiscent of Nkx2-5 in vertebrates, which is required for establishing heart function in the developing mouse embryo, for example, and also later for its maintenance in the adult (Pashmforoush et al., 2004). The genetic networks that Nkx2-5 participates in and how these interactions regulate adult heart functions are not well understood. A major reason for this is that it is often difficult to identify and study polygenic modifiers of (cardiac) disease in a vertebrate system, in part because the organism viability is critically dependent on a functional heart.
In a previous study, we identified a genetic interaction between neuromancer (nmr)/Tbx20 and tinman/Nkx2-5 in the adult Drosophila heart (Qian et al., 2008b). Here, we performed a deletion screen in a reduced tinman genetic background to identify genes that interact with tinman to maintain normal heart function in Drosophila. One of the strong genetic interactions causing cardiac dysfunction was between tinman and the gene Cdc42, a Rho family member of the small GTPase family, suggesting a role for Cdc42 in maintaining heart function. Indeed, dominant-negative interference with Cdc42 function alone in the adult fly heart compromises cardiac performance. We also identified potential downstream effectors of the Cdcd42–tinman interaction, including dSUR and slowpoke, which code for two K+ channels that are known to be required for heart function.
Cdc42 has a conserved, essential role in regulating cell cycle, directed migration, epithelial polarity, and cell fate specification (Brown et al., 2006; Heasman and Ridley, 2008); and in adult mouse cardiomyocytes Cdc42 is involved in growth and prevention of hypertrophy (Maillet et al., 2009). Here, we find that the genetic interaction between Cdc42 and Nkx2-5 is conserved in the mouse, and that Cdc42 and Nkx2-5 synergize to regulate heart function and physiology, which is likely mediated by miR-1. These data suggest that Cdc42 plays a conserved role in establishing and maintaining normal heart function downstream of, and potentially in concert with, the cardiac determinant tinman/Nkx2-5.
Results
Genetic interactions between Cdc42 and tinman in regulating heart performance in Drosophila
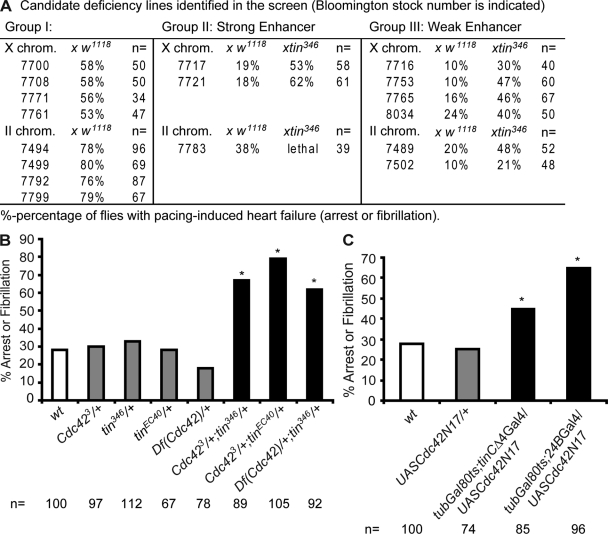
Increasing evidence suggests that the conserved cardiogenic transcription factor network that regulates heart formation also functions to establish and maintain cardiac performance in the adult (Zaffran et al., 2006; Ocorr et al., 2007; Qian et al., 2008b; Qian and Bodmer, 2009). To uncover genes that interact with this transcription factor network in adult heart function, we conducted a screen for mutations that aggravate the cardiac stress response of flies heterozygous for tinman. We examined cardiac performance of double-heterozygous flies for tinman and deficiency lines (97 in total) covering the X chromosome and part of the second chromosome (34% of the genome, see Table S1). We identified several categories of candidate genes suggestive of a developmental interaction: for example, the trans configuration of tinman with the deficiency line 7783 is lethal (Fig. 1 A, group II bottom), possibly due to a vital interaction with the dpp gene (contained within 7783) that is critically involved in cardiac induction during early embryogenesis, along with tinman (Qian et al., 2008a). In another category, the heterozygous candidate lines by themselves exhibit cardiac defects (increased pacing-induced heart failure rate, see Materials and methods; e.g., lines 7708 and 7499; Fig. 1 A, group I), indicating that haploinsufficiency in one or more genes covered by these deficiencies results in cardiac deficits. Of particular interest, the “strong enhancer” category of deficiency lines shows an interaction when in trans with tinman (>50%), but when crossed to wild type (w1118) exhibits a low rate of heart failure (<20%) that is similar to wild-type flies (e.g., 7717, 7721; Fig. 1 A, group II top). We chose for further analysis line 7721, which exhibits a robust interaction with tinman, in that the trans combination shows a threefold increase in susceptibility to pacing-induced heart failure (7721 x tin346) compared with single heterozygotes (e.g., 7721 x w1118).
Figure 1.
Genetic screen identifying Cdc42. (A) Candidate deficiency lines interacting with tin346 (susceptibility to pacing-induced heart arrest or fibrillation). (B) Genetic interaction between tinman, Cdc42, and deficiency 7721. Cdc42+/−;tin+/− flies show dramatic increase in pacing-induced heart arrest or fibrillation rate compared with Cdc42+/− or tin+/− (Chi square analysis: *, P < 0.01). (C) TARGET-mediated overexpression (see Materials and methods) of dominant-negative Cdc42 (Cdc42N17) in the adult heart results in elevated heart dysfunction (*, P < 0.01). Each bar in B and C represents a single experiment. n, number of flies tested.
Among the nine genes deleted by the deficiency 7721 (Df(1)Exel6253, referred to as Df(Cdc42)) is the small GTPase encoded by Cdc42, which is known for its role in cell cycle regulation and actin cytoskeletal rearrangement (Eaton et al., 1995; Genova et al., 2000; Jacinto et al., 2000; Hurd et al., 2003; Heasman and Ridley, 2008) and has been implicated in cardiomyocyte growth and hypertrophy (Carè et al., 2007; Maillet et al., 2009). We took a candidate gene approach and examined whether known Cdc42 loss-of-function mutants also interacted with tinman. Both tinEC40 (Bodmer, 1993) and tin346 (Azpiazu and Frasch, 1993) heterozygotes show a dramatic increase in heart failure rate in a Cdc423 heterozygous background (Fehon et al., 1997; Genova et al., 2000), similar to that observed for tinman, Df(1)Exel6253 double heterozygotes (Fig. 1 B). This suggests that the interaction between tinman and Cdc42 is required for the maintenance of proper adult heart function.
Cdc42-encoded RhoGTPase is required for adult cardiac function in Drosophila
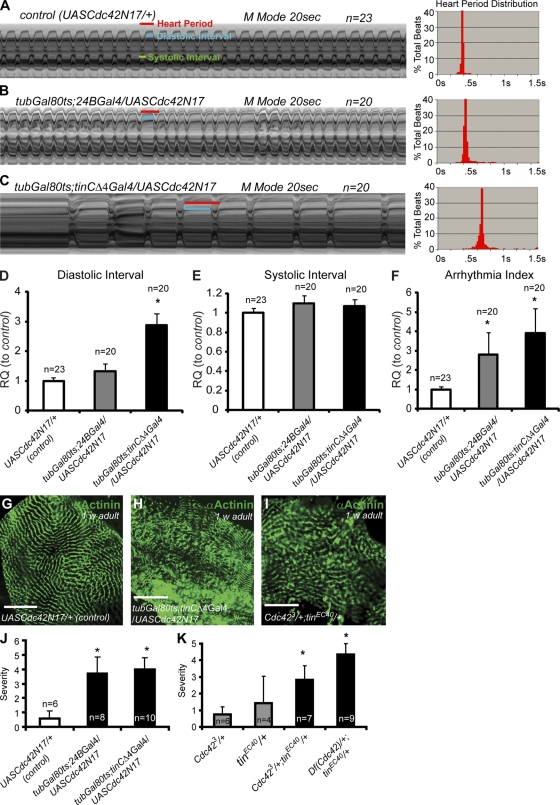
To examine a possible role for Cdc42 in the fly heart, we asked whether Cdc42 plays a muscle- or cardiac-autonomous role. For this purpose, we expressed a dominant-negative form of Cdc42 (UAS-Cdc42N17; Luo et al., 1994) specifically in the adult heart using tinCΔ4Gal4 (a UAS-Gal4 system that drives gene expression only in myocardial cells; Qian et al., 2005a), or in all mesoderm using 24BGal4 (drives gene expression pan-mesodermally; Brand and Perrimon, 1993). The UAS-GAL4 system has two parts: the GAL4 gene that encodes the yeast transcriptional activator protein Gal4, and the upstream activation sequence (UAS), which is a short section of the promoter region to which Gal4 specifically binds to activate gene transcription (Brand and Perrimon, 1993). By using the TARGET system (Gal4/Gal80, see Materials and methods; McGuire et al., 2004) in combination with UASCdc42N17, we specifically interfered with Cdc42 function in the adult heart without disturbing its function in the embryo. Interference with Cdc42 function specifically in the heart within the first week of adult life causes a dramatic increase in pacing-induced heart failure (Fig. 1 C), suggesting that Cdc42 is essential for maintaining normal heart function. These Cdc42N17 mutant hearts, when assessed for their beating pattern, also show increased incidence of arrhythmias and an overall lower heart rate, due to lengthening of diastolic intervals, compared with controls (Fig. 2, A–F). Using the mean standard deviation of heart period as an indicator of heart rhythm disturbance (“arrhythmia index”; Ocorr et al., 2007; Fink et al., 2009), we found a dramatic increase in arrhythmias in Cdc42N17 hearts (Fig. 2 F). These effects are more dramatic than in Cdc42 heterozygotes and together strongly support a crucial requirement for Cdc42 in regulating adult heart function.
Figure 2.
