Abstract
By using a mini-transposon, we obtained two mutated strains of a diarrheal isolate, SSU, of Aeromonas hydrophila that exhibited a 50 to 53% reduction in the hemolytic activity and 83 to 87% less cytotoxic activity associated with the cytotoxic enterotoxin (Act). Act is a potent virulence factor of A. hydrophila and has been shown to contribute significantly to the development of both diarrhea and septicemia in animal models. Subsequent cloning and DNA sequence analysis revealed that transposon insertion occurred at different locations in these two mutants within the same 1,890-bp open reading frame for the glucose-inhibited division gene (gidA). A similar reduction in hemolytic (46%) and cytotoxic (81%) activity of Act was noted in the gidA isogenic mutant of A. hydrophila that was generated by marker exchange mutagenesis. Northern blot analysis revealed that the transcription of the cytotoxic enterotoxin gene (act) was not altered in the gidA transposon and isogenic mutants. However, by generating a chromosomal act::alkaline phosphatase gene (phoA) reporter construct, we demonstrated significantly reduced phosphatase activity in these mutants, indicating the effect of glucose-inhibited division (GidA) protein in modulating act gene expression at the translational level. The biological effects of Act in the gidA mutants were restored by complementation. The virulence of the gidA mutants in mice was dramatically reduced compared to the those of the wild-type (WT) and complemented strains of A. hydrophila. The histopathological examination of lungs, in particular, indicated severe congestion, alveolar hemorrhage, and acute inflammatory infiltrate in the interstitial compartment and the alveolar spaces when mice were infected with the WT and complemented strains. Minimal-to-mild changes were noted in the lungs with the gidA mutants. Taken together, our data indicate for the first time that GidA regulates the most-potent virulence factor of A. hydrophila, Act.
Aeromonas hydrophila is an emerging human pathogen that causes septicemia and gastroenteritis (13). The organism is increasingly becoming resistant to chlorination in water and to multiple antibiotics (3, 9). As a result, the Environmental Protection Agency has placed this organism on the “Candidate Contaminant List,” and the monitoring of U.S. water supplies for this organism was started in 2002. Epidemiological studies implicated Aeromonas species in causing food-borne outbreaks and traveler's diarrhea (13). Pathogenesis of A. hydrophila infections is multifactorial and complex (1, 4). Two categories of enterotoxins, namely, cytotonic and cytotoxic, were molecularly characterized in our laboratory (12, 14-16), and we demonstrated their role in causing gastroenteritis and/or septicemia by developing isogenic mutants (52, 57). In addition to these enterotoxins, Merino et al. (34, 35) reported that many strains of Aeromonas have an S-layer that resists complement-mediated killing by impeding complement activation. Aeromonas spp. produce a wide range of proteases which could cause tissue damage and aid in establishing an infection by overcoming host defenses and providing nutrients for cell proliferation (44). Other virulence factors, such as lectins, adhesins, type IV pili, and flagella, are also associated with the pathogenesis of Aeromonas infections (13). Recently, we characterized A. hydrophila surface-expressed enolase, a glycolytic enzyme, which bound human plasminogen and could be involved in disseminating bacteria to different organs by dissolving fibrin clots (51). Interestingly, the enolase gene was differentially expressed in vivo (51).
Among the three enterotoxins of A. hydrophila characterized in our laboratory (52), the cytotoxic enterotoxin (Act) is the most potent and has hemolytic, cytotoxic, and enterotoxic activities (14, 21, 57). In addition, the toxin is lethal to mice when injected intravenously, with a 50% lethal dose of 27 ng (21). Act is a single-chain polypeptide, and in its mature form the toxin is 49 to 52 kDa. The culture filtrate from an act isogenic mutant from A. hydrophila SSU was devoid of hemolytic and cytotoxic activities and evoked a minimal enterotoxic response over an observation period of 4 to 6 h. Further, the mutant was significantly attenuated in causing infection in a mouse model (57). We demonstrated that Act activated proinflammatory cytokine and eicosanoid cascades in murine macrophages and in rat intestinal epithelial cells, leading to tissue damage and a fluid secretory response (17, 49). We recently published detailed studies in which we used microarrays to examine Act-induced cell signaling, leading to the activation of cytokine and eicosanoid cascades in macrophages (24, 49). The present study was undertaken to identify potential regulatory gene(s) that could control act gene expression.
Previously, we demonstrated that Act-associated hemolytic activity in A. hydrophila was affected by a number of environment stimuli, such as iron, calcium, pH, temperature, and glucose (53). Subsequently, we molecularly characterized a ferric uptake regulatory gene (fur) that repressed act gene expression in the presence of high amounts of iron (53). In searching for other regulatory genes, we used transposon mutagenesis and obtained A. hydrophila mutants with significantly lower hemolytic and cytotoxic activities. Our subsequent cloning and sequence analysis revealed that transposition in two studied mutants occurred within the glucose-inhibited division gene (gidA). A similar and significant reduction in the hemolytic and cytotoxic activity was also noted in the gidA isogenic mutant of A. hydrophila. Both gidA transposon and isogenic mutants were avirulent in mice, with minimal to mild pathology, compared to the wild-type (WT) and complemented strains of A. hydrophila, which were lethal to mice and caused severe pathology in the lungs, liver, and spleen.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The sources of A. hydrophila SSU and Escherichia coli strains, as well as