Cdc42 is required for adult heart function in flies. (A–C) Cardiac M-mode traces prepared from high speed movies of semi-intact flies. (A) M-mode from control flies shows a regular beating pattern. (B and C) Arrhythmic heart beats are evident in flies overexpressing Cdc42N17 using tinCΔ4-Gal4 (G) or 24BGal4 (H). (A′–C′) Heart period histograms. (D–F) Statistical analysis of Cdc42N17 mutant heart contractions (relative quantification [RQ] compared with control). Disruption of Cdc42 in the adult heart results in longer diastolic but not systolic intervals (D and E) and in increased incidence of arrhythmias (F). Error bars represent SEM. (G–I) α-Actinin labeling of the adult heart (one segment). Cdc42N17 adult heart or Cdc42, tinman double heterozygotes show disruption of cardiomyocyte myofibrillar alignment. Bars, 50 µm. (J and K) Quantification of abnormalities in Cdc42N17 and Cdc42+/−;tin+/− hearts. Severity is determined by the average number of segments with significant abnormal myofiber structure. n, number of flies tested. Unpaired Student's t test: *, P < 0.05.
The defective Cdc42N17 adult hearts also showed abnormalities in the cellular architecture of the cardiomyocytes, as manifest by severely misaligned myofibrils and irregularities in the repeated Z-line pattern (Fig. 2, G–I). These structural abnormalities in Cdc42 mutant hearts are consistent with Cdc42's role in actin filament assembly, also observed in Drosophila nurse cells, neurons, and epithelial cells (Genova et al., 2000; Jacinto et al., 2000; Jhaveri and Rodrigues, 2002). During Drosophila dorsal closure, Cdc42 activates the serine/threonine protein kinase Pak to regulate dynamic actin structure and epithelial plasticity (Harden et al., 1996; Bahri et al., 2010). Therefore, we reasoned that Cdc42 may act through the Pak pathway to regulate actin filament assembly and thus myofibrillar structure in the fly heart. To test this hypothesis, we generated Cdc42, Pak transheterozygotes and examined their cardiac structure and function (Fig. S1). Indeed, the majority of these transheterozygous hearts exhibited heart tube abnormalities where the myofibrils were misaligned or had gaps between the fibrils, whereas the hearts from single heterozygotes appear wild type (Fig. S1, A–C). Furthermore, functional analyses revealed that Cdc42, Pak double heterozygotes also exhibited other contractile abnormalities, which are likely due to the observed defects in myofibrillar organization. In contrast, the heart beat intervals and overall rhythmicity are not affected (Fig. S1, D–F), compared with Cdc42's interaction with tinman (Fig. 2, A–F). These data are consistent with a Cdc42 interaction with Pak in regulating cardiac actin alignment and thus the myofibrillar organization of the heart, but perhaps less so in regulating the heart rate and rhythm.
Cdc42–tinman interaction contributes to adult heart structure and function
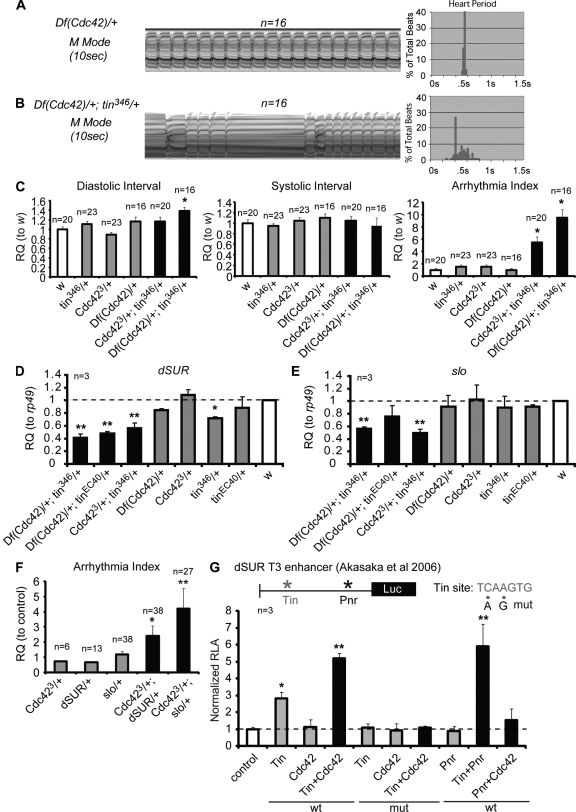
To further explore the Cdc42–tinman interaction, we examined the contractility and beating patterns in double-heterozygous hearts. Compared with the single heterozygotes, both tin346/Cdc423 and tin346/Df(Cdc42) flies exhibit an irregular beating pattern and a tendency toward longer diastolic intervals (Fig. 3, A–C), similar to Cdc42N17 hearts (Fig. 2, A–F). The elevated arrhythmias are also illustrated in the broadening of the heart period distribution in the double heterozygotes (Fig. 3, A′ and B′). Thus, combined partial loss of Cdc42 and tinman function synergistically compromises heart function. In addition, Cdc42 also shows a synergistic interaction with tinman in establishing a regular myofibrillar structure of the adult heart (Fig. 2, I and K). In contrast, a transheterozygous combination of Cdc42 with Df(2L)Exel6012, a deficiency covering the nmr/Tbx20 locus that also interacts with tinman (Qian et al., 2008b), fails to exhibit such an interaction (Fig. S1, G–L). Thus, Cdc42 seems to engage in a specific genetic interaction with tinman to regulate adult heart function in the adult fly.
Figure 3.
Cdc42 and tinman interact to maintain normal cardiac contraction. (A and B) Representative M-mode traces showing arrhythmic heart contractions in Df(Cdc42)/+;tin346/+ (B), compared with single heterozygotes Df(Cdc42)/+ (A). (A′–B′) Heart period histograms. (C) Statistical analysis of heart contraction in controls (w, tin346/+, Cdc423/+, Df(Cdc42)/+) and Cdc423/+;tin346/+ and Df(Cdc42)/+;tin346/+ flies, shown as relative quantification (RQ). Error bars represent SEM; *, P < 0.05 (one-way ANOVA). (D and E) RQ of dSUR (A) and slowpoke (B) mRNA in 1-wk-old adult hearts (normalized to rp49, ribosomal protein 49) relative to control (w). Double heterozygotes Df(Cdc42)/+;tin346/+, Df(Cdc42)/+;tinEC40/+, and Cdc423/+;tin346/+ showed lower expression than tinman or Cdc42 heterozygotes, or w controls. (F) Functional analyses of Cdc42,dSUR and Cdc42,slo double heterozygotes showed a significant increase in the arrhythmia index. (G) Luciferase assay using the dSUR T3 enhancer (Akasaka et al., 2006). Note the increased relative luciferase activity (RLA) by transfection of Tin was further enhanced by cotransfection with Cdc42 or Pnr, but not when Pnr and Cdc42 were cotransfected. Mutation of the Tin site abolished the responsiveness. Experiments were repeated three times. Error bars represent SEM. Unpaired Student’s t test: *, P < 0.05; **, P < 0.01.
To determine how Cdc42 and tinman interact to maintain heart function, we examined the gene expression levels of a cadre of cardiac-specific potential targets or effectors of the Cdc42–tinman interaction by quantitative RT-PCR (qPCR; see Materials and methods for complete list). In particular, we focused on ion channel genes that have previously been shown to regulate heart function in the adult fly (Bodmer, 2005; Akasaka et al., 2006; Ocorr et al., 2007; Qian et al., 2008a). Of these candidates, the two potassium channels dSUR and slowpoke were most severely down-regulated (Fig. 3, D and E; see Fig. S2, A and B, for examples of genes showing no changes), and they have previously been shown to be required for normal cardiac performance (Johnson et al., 1998; Akasaka et al., 2006). To determine whether Cdc42 genetically interacted with these two potential downstream effectors, we generated Cdc42+/−;dSUR+/− and Cdc42+/−;slo+/− flies. These transheterozygotes showed a defective cardiac structure as determined by α-Actinin staining (Fig. S2, C–H). These hearts do not show dramatically altered heart beat intervals (Fig. S2, I and J), but they do beat more arrhythmically than single heterozygous controls, as indicated by an increased arrhythmia index (Fig. 3 I). This suggests that Cdc42 indeed interacts with dSUR and slo to ensure normal heart structure and function. This interaction phenotype also suggests the possibility that malfunctioning ion channels may contribute to the structural remodeling (see Discussion). Interestingly, expression of dSUR is also down-regulated in the heart with age, and old fly hearts exhibit functional abnormalities, including an increased incidence of arrhythmias and pacing-induced heart failure (Akasaka et al., 2006; Ocorr et al., 2007), similar to what we observed in the (young) Cdc42+/−;tin+/− adult flies.
To further explore how Cdc42 interacts with tinman in regulating dSUR expression, we took advantage of a previously published enhancer element of dSUR (Akasaka et al., 2006) and performed luciferase assays in Drosophila S2 cells transfected with Cdc42 and/or tinman expression constructs. Consistent with the previous results, the dSUR enhancer element is responsive to cotransfection with a tinman expression construct (Fig. 3 G). Importantly, luciferase activity is dramatically enhanced by addition of a Cdc42 construct whereas transfection of Cdc42 by itself did not exert any effect on this enhancer (Fig. 3 G). Mutating the Tinman binding site in this enhancer abolished the responsiveness (Fig. 3 G). Conversely, addition of Cdc42 did not enhance the transcriptional activity of another transcription factor, the GATA factor Pannier, which in turn synergizes dramatically with Tinman in this assay (Fig. 3 G), as well as in vivo (Akasaka et al., 2006). These data suggest that Cdc42 may cooperate with Tinman (but not with Pannier) to activate the dSUR enhancer directly rather than by acting indirectly through separate pathways in the regulation of dSUR expression. This is consistent with the idea that the changes in expression of these ion channel genes may not be secondary to the sarcomeric defects in Cdc42+/−;tin+/− hearts. However, indirect mechanisms of Cdc42 interaction with Tinman’s transcriptional activity cannot be ruled out, as it is also possible that the myofibril alignment defects are secondary to the contractility defects, but all these considerations have to await future analysis. Based on the genetic interaction of Cdc42 with tinman presented here, it is suggested that Rho GTPases may represent a novel class of cardiac disease genes and/or act as polygenic interactors with known heart disease genes, such as tinman/Nkx2-5.
Cdc42 interacts with Nkx2-5 in modulating adult mouse heart function
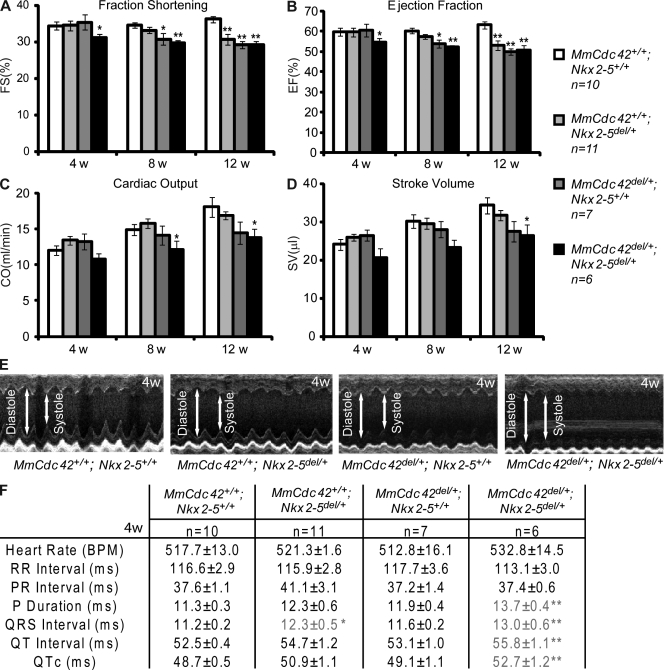
Because Cdc42 and tinman are conserved in vertebrates, we tested whether the interaction between them is also conserved in mice. We first generated compound Mm Cdc42del/+;Nkx2-5del/+ mice, which are viable at normal Mendelian ratios (unpublished data), and compared them to single-heterozygous littermates. We used high-resolution echocardiography to monitor structural and functional aspects of the postnatal mouse heart (Fig. 4, A–E). Mm Cdc42+/+;Nkx2-5+/+, Mm Cdc42+/+;Nkx2-5del/+, Mm Cdc42del/+;Nkx2-5+/+, and Mm Cdc42del/+;Nkx2-5del/+ adult mice were subjected to serial imaging at 4–12 wk after birth. At 4 wk of age, cardiac function in single heterozygotes (MmCdc42del/+ or Nkx2-5del/+) was not significantly different from wild-type littermates, whereas double heterozygotes showed declining heart function as indicated by decreased fraction of blood ejected from the left ventricle with each contraction (ejection fraction, EF), the fractional shortening of the ventricular chamber (fractional shortening, FS), volume of blood ejected with each heart beat (stroke volume, SV), and cardiac output (CO; see Fig. 4, A–D for quantification and Fig. 4 E for representative M-modes of the four genotypes). As the mice aged, single heterozygotes progressively exhibited weakened pumping ability, whereas double heterozygotes always performed worse than their single-heterozygous littermates (Fig. 4, A–D). Collectively, these results demonstrate that cardiac output, as assessed by multiple imaging parameters, is perturbed in Cdc42 heterozygous mice, and as expected in Nkx2-5 heterozygotes. Compellingly, these defects are markedly worse when the two alleles are combined in trans, suggesting that Cdc42 and Nkx2-5 synergistically regulate cardiac output and function in the mouse adult heart.
Figure 4.
Compound haploinsufficiency of mouse Cdc42 (MmCdc42) and Nkx2-5 resulted in defective cardiac contraction and electrophysiological function. (A–D) Histograms showing echocardiographic parameters of MmCdc42+/+;Nkx2-5+/+, MmCdc42+/+;Nkx2-5del/+, MmCdc42del/+;Nkx2-5+/+, and MmCdc42del/+;Nkx2-5del/+ adult mice at 4, 8, and 12 wk. Combined reduction of MmCdc42 and Nkx2-5 worsened heart function shown as (A) lower fraction shortening (FS), (B) ejection fraction (EF), (C) stroke volume (SV), and (D) cardiac output (CO) at all time points. Unpaired Student’s t test (relative to wild type): *, P < 0.05; **, P < 0.01. (E) Representative M-mode echocardiograms illustrate decrease in contractility of MmCdc42del/+;Nkx2-5del/+ hearts (right panel, larger systole) compared with controls. (F) Table with electrocardiographic parameters of MmCdc42 and Nkx2-5 combinations in adult mice (± SEM). One-way ANOVA: *, P < 0.05; **, P < 0.01.
Mouse models of Nkx2-5 deficiency revealed a critical role for Nkx2-5 in establishing and maintaining proper cardiac conduction system function (Pashmforoush et al., 2004; Briggs et al., 2008; Takeda et al., 2009), and human mutations in Nkx2-5 cause conduction-system dysfunction (Schott et al., 1998). Given the interaction between tinman and Cdc42 that we identified in the fly, we hypothesized that Nkx2-5 also interacts with Cdc42 in regulating the electrical activity of adult mouse heart. Therefore, we performed surface electrocardiography (ECG) and found that the average heart rate of single and double heterozygotes is not significantly different from that of wild-type mice, nor is there any difference in the delay between atrial and ventricular depolarization as indicated by the PR interval (Fig. 4 F). However, combined reduction of Cdc42 and Nkx2-5 resulted in prolonged atrial depolarization manifested by increased P duration (Fig. 4 F). The QRS complex, the portion of an ECG that corresponds to the depolarization of the right and left ventricles, was also significantly prolonged in double heterozygotes and was associated with prolongation of the QT and QTc intervals (Fig. 4 F). We also noticed that the double heterozygotes showed signs of right bundle branch block based upon S wave depression and the delays, as calculated in Fig. 4 F, which was not observed in single heterozygotes or wild-type controls. We then examined cardiac structure and fibrosis of these adult mice by performing the standard hematoxylin and eosin (H&E) and Masson Trichrome stainings, but did not observe any consistent differences among different genotypes (unpublished data and Fig. S3). Thus, compound heterozygosity for Nkx2-5 and Cdc42 does not alter cardiac morphology in the mouse as significantly as in the fly. This could be due to compensatory or redundant mechanisms that may be more prevalent in mammals. Nevertheless, our data show that Mm Cdc42 interacts strongly with Nkx2-5 in the mouse heart and that this genetic interaction is essential for proper cardiac contraction, electrical conduction, and rhythmicity.
Screening for CDC42 gene variants in human cohorts with heart disease
The strong evidence in fly and mouse that implicates Cdc42 as an important determinant of normal heart function points to CDC42 as a candidate gene for human heart disease. Mutations in the human homologue of tinman, NKX2-5, or other participants in the cardiogenic transcriptional network, TBX5, TBX20, and GATA4, have been associated with a variety of congenital and adult-onset heart disease. In humans, the CDC42 locus maps to chromosome 1p36. Monosomy 1p36 is the most common terminal deletion syndrome and results in mental retardation and multiple congenital anomalies, including atrial septal defects (ASD), ventricular septal defects (VSD), patent ductus arteriosus (PDA), and dilated cardiomyopathy (DCM). Loss of CDC42 could possibly account for the cardiac defects of this syndrome, as well as for cases in which these defects occur in isolation. We performed mutation screening of the CDC42 gene in two heart disease patient populations from Houston (J. Towbin) and Sydney (D. Fatkin).
In the Houston study, 331 subjects with heart disease, including 96 probands with dilated cardiomyopathy (DCM), 48 with hypertrophic cardiomyopathy (HCM), 91 with left ventricular noncompaction (LVNC), and 96 with congenital heart disease (CHD), as well as over 400 controls (over 800 chromosomes), were examined for potential variants in the human CDC42 gene (GenBank/EMBL/DDBJ accession no. NM_001791; Table I). Seven nonsynonymous and seven synonymous variants and four intronic variants were identified. Six out of the seven nonsynonymous variants were polymorphic and identified in subjects and controls. A single rare nonsynonymous variant, c.539A>G, which is predicted to result in an amino acid change of T125A, was not detected in over 300 Caucasian and over 100 Hispanic controls (Table I). This variant was found in a sporadic CHD patient with an atrial septal defect (ASD) and a patent ductus arteriosus (PDA). The parents did not participate in the study. In the Sydney study, over 300 heart disease patients (AF, CHD, LVNC) and 100 controls were examined (Table II), with no apparent disease-causing mutations identified. Some X synonymous variants were identified and no nonsynonymous variants were found.
Table I.
Nonsynonymous or synonymous variants identified in CDC42 (NM_001791) in probands either with DCM, HCM, LVNC, or congenital heart diseases (Houston study)
| Location | Nucleotide change | Amino acid change | Type of variant | Patient group | Frequency | |||
| DCM (n = 96) | HCM (n = 48) | CHD (n = 96) | LVNC (n = 91) | |||||
| Amplicon1 (Exon 1) | c.188G>T | V8F | NP | 6 | 0 | 0 | 0 | 6/331 |
| (1.8%) | ||||||||
| c.242A>C | N26N | S | 1 | 0 | 0 | 0 | 1/331 | |
| (0.3%) | ||||||||
| c.256G>A | S30S | S | 2 | 0 | 1 | 0 | 3/331 | |
| (0.9%) | ||||||||
| c.257G>C | E31Q | NP | 46 | 11 | 52 | 29 | 138/331 | |
| (41.7%) | ||||||||
| c.271+6 T>G | Intron 3 splice ds | NP | 2 | 5 | 0 | 0 | 7/331 | |
| (2.1%) | ||||||||
| Amplicon2 (Exon2) | c.272-9 insT | Intron3 splice as | NP | 4 | 0 | 0 | 0 | 4/331 |
| (1.2%) | ||||||||
| c.272-7 T>C | Intron3 splice as | NP | 5 | 2 | 0 | 8 | 15/331 | |
| (4.5%) | ||||||||
| c.272-2 T>C | Intron3 splice as | NP | 6 | 0 | 4 | 3 | 13/331 | |
| (3.9%) | ||||||||
| c.289 A>C | A41A | S | 4 | 0 | 2 | 1 | 7/331 | |
| (2.1%) | ||||||||
| c.295 A>C | T43T | S | 5 | 0 | 0 | 0 | 5/331 | |
| (1.5%) | ||||||||
| c.317 T>C | Y51H | NP | 1 | 22 | 14 | 10 | 47/331 | |
| (14.2%) | ||||||||
| Amplicon4 (Exon 4) | c.502T>A | L112L | S | 0 | 0 | 0 | 2 | 2/331 |
| (0.6%) | ||||||||
| c.513A>C | Q116P | NP | 5 | 0 | 0 | 1 | 6/331 | |
| (1.8%) | ||||||||
| c.523 C>T | L119L | S | 7 | 0 | 1 | 2 | 10/331 | |
| (3.0%) | ||||||||
| c.525 G>A | R120K | NP | 2 | 0 | 49 | 0 | 51/331 | |
| (15.4%) | ||||||||
| c.628 T>C | Y154Y | S | 5 | 1 | 2 | 0 | 8/331 | |
| (2.4%) | ||||||||
| (rs16826536) | ||||||||
| c.539 A>G | T125A | Rare variant | 0 | 0 | 1 | 0 | 1/331 | |
| (0.3%) | ||||||||
| Amplicon5 (Exon5) | c.730 T>G | C188W | NP | 3 | 0 | 10 | 12 | 25/331 |
| (7.5%) | ||||||||
DCM, dilated cardiac myopathy; HCM, hypertrophic cardiac myopathy; LVNC, left ventricular noncompaction; CHD, congenital heart disease; S, synonymous change; NP, nonsynonymous polymorphism.
Table II.
Cdc42 variants identified in 328 individuals with adult and congenital heart disease (Sydney study)
| Location | Variant | Amino acid change | Patient group | Published SNP frequency | Reference | ||
| AF (n = 100) | CHD (n = 200) | LVNC (n = 28) | |||||
| Intron 3 | −22G>T | NA | 3 (0.03%) | 1 (0.005%) | 0 | NA | Rs41300114 |
| Intron 3 | −8insT | NA | 3 (0.03%) | 8 (0.04%) | 0 | NA | Novel |
| Intron 3 | −3T>C | NA | 0 | 0 | 1 (0.04%) | 3/90 (0.03%) | Rs17837976 |
| Exon 5 | 180G>A | G60G | 0 | 1 (0.005%) | 0 | ||
| Exon 5 | 276C>T | N92N | 1 (0.01%) | 0 | 0 | 1/90 (0.01%) | Rs16826534 |
| Exon 6 | 462T>C | Y154Y | 1 (0.01%) | 0 | 0 | 4/85 (0.05%) | Rs16826536 |
| Exon 8 | 546G>A | P182P | 0 | 0 | 1 (0.04%) | 1/85 (0.01%) | Rs16826536 |
AF, atrial fibrillation; CHD, congenital heart disease; LVNC, left ventricular noncompaction; NA, not available; SNP, single nucleotide polymorphism.
These data indicate that despite the clear biological significance of Cdc42 in the heart, coding sequence variants in the CDC42 gene are not a common cause of CHD or adult heart disease. A possible reason is that this small GTPase is extremely conserved, which does not allow for any sequence variation resulting in an amino acid change to be tolerated, because most may result in lethality. Future mutation screening in intronic and regulatory regions of this gene, resulting in only partial loss of function, and studies in larger patient cohorts will likely provide further insights into the potential role of CDC42 as a new human heart disease gene.
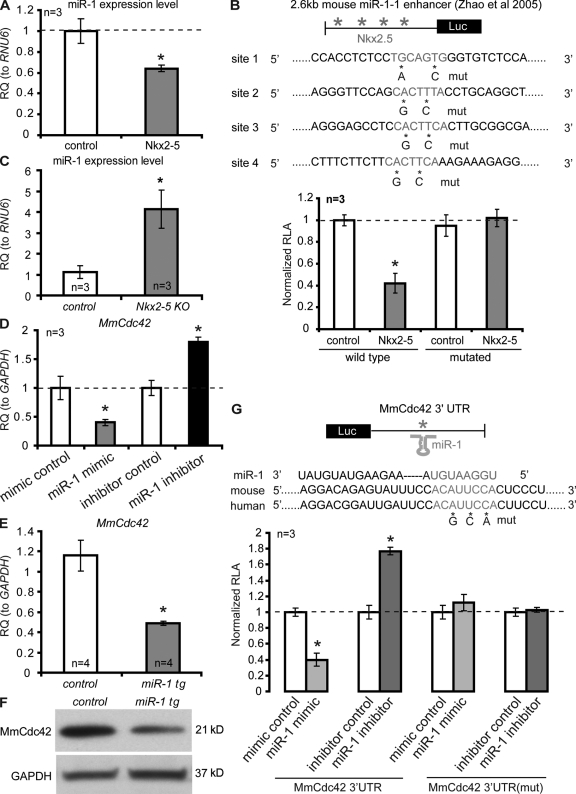
Cdc42 is a direct target of miR-1 that is negatively regulated by Nkx2-5
Examination of the upstream regulatory region of mouse miR-1 revealed the presence of four Nkx2-5 consensus binding sites (Fig. 5 B). This made us wonder whether Cdc42 is regulated by miR-1, which would provide a possible mechanism for the genetic interaction between Tinman/Nkx2-5 and Cdc42. Indeed, we identified a miR-1 consensus target site in the 3′-UTR of Cdc42 (Fig. 5 G, see below).
Figure 5.
MmCdc42 is a direct target of miR-1 that is negatively regulated by Nkx2-5. (A) Relative levels (RQ) of miR-1 expression determined by qPCR in HL-1 cells electroporated with Nkx2-5 compared to control (GFP). (B) A 2.6-kb miR-1-1 enhancer luciferase construct containing four wild-type or mutated Nkx2-5 sites (Zhao et al., 2005) was cotransfected with Nkx2-5. (C) qPCR of miR-1 from Nkx2-5del/del mouse embryos (E9.5) relative to wild-type littermate controls. (D) qPCR of Cdc42 mRNA levels from HL-1 cells transfected with miR-1 mimic or miR-1 inhibitor. Experiments were done in three biological triplicates and technical quadruplicates. (E) qPCR of MmCdc42 mRNA from 4 miR-1 transgenic mouse hearts (αMHC-miR-1) compared with four littermate controls. (F) Western blot of MmCdc42 protein from αMHC-miR-1 transgenic and control mice. (G) RLA from cells cotransfected with a luciferase reporter—containing wild-type or mutated miR-1 target site in the Cdc42 3′UTR—and a miR-1 mimic or inhibitor. Experiments (each condition) were repeated three times with technical quadruplicates. Unpaired Student’s t test: *, P < 0.05; **, P < 0.01.
To determine whether Nkx2-5 is capable of regulating miR-1 transcription we transfected an Nkx2-5 expression construct into HL-1 cells, a line derived from atrial cardiomyocytes (Claycomb et al., 1998). We found a decrease in miR-1 levels detected by qPCR (Fig. 5 A). Similar results were obtained when we transfected Nkx2-5 into primary cardiomyocytes that were directly isolated from neonatal hearts (not depicted). This suggests that Nkx2-5 negatively regulates miR-1 in cardiomyocytes. Furthermore, we identified four conserved Nkx2-5 binding sites in the 2.6-kb minimal enhancer region of miR-1 (Fig. 5 B). To test whether Nkx2-5 represses miR-1 by binding to these sites, the miR-1 enhancer was cloned upstream of the coding region of a luciferase reporter. A decrease in luciferase activity was observed when the constitutively active reporter was cotransfected with Nkx2-5 into HL-1 cells (Fig. 5 B). Mutations of the Nkx2.5 binding sites in the miR-1 enhancer abolished Nkx2-5 responsiveness (Fig. 5 B), suggesting that Nkx2-5 represses miR-1 by directly binding to this enhancer.
Next, we tested whether miR-1 directly regulates Cdc42 by transfecting HL-1 cells with miR-1 mimics (chemically synthesized double-stranded oligonucleotides that mimic the function of endogenous mature miR-1) and miR-1 inhibitors (modified antisense oligoribonucleotides that inhibit miR-1 function). Overexpression of miR-1 resulted in decreased endogenous Cdc42 expression, whereas inhibition of miR-1 caused an increased expression of Cdc42 (Fig. 5 D), which is consistent with the presence of a conserved miR-1 binding site in the Cdc42 3′-UTR (Fig. 5 G). To test whether miR-1 is capable of directly repressing Cdc42 via this site, the Cdc42 3′-UTR was inserted into the 3′-UTR of a luciferase reporter, downstream of the coding region. Transfection of the Cdc42 3′-UTR luciferase reporter with miR-1 mimic into HL-1 cells results in a decrease in luciferase activity, whereas cotransfection with miR-1 inhibitor increases luciferase activity (Fig. 5 G). A gradient of miR-1 was used to confirm the luciferase activity was dose responsive (Fig. S4 A). Mutations of the miR-1 binding site in the Cdc42 3′-UTR abolished miR-1 responsiveness (Fig. 5 G), suggesting that miR-1 represses Cdc42 by physically binding to its 3′-UTR.
We extracted RNA and protein from transgenic mice in which miR-1 was overexpressed in the adult heart via the α-MHC promoter (α-MHC–miR-1; unpublished mice from the Srivastava laboratory) and examined Cdc42 mRNA and protein levels. Consistently, miR-1 transgenic hearts showed significantly reduced Cdc42 mRNA and protein levels (Fig. 5, E and F), suggesting a repressive relationship between miR-1 and Cdc42 in vivo. We did not observe increased Cdc42 expression in miR-1-2 knockout hearts, which might be due to the incomplete depletion of miR-1 expression because of the intact miR-1-1 locus (Zhao et al., 2007; unpublished data). Furthermore, we determined miR-1 levels in Nkx2.5 knockout embryos (E9.5) by qPCR and found an increase in miR-1 levels compared with littermate controls (Fig. 5 C). These data provide further evidence that the genetic interaction between Nkx2-5 and Cdc42 is mediated by miR-1.
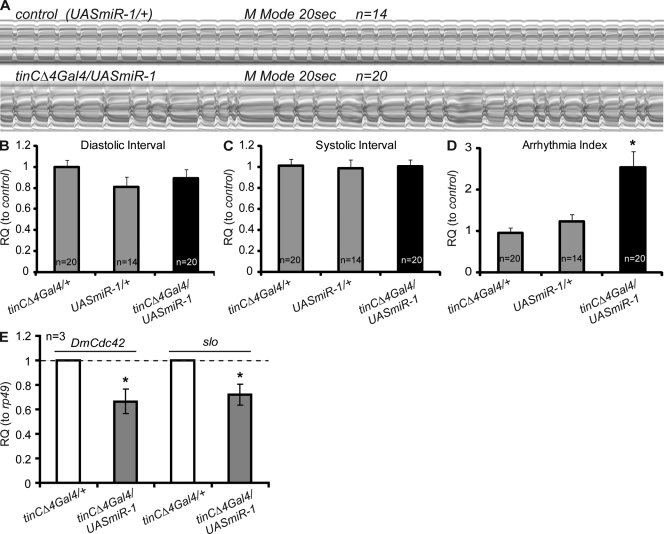
We then wondered whether the genetic interaction between tinman and Cdc42 in Drosophila is also governed by a double-inhibitory mechanism involving the fly homologue of miR-1. We found that cardiac-specific overexpression of miR-1 causes an increase in arrhythmias (Fig. 6, A–D), comparable to cardiac expression of dominant-negative Cdc42N17 (Fig. 2). In addition, fly hearts with excessive cardiac miR-1 expression exhibit reduced levels of Cdc42 and slo mRNA in the heart (Fig. 6 E), suggesting that miR-1’s influence on heart function includes inhibition of Cdc42 and one or more potassium channels. In contrast to the mouse, however, we did not identify any miR-1 binding sites in the 3′-UTR of fly Cdc42. To test whether fly Cdc42 regulation by miR-1 is through binding to “seedless” 3’-UTR sequences that lack predicted microRNA recognition elements (Bamshad et al., 1997), we cloned fly Cdc42 3′-UTR into the 3′-UTR of a luciferase reporter and performed luciferase assays in Drosophila S2 cells. However, we did not observe consistent and reproducible repression in the presence of miR-1 (Fig. S4 B). Recent evidence suggests that microRNA target sites may not always be conserved (Hyun et al., 2009; unpublished data), even though the pathway overall is likely conserved, as in this case and in a recent report (King et al., 2011). Thus, it remains to be determined how microRNAs regulate downstream effectors (such as Cdc42; Fig. 6 E), directly by cryptic sites or via an unknown possibly indirect mechanism. However, we did find two consensus binding sites for Tinman within the enhancer of fly miR-1, and luciferase assays in S2 cells revealed that these two sites are responsible for the regulation of fly miR-1 by Tinman (Fig. S5). Taken together, these data suggest a functionally conserved regulatory pathway involving tinman–miR-1–Cdc42 in establishing cardiac functionality, although some aspects of the molecular mechanisms involved may be different between flies and vertebrates, which could also explain the different severity in phenotypes between flies and mice in our study.
Figure 6.
Overexpression of miR-1 causes arrhythmias in the adult fly heart. (A) M-mode traces illustrate arrhythmic heart contractions with cardiac miR-1 overexpression (tinCΔ4Gal4/UASmiR-1). (B–D) Statistical analysis of heart contraction in miR-1–overexpressing adult hearts. (E) Cardiac qPCR of fly Cdc42 (DmCdc42) and potassium channels slowpoke (slo) with tinCΔ4Gal4/UASmiR-1. Heart-specific overexpression of miR-1 resulted in reduced cardiac expression of Cdc42 and slo. *, P < 0.05.
Discussion
Drosophila has been successfully used to study regulatory genetic networks of inductive signals and transcription factors that determine cardiac specification and morphogenesis, but the genetic control of cardiac physiology is only beginning to be investigated. Using a cardiac performance-based genetic screen, we discovered a novel genetic interaction between Cdc42 and the NK homeobox gene tinman in regulating cardiac function in the fly. We have identified a heretofore-unknown requirement for the Rho GTPase Cdc42 in regulating the establishment of cardiac function in the adult fly. Significantly, this interaction is conserved in mice, where combined reduction of Cdc42 and Nkx2-5 caused defects in heart contraction, rhythm, and electrophysiological function. Based on the discovery of the interaction between this Rho GTPase with a cardiogenic transcription factor in maintaining proper cardiac function, it is conceivable that other genetic interactors and potential polygenic contributors to heart disease traits can be identified using the fly as a screening platform.
Cdc42 is a well-characterized GTPase, and has been widely studied in different organs and tissues, ranging from worms to humans. Because Cdc42 was shown to be involved in regulation of the actin/myosin cytoskeleton in the cells of various organisms, we reasoned that Cdc42 might also control cardiac cell morphogenesis. In Drosophila and mouse we found that complete loss of Cdc42 in the heart leads to defects in cardiac morphogenesis (unpublished data). In the adult heart, Cdc42 was previously shown to be associated with cardiac myocyte hypertrophy (Clerk and Sugden, 2000; Pandur et al., 2002; Sellin et al., 2006). Recent work shows that Cdc42 plays a protective role against hypertrophy, as conditional deletion of Cdc42 in the adult heart led to greater cardiac hypertrophy and increased incidence of heart failure (Maillet et al., 2009). Interestingly, our findings show that interference with Cdc42 function in the adult fly heart via expression of a dominant-negative form of Cdc42 is sufficient to induce functional and morphological defects, suggesting that, in addition to interacting with tinman, Cdc42 itself is essential for maintaining normal adult heart structure and function. The specific differences between the fly and the mouse phenotypes in these studies may be attributed to differences in assays as well as to evolutionary changes of Cdc42 function from insects to vertebrates. However, the fact that Cdc42 adult-specific mutant fly hearts are defective in Z-line/myofibril assembly is consistent with a previous report in a tissue culture model for cardiac hypertrophy showing that sarcomere units are misassembled in cultured rat cardiomyocytes when Cdc42 expression is disrupted (Nagai et al., 2003; Brown et al., 2006). In addition, Cdc42 in Drosophila interacts with a known downstream effector of Cdc42, Pak, which participates in actin filament assembly and dynamics (Harden et al., 1996; Bahri et al., 2010), thus consistent with a possible role for Cdc42 in myofibrillar organization.
Similar to our Cdc42;Nkx2-5 compound heterozygote analysis, it has been shown that mice transheterozygous for loss-of-function alleles of Nkx2-5 and Tbx5 also exhibit a synergistic interaction, in particular with respect to the development of the cardiac conduction system (Moskowitz et al., 2007). It is therefore likely that other cardiac transcription factors in combination with new genes will reveal similar patterns of interactions in flies and mammals. Although different model organisms have been used to study cardiac disease, the ability to investigate polygenic traits underlying these diseases in a systematic fashion in the adult is limited. Here, we provide evidence that rigorous screening for modifiers of heart disease can be achieved using Drosophila, leading to identification of similar polygenic trait interactions in mammals.
The genetic relationship between fly Cdc42 with the cardiac determinant tinman in regulating adult heart contraction may occur, in part, via modulation of potassium channel gene expression, dSUR and slo, which have previously been shown to be critical regulators of heart function (Johnson et al., 1998; Akasaka et al., 2006). Indeed, Cdc42-dSUR or –slo transheterozygotes show a strong interaction reflected in increased arrhythmias. Interestingly, these transheterozygotes also exhibit disturbances in myofibrillar organization, suggesting these ion channel abnormalities somehow also produce sarcomeric defects, perhaps via a secondary structural remodeling effect. Although there is increasing evidence of such remodeling processes being affected by ion channel defects (unpublished data), the underlying mechanism is not yet understood.
A strong interaction was also observed between the mouse homologues of Cdc42 and Nkx2-5 in adult heart function. Most interestingly, double haploinsufficiency of Cdc42 and Nkx2-5 led to elongated QRS intervals, which is typical of abnormal conduction along the atrio-ventricular bundle, bundle branches, and Purkinje fibers. The observed elongated QT intervals in compound heterozygotes are also characteristic of long-QT syndrome in humans. These findings suggest a possible contribution of compound haploinsufficiency of Cdc42 and Nkx2-5 in human cardiac conduction diseases.
While attempting to elucidate the mechanistic link between Cdc42 and Nkx2-5, we found that miR-1 directly represses Cdc42 and that miR-1 itself can be repressed by Nkx2.5 expression. miR-1 is the first microRNA identified in the heart and plays a conserved role in cardiac morphogenesis, cell cycle, and conduction (Zhao et al., 2005, 2007). miR-1 null mutant mice showed cardiac conduction defects including widened QRS complex and prolonged QT intervals (Zhao et al., 2007), which is also affected in Cdc42;Nkx2-5 double-heterozygous mice. We speculate that loss or gain of miR-1 function causes an imbalance in gene expression in the heart, including that of Cdc42. Decreasing the copy number of the direct miR-1 target Cdc42, and that of a direct transcriptional miR-1 repressor, Nkx2-5, apparently destroys a delicate balance in the adult heart, tipping it toward deregulated cardiac function. This does not rule out that other mechanisms are also involved in the genetic interaction between Cdc42 and Nkx2-5. Because Cdc42 is a direct target of miR-1 and has a crucial function in modulating hypertrophic responses in the mouse, it will be interesting to test whether miR-1 and other microRNAs play a direct role in adult cardiac hypertrophy. Interestingly, Cdc42 was also found to be a target of miR-133, which is involved in cardiac hypertrophy (Carè et al., 2007). Thus, the RhoGTPase Cdc42 appears to be directly regulated by two major cardiac microRNAs that are involved in controlling cardiac function and hypertrophy. The synergistic transcriptional activation of the dSUR enhancer by Tinman and Cdc42 in vitro (Fig. 3 G) is consistent with the idea that Cdc42 can act in a coactivating mechanism with Tinman, but this remains to be further investigated.
In conclusion, we have used the power of Drosophila genetics to uncover a new, conserved genetic pathway, linking genes with critical heart functions, tinman/Nkx2-5, miR-1, and Cdc42, to a novel network regulating adult heart physiology and performance. With the fly as a screening platform, it is thus possible to identify new polygenic contributors that ensure normal cardiac function relevant to human heart disease.
Materials and methods
Drosophila stocks
The following mutant stocks were used: Cdc423/FM6 (Boutros et al., 1998); tin346/TM3-ftz-lacZ (Azpiazu and Frasch, 1993); tinEC40/TM3, Df(3R)tinGC14 (Bodmer, 1993); Pak6/TM3,Sb (Pehlivan et al., 1999); Df(2L)Exel9032/Cyo (indicated as dSUR/+), Df(3R)BSC397/TM3,Sb (indicated as slo/+), Df(2L)Exel6012 (Qian et al., 2005b; Exelixis deficiency deleting both nmr1 and nmr2, also indicated as Df(nmr)), and Df(1)Exel6253 (Exelixis deficiency deleting the whole genomic region of Cdc42, also indicated as Df(Cdc42)). Wild-type control flies are w1118. Overexpression of transgenes was achieved using the UAS-Gal4 system (Brand and Perrimon, 1993). The following Gal4 and UAS lines were used: twi-Gal4 (twi>; Greig and Akam, 1993), 24B-Gal4 (24B>; Brand and Perrimon, 1993), the double combination twi-Gal4;24B-Gal4 (twi24B>; pan-mesodermal expression; Lockwood and Bodmer, 2002), tinCΔ4-Gal4 (Lo and Frasch, 2001), and UAS-Cdc42N17 (Luo et al., 1994). We used TARGET to temporally and spatially activate gene expression (McGuire et al., 2004). In brief, we raised flies (with Gal4, Gal80-ts, and UAS constructs) at 18°C before eclosion; after eclosion we raised adult flies at 29°C for 1 wk and then performed heart function assays.
Mouse stocks
Mice harboring a loss-of-function allele for Nkx2-5 (Nkx2-5del/+/Nkx2.5tm1Siz) have been described previously (Tanaka et al., 1999). Mice harboring a floxed allele of Cdc42 (Cdc42tm1Yizh) have also been described previously (Chen et al., 2006). Cdc42 males were bred to Mef2c-AHF-cre (Verzi et al., 2005) females to enable germ line deletion of Cdc42 (Cdc42del/+), and offspring that did not inherit the Cre allele were then mated to Nkx2.5 heterozygous animals to generate compound heterozygous and single heterozygous offspring. For all functional studies detailed below, littermates of each indicated genotype were analyzed.
Immunohistochemistry
For fly heart immunohistochemistry, antibody staining was performed as described previously (Han et al., 2002). In brief, after micro-dissection, fly hearts were fixed in 4% PFA overnight at 4°C. After blocking in 10% BSA (Sigma-Aldrich)/PBS for 1 h, fly hearts were incubated in primary antibodies for 1 h, washed three times (15 min each) with PBT (PBS + 0.5% Triton), and then incubated in secondary antibodies for 1 h. After washing 3 × 15 min in PBT, fly hearts were mounted in VectaShield (Vector Laboratories). Cy3- or FITC-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used for fluorescent confocal microscopy. All the secondary antibodies were used at 1:200. Fly heart preparations were analyzed using a confocal microscope (model LSM510; Carl Zeiss). Mouse anti–α-Actinin (Saide et al., 1989) was used in this study. For mouse heart immunohistochemistry, after perfusion fix the heart was removed and fixed by immersion in 4% PFA in PBS (diluted from 20% PFA stock; Electron Microscopy Sciences) and routinely processed and paraffin embedded. 5-µm sections were stained with hematoxylin and eosin (H&E) and Masson Trichrome and analyzed for regular morphology and fibrosis. Images were acquired by Zeiss LSM software (for confocal images) or Image-Pro Plus 5.1 (for regular IHC images). Adobe Photoshop was used for image processing; contrast and brightness were adjusted for better visualization.
Electrical pacing in flies
The electrical pacing was conducted as described previously (Wessells and Bodmer, 2004; Wessells et al., 2004). In brief, 50–100 flies were paced with a square wave stimulator at 40 V and 6 Hz for 30 s and scored for heart failure rate, defined as the percentage of flies that either exhibit cardiac arrest or continuous fibrillation immediately after pacing.
Image analysis and M-mode traces on semi-intact fly preparations
Procedures used were as described previously (Ocorr et al., 2007; Fink et al., 2009). In brief, flies were anesthetized and the head, ventral thorax, and ventral abdominal cuticle were removed. All internal organs except the heart were removed as well as any abdominal fat. Dissections were done under oxygenated saline (108 mM Na+, 5 mM K+, 2 mM Ca2+, 8 mM MgCl2, 1 mM NaH2PO4, 4 mM NaHCO3, 10 mM sucrose, 5 mM trehalose, and 5 mM Hepes, pH 7.1). These semi-intact preparations (Ocorr et al., 2009; Vogler and Ocorr, 2009) were allowed to equilibrate with oxygenation for 10–20 min before filming. These procedures were performed at room temperature. Image capture of heart contractions was performed using an EM-CCD digital camera (Hamamatsu Photonics) on a microscope (model DM LFSA; Leica) using direct immersion lenses. High-speed movies taken at rates of 100–200 frames per second were acquired and contrast enhanced using Simple PCI imaging software (Compix, Inc.). Movie analysis and M-mode generation was performed using a MatLab-based image analysis program (Ocorr et al., 2007, 2009; Fink et al., 2009; Vogler and Ocorr, 2009).
Mouse echocardiography
The mouse protocol was approved by institutional guidelines (UCSF). All analyses were performed in a blinded fashion for genotype and intervention. Mice were anesthetized with 2.4% isoflurane/97.6% oxygen and placed in a supine position on a heating pad (37°C). Echocardiography was performed by a High-Resolution Micro-Imaging System (Vevo 770; VisualSonics) with a 15-MHz linear array ultrasound transducer. The left ventricle (LV) was assessed in both parasternal long-axis and short-axis views at a frame rate of 120 Hz. End-systole or end-diastole was defined as the phase in which the smallest or largest area of LV, respectively, was obtained. Left ventricular end-systolic diameter (LVESD) and left ventricular end-diastolic diameter (LVEDD) were measured from the LV M-mode tracing with a sweep speed of 50 mm/s at the papillary muscle level.
Mouse electrocardiography
For surface electrocardiography, mice were anesthetized with 1.75% isoflurane at a core temperature of 37–38°C. Four needle electrodes (AD Instruments) were placed subcutaneously in standard limb lead configurations. For each mouse, 10–20 s of continuous signals were sampled at 10 KHz in each lead configuration using a PowerLab4/30 interface (AD Instruments). Data analysis was performed offline using electronic calipers on averaged beats (Chart5Pro v5.4.2; AD Instruments). The QRS interval was measured from the onset of the Q-wave to the isoelectric point preceding the first, rapid repolarization wave. The QT interval was measured from the onset of the Q-wave to the end of the second, slower repolarization wave. The measured QT interval was corrected for heart rate using the rodent correction formula, QTc = QT/(RR/100)0.5.
Quantitative real-time PCR (qPCR)
For fly qPCR, total RNA was extracted from 20 dissected hearts of 1-wk-old adult females by using TRIzol (Invitrogen). After lysis with TRIzol followed by phase separation, the aqueous phase was subjected to RNA extraction (Mini RNA Isolation kit; Zymo Research) with on-column DNaseI treatment (QIAGEN). After RNA extraction with RNase-free water, the first-strand cDNA was immediately transcribed with SuperScript III (Invitrogen) by using oligo(dT) primer, followed by the second-strand synthesis with DNA polymerase I, E. coli DNA ligase, and RNase H. Quantitative PCR was performed using the LightCycler FastStart DNA Master PLUS SYBR Green I kit (Roche). The primer sets were as follows: for rp49, 5′-GACGCTTCAAGGGACAGTATCTG-3′ and 5′-AAACGCGGTTCTGCATGAG-3′; for dSUR, 5′-CATCACCATTGCCCATCGTCT-3′ and 5′-CGCCCTTCTCCAGTAAACC-3′; for slowpoke, 5′-GCCAACAGATCAGGTATTCGT-3′ and 5′-GCGCTACACAGTAACAATCA-3′; for cdc42, 5′-CTACACACTGGGCCTGTTC-3′ and 5′-TTTGGCAATGGTGTGTAATC-3′. The following gene expressions did not show any consistent and/or significant difference among the genotypes we tested: potassium channel genes: shaker, shaker-like, seizure, Inwardly rectifying potassium channel (Ir), and KCNQ; calcium channel and exchanger genes: cacophony, Na/Ca-exchange protein (calx), Inositol 1,4,5,-tris-phosphate receptor (Itp-r83A), and Na+-driven anion exchanger 1 (Ndae1).
For mouse qPCR, RNA was extracted by TRizol method (Invitrogen) from individual hearts. RT-PCR was performed using the Superscript III First-Strand Synthesis System (Invitrogen). qPCR was performed using the ABI 7900HT system (TaqMan; Applied Biosystems), per the manufacturer’s protocols. Optimized primers from Taqman Gene Expression Array were used. miRNA RT was conducted using the Taqman MicroRNA Reverse Transcription kit (Applied Biosystems). miRNA real-time PCR (qRT-PCR) was performed per the manufacturer’s protocols by using primers from Taqman MicroRNA Assays (Applied Biosystems). Expression levels were normalized to GAPDH expression and miR-16 (microRNA qPCR).
Cell culture, transfection, and luciferase assay
HL-1 cells were maintained in Calycomb Medium supplemented with 100 µM norepinephrine, 10% FBS, and 4 mM l-glutamine. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2/95% air. Lipofectamine 2000 (Invitrogen)–mediated transfection was performed according to Invitrogen’s protocol. miR-1 mimic and inhibitor were purchased from Thermo Fisher Scientific. For each transfection in one well of a six-well plate, 40 pmol of mimic or inhibitor was used. For plasmid transfection, 100 ng of plasmid was transfected in one well of a six-well plate using the Amaxa Nucleofection kit (Lonza) per the manufacturer’s protocol. Luciferase assays were performed as described previously (Zhao et al., 2005) with the Dual-Luciferase Reporter Assay System (Promega). Cdc42 3′-UTR was cloned from mouse genomic DNA with the following primers: 5′-ACTAGTACGCATCTCCAGAGCCCTTTCTGC-3′ and 5′-AAGCTTTAGCCCTGACTGGTCCCCATGTTGG-3′. Amplified DNA was cloned into pCRII vector and subsequently cloned into pMIR-REPORT vector (Invitrogen). Site-directed PCR-mediated mutagenesis was performed using the QuickChange II XL Site-Directed Mutagenesis kit (Agilent Technologies). Plasmids containing Tinman and Nkx2.5 have been described previously (Akasaka et al., 2006; Takeuchi and Bruneau, 2009). Tinman cDNA was finally inserted in pUAST vector; Nkx2-5 cDNA was finally inserted in pCDNA3.1 vector. Luciferase constructs containing miR-1 enhancer and dSUR enhancer were described previously (Zhao et al., 2005; Akasaka et al., 2006).
Western blots
Western blots were performed as described previously (Zhao et al., 2005). Rabbit anti-Cdc42 (Cell Signaling Technology) and rabbit anti-GAPDH (Santa Cruz Biotechnology, Inc.) were both used at a 1:1,000 dilution for Western blots. All Western blots were quantified using AlphaImager software from Alpha Innovations.
Statistics
Mean, standard deviation, and standard error of the mean were calculated. Error bars are represented as standard error of the mean. Sample numbers were indicated in corresponding figures. Comparisons between groups were made by one-way ANOVA or unpaired two-tails assuming equal variance t test, as applicable. For fly pacing experiments, χ2 test was applied.
Human subjects and DNA sequence analysis
Houston study.
The study population from Baylor College of Medicine included 331 unrelated pediatric probands either sporadic or familial cases of heart disease including 96 individuals with dilated cardiomyopathy (DCM), 48 individuals with hypertrophic cardiomyopathy (HCM), 96 individuals with congenital heart disease (CHD), and 91 individuals with either isolated or nonisolated left ventricular noncompaction (LVNC; Table I). All patients were evaluated by physical examination, chest radiography, electrocardiography, echocardiography, cardiac magnetic resonance imaging (cMRI), and angiography. Left ventricular size and function was evaluated by M-mode and two-dimensional echocardiography, Doppler, and color Doppler analyses. Further details on heart function measures are as described in Qian et al. (2008b). All subjects or their guardians provided written informed consent. Blood for EBV-transformed lymphoblastoid cell lines and DNA extraction was obtained, as regulated by the Baylor College of Medicine Institutional Review Board. DNA was extracted either directly from blood or from cultured cells using the Puregene DNA Isolation kit (Genetra Systems) according to the manufacturer’s protocol. Primers were designed in to the intron sequences flanking coding exons of CDC42 (Genbank/EMBL/DDBJ accession no. NM_001791). Protein encoding amplicons were amplified by polymerase chain reaction using genomic DNA, as described previously (Qian et al., 2008b). PCR products were purified and sequenced by automated sequencing using a DNA sequencer (ABI 3730; Applied Biosystems) and Big Dye terminator chemistry (version 3.1) according to the manufacturer’s instructions. The only rare variant we found was T125A in a CHD patient. Over 300 Caucasian and >100 Hispanic controls were analyzed for all variants identified.
Sydney study.
The population study in Australia was comprised of 100 individuals with adult-onset familial atrial fibrillation (AF), 200 individuals with CHD, comprised of ventricular septal defect (VSD), n = 113; atrial septal defect (ASD), n = 59; patent ductus arteriosis (PDA), n = 28, and 28 individuals with isolated or syndromic LVNC. Seven sequence variants were identified, including four synonymous amino acid substitutions and three intronic variants. Five of these variants have been reported previously and two were novel (Table II). Of note, the T125A variant from the Houston study was not detected in any DNA samples from the 328 heart disease patients or in samples from 100 healthy Caucasian volunteers (Table II). These patient populations were recruited from St. Vincent’s Hospital, Darlinghurst, the Kids Heart Research DNA Bank at The Children’s Hospital at Westmead, the Royal Prince Alfred Hospital, Camperdown, and the National Australian Childhood Cardiomyopathy Study. 100 healthy Caucasian volunteers comprised a control group. All participants gave informed written consent and study protocols were approved by the relevant institutional Human Research Ethics Committees.
DNA sequencing
DNA was isolated from patient blood samples using conventional techniques. Protein-coding sequences and adjacent intronic sequences of the CDC42 gene were amplified by polymerase chain reaction from genomic DNA using primers derived from intron sequences. Amplimers were purified and sequenced using the Big Dye terminator and were analyzed on a DNA Analyzer (ABI PRISM 3700; Applied Biosystems) at the Ramaciotti Centre for Gene Function Analysis (University of New South Wales, Kensington, New South Wales, Australia).
Online supplemental material
Fig. S1 describes how Cdc42 interacts with Pak to regulate actin filament alignment and cardiac function in fly adult heart and Cdc42 does not interact with nmr/Tbx20 in maintaining proper heart function. Fig. S2 shows regulation of ion channel genes dSUR and slowpoke by Cdc42 and tinman. Fig. S3 shows cardiac morphology and fibrosis of MmCdc42+/+;Nkx2-5+/+, MmCdc42+/+;Nkx2-5del/+, MmCdc42del/+;Nkx2-5+/+, and MmCdc42del/+;Nkx2-5del/+ adult mice. Fig. S4 shows additional luciferase assays to test regulation of Cdc42 by miR-1. Fig. S5 shows regulation of fly miR-1 by Tinman. Table S1 shows the cardiac arrest or fibrillation rate for deficiency lines covering chromosome X and part of chromosome II. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006114/DC1.
Acknowledgments
We thank the Bloomington stock center and Developmental Studies Hybridoma Bank for sending fly-stocks and antibodies. We are grateful for technique support from T. Akasaka and L. Elmén, and to Dr. Philip Ursell (Anatomical Pathology, UCSF) for assistance with interpretation of mouse histology.
L. Qian and J. Liu were supported by a predoctoral fellowship from the American Heart Association. L. Qian is supported by a postdoctoral scholarship from the California Institute for Regenerative Medicine and a fellowship from the Lynda and Stewart Resnick Foundation. K. Ocorr is supported by the American Heart Association. J.D. Wythe is supported by a postdoctoral fellowship from the National Institutes of Health (T32HL007731). This work was funded by grants from the NHLBI of the National Institutes of Health to J.A. Towbin, J.D. Molkentin, B.G. Bruneau, D. Srivastava, and R. Bodmer, and from the National Health and Medical Research Council (Australia) to D. Fatkin, R.P. Harvey, and C. Semsarian.
Footnotes
Abbreviations used in this paper:
- CHD
- congenital heart disease
- UAS
- upstream activation sequence
References
- Adams M.D., Sekelsky J.J. 2002. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat. Rev. Genet. 3:189–198 10.1038/nrg752 [DOI] [PubMed] [Google Scholar]
- Akasaka T., Klinedinst S., Ocorr K., Bustamante E.L., Kim S.K., Bodmer R. 2006. The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc. Natl. Acad. Sci. USA. 103:11999–12004 10.1073/pnas.0603098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N., Frasch M. 1993. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7(7B):1325–1340 10.1101/gad.7.7b.1325 [DOI] [PubMed] [Google Scholar]
- Bahri S., Wang S., Conder R., Choy J., Vlachos S., Dong K., Merino C., Sigrist S., Molnar C., Yang X., et al. 2010. The leading edge during dorsal closure as a model for epithelial plasticity: Pak is required for recruitment of the Scribble complex and septate junction formation. Development. 137:2023–2032 10.1242/dev.045088 [DOI] [PubMed] [Google Scholar]
- Bamshad M., Lin R.C., Law D.J., Watkins W.C., Krakowiak P.A., Moore M.E., Franceschini P., Lala R., Holmes L.B., Gebuhr T.C., et al. 1997. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet. 16:311–315 10.1038/ng0797-311 [DOI] [PubMed] [Google Scholar]
- Bier E. 2005. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6:9–23 10.1038/nrg1503 [DOI] [PubMed] [Google Scholar]
- Bier E., Bodmer R. 2004. Drosophila, an emerging model for cardiac disease. Gene. 342:1–11 10.1016/j.gene.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Bodmer R. 1993. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 118:719–729 [DOI] [PubMed] [Google Scholar]
- Bodmer R. 1995. Heart development in Drosophila and its relationship to vertebrates. Trends Cardiovasc. Med. 5:21–28 10.1016/1050-1738(94)00032-Q [DOI] [PubMed] [Google Scholar]
- Bodmer R. 2005. Development of the Cardiac Musculature. Muscle Development in Drosophila. Sink H., editor [Google Scholar]
- Boutros M., Paricio N., Strutt D.I., Mlodzik M. 1998. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 94:109–118 10.1016/S0092-8674(00)81226-X [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415 [DOI] [PubMed] [Google Scholar]
- Briggs L.E., Takeda M., Cuadra A.E., Wakimoto H., Marks M.H., Walker A.J., Seki T., Oh S.P., Lu J.T., Sumners C., et al. 2008. Perinatal loss of Nkx2-5 results in rapid conduction and contraction defects. Circ. Res. 103:580–590 10.1161/CIRCRESAHA.108.171835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.H., Del Re D.P., Sussman M.A. 2006. The Rac and Rho hall of fame: a decade of hypertrophic signaling hits. Circ. Res. 98:730–742 10.1161/01.RES.0000216039.75913.9e [DOI] [PubMed] [Google Scholar]
- Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D., et al. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13:613–618 10.1038/nm1582 [DOI] [PubMed] [Google Scholar]
- Chen L., Liao G., Yang L., Campbell K., Nakafuku M., Kuan C.Y., Zheng Y. 2006. Cdc42 deficiency causes Sonic hedgehog-independent holoprosencephaly. Proc. Natl. Acad. Sci. USA. 103:16520–16525 10.1073/pnas.0603533103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb W.C., Lanson N.A., Jr, Stallworth B.S., Egeland D.B., Delcarpio J.B., Bahinski A., Izzo N.J., Jr 1998. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 95:2979–2984 10.1073/pnas.95.6.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerk A., Sugden P.H. 2000. Small guanine nucleotide-binding proteins and myocardial hypertrophy. Circ. Res. 86:1019–1023 [DOI] [PubMed] [Google Scholar]
- Cripps R.M., Olson E.N. 2002. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 246:14–28 10.1006/dbio.2002.0666 [DOI] [PubMed] [Google Scholar]
- Eaton S., Auvinen P., Luo L., Jan Y.N., Simons K. 1995. CDC42 and Rac1 control different actin-dependent processes in the Drosophila wing disc epithelium. J. Cell Biol. 131:151–164 10.1083/jcb.131.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon R.G., Oren T., LaJeunesse D.R., Melby T.E., McCartney B.M. 1997. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics. 146:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink M., Callol-Massot C., Chu A., Ruiz-Lozano P., Izpisua Belmonte J.C., Giles W., Bodmer R., Ocorr K. 2009. A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. Biotechniques. 46:101–113 10.2144/000113078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova J.L., Jong S., Camp J.T., Fehon R.G. 2000. Functional analysis of Cdc42 in actin filament assembly, epithelial morphogenesis, and cell signaling during Drosophila development. Dev. Biol. 221:181–194 10.1006/dbio.2000.9671 [DOI] [PubMed] [Google Scholar]
- Greig S., Akam M. 1993. Homeotic genes autonomously specify one aspect of pattern in the Drosophila mesoderm. Nature. 362:630–632 10.1038/362630a0 [DOI] [PubMed] [Google Scholar]
- Han Z., Fujioka M., Su M., Liu M., Jaynes J.B., Bodmer R. 2002. Transcriptional integration of competence modulated by mutual repression generates cell-type specificity within the cardiogenic mesoderm. Dev. Biol. 252:225–240 10.1006/dbio.2002.0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden N., Lee J., Loh H.Y., Ong Y.M., Tan I., Leung T., Manser E., Lim L. 1996. A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol. Cell. Biol. 16:1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Hurd T.W., Gao L., Roh M.H., Macara I.G., Margolis B. 2003. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 5:137–142 10.1038/ncb923 [DOI] [PubMed] [Google Scholar]
- Hyun S., Lee J.H., Jin H., Nam J., Namkoong B., Lee G., Chung J., Kim V.N. 2009. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 139:1096–1108 10.1016/j.cell.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Jacinto A., Wood W., Balayo T., Turmaine M., Martinez-Arias A., Martin P. 2000. Dynamic actin-based epithelial adhesion and cell matching during Drosophila dorsal closure. Curr. Biol. 10:1420–1426 10.1016/S0960-9822(00)00796-X [DOI] [PubMed] [Google Scholar]
- Jhaveri D., Rodrigues V. 2002. Sensory neurons of the Atonal lineage pioneer the formation of glomeruli within the adult Drosophila olfactory lobe. Development. 129:1251–1260 [DOI] [PubMed] [Google Scholar]
- Johnson E., Ringo J., Bray N., Dowse H. 1998. Genetic and pharmacological identification of ion channels central to the Drosophila cardiac pacemaker. J. Neurogenet. 12:1–24 10.3109/01677069809108552 [DOI] [PubMed] [Google Scholar]
- King I.N., Qian L., Liang J., Huang Y., Shieh J.T., Kwon C., Srivastava D. 2011. A genome-wide screen reveals a role for microRNA-1 in modulating cardiac cell polarity. Dev. Cell. 20:497–510 10.1016/j.devcel.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo P.C., Frasch M. 2001. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech. Dev. 104:49–60 10.1016/S0925-4773(01)00361-6 [DOI] [PubMed] [Google Scholar]
- Lockwood W.K., Bodmer R. 2002. The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech. Dev. 114:13–26 10.1016/S0925-4773(02)00044-8 [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y.J., Jan L.Y., Jan Y.N. 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8:1787–1802 10.1101/gad.8.15.1787 [DOI] [PubMed] [Google Scholar]
- Maillet M., Lynch J.M., Sanna B., York A.J., Zheng Y., Molkentin J.D. 2009. Cdc42 is an antihypertrophic molecular switch in the mouse heart. J. Clin. Invest. 119:3079–3088 10.1172/JCI37694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S.E., Mao Z., Davis R.L. 2004. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004:pl6 10.1126/stke.2202004pl6 [DOI] [PubMed] [Google Scholar]
- Moskowitz I.P., Kim J.B., Moore M.L., Wolf C.M., Peterson M.A., Shendure J., Nobrega M.A., Yokota Y., Berul C., Izumo S., et al. 2007. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 129:1365–1376 10.1016/j.cell.2007.04.036 [DOI] [PubMed] [Google Scholar]
- Nagai T., Tanaka-Ishikawa M., Aikawa R., Ishihara H., Zhu W., Yazaki Y., Nagai R., Komuro I. 2003. Cdc42 plays a critical role in assembly of sarcomere units in series of cardiac myocytes. Biochem. Biophys. Res. Commun. 305:806–810 10.1016/S0006-291X(03)00838-6 [DOI] [PubMed] [Google Scholar]
- Neely G.G., Kuba K., Cammarato A., Isobe K., Amann S., Zhang L., Murata M., Elmén L., Gupta V., Arora S., et al. 2010. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 141:142–153 10.1016/j.cell.2010.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K., Reeves N.L., Wessells R.J., Fink M., Chen H.S., Akasaka T., Yasuda S., Metzger J.M., Giles W., Posakony J.W., Bodmer R. 2007. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc. Natl. Acad. Sci. USA. 104:3943–3948 10.1073/pnas.0609278104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K., Fink M., Cammarato A., Bernstein S., Bodmer R. 2009. Semi-automated optical heartbeat analysis of small hearts. J. Vis. Exp. pii: 1435 10.3791/1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N. 2006. Gene regulatory networks in the evolution and development of the heart. Science. 313:1922–1927 10.1126/science.1132292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur P., Läsche M., Eisenberg L.M., Kühl M. 2002. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 418:636–641 10.1038/nature00921 [DOI] [PubMed] [Google Scholar]
- Pashmforoush M., Lu J.T., Chen H., Amand T.S., Kondo R., Pradervand S., Evans S.M., Clark B., Feramisco J.R., Giles W., et al. 2004. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 117:373–386 10.1016/S0092-8674(04)00405-2 [DOI] [PubMed] [Google Scholar]
- Pehlivan T., Pober B.R., Brueckner M., Garrett S., Slaugh R., Van Rheeden R., Wilson D.B., Watson M.S., Hing A.V. 1999. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am. J. Med. Genet. 83:201–206 [DOI] [PubMed] [Google Scholar]
- Prall O.W., Elliott D.A., Harvey R.P. 2002. Developmental paradigms in heart disease: insights from tinman. Ann. Med. 34:148–156 [PubMed] [Google Scholar]
- Qian L., Bodmer R. 2009. Partial loss of GATA factor Pannier impairs adult heart function in Drosophila. Hum. Mol. Genet. 18:3153–3163 10.1093/hmg/ddp254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Liu J., Bodmer R. 2005a. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev. Biol. 279:509–524 10.1016/j.ydbio.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Qian L., Liu J., Bodmer R. 2005b. Slit and Robo control cardiac cell polarity and morphogenesis. Curr. Biol. 15:2271–2278 10.1016/j.cub.2005.10.037 [DOI] [PubMed] [Google Scholar]
- Qian L., Liu J., Bodmer R. 2008a. Heart development in Drosophila. Advances in Developmental Biology. Vol. 18 Bodmer R., editor Elsevier; 1–29 [Google Scholar]
- Qian L., Mohapatra B., Akasaka T., Liu J., Ocorr K., Towbin J.A., Bodmer R. 2008b. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc. Natl. Acad. Sci. USA. 105:19833–19838 10.1073/pnas.0808705105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saide J.D., Chin-Bow S., Hogan-Sheldon J., Busquets-Turner L., Vigoreaux J.O., Valgeirsdottir K., Pardue M.L. 1989. Characterization of components of Z-bands in the fibrillar flight muscle of Drosophila melanogaster. J. Cell Biol. 109:2157–2167 10.1083/jcb.109.5.2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J.J., Benson D.W., Basson C.T., Pease W., Silberbach G.M., Moak J.P., Maron B.J., Seidman C.E., Seidman J.G. 1998. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 281:108–111 10.1126/science.281.5373.108 [DOI] [PubMed] [Google Scholar]
- Sellin J., Albrecht S., Kölsch V., Paululat A. 2006. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expr. Patterns. 6:360–375 10.1016/j.modgep.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Srivastava D. 2006. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 126:1037–1048 10.1016/j.cell.2006.09.003 [DOI] [PubMed] [Google Scholar]
- St Johnston D. 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3:176–188 10.1038/nrg751 [DOI] [PubMed] [Google Scholar]
- Takeda M., Briggs L.E., Wakimoto H., Marks M.H., Warren S.A., Lu J.T., Weinberg E.O., Robertson K.D., Chien K.R., Kasahara H. 2009. Slow progressive conduction and contraction defects in loss of Nkx2-5 mice after cardiomyocyte terminal differentiation. Lab. Invest. 89:983–993 10.1038/labinvest.2009.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J.K., Bruneau B.G. 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 459:708–711 10.1038/nature08039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Wechsler S.B., Lee I.W., Yamasaki N., Lawitts J.A., Izumo S. 1999. Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development. 126:1439–1450 [DOI] [PubMed] [Google Scholar]
- Verzi M.P., McCulley D.J., De Val S., Dodou E., Black B.L. 2005. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 287:134–145 10.1016/j.ydbio.2005.08.041 [DOI] [PubMed] [Google Scholar]
- Vogler G., Ocorr K. 2009. Visualizing the beating heart in Drosophila. J. Vis. Exp. pii: 1425 10.3791/1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells R.J., Bodmer R. 2004. Screening assays for heart function mutants in Drosophila. Biotechniques. 37:58–60: 62: 64 passim [DOI] [PubMed] [Google Scholar]
- Wessells R.J., Fitzgerald E., Cypser J.R., Tatar M., Bodmer R. 2004. Insulin regulation of heart function in aging fruit flies. Nat. Genet. 36:1275–1281 10.1038/ng1476 [DOI] [PubMed] [Google Scholar]
- Zaffran S., Reim I., Qian L., Lo P.C., Bodmer R., Frasch M. 2006. Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development. 133:4073–4083 10.1242/dev.02586 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 436:214–220 10.1038/nature03817 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. 2007. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 129:303–317 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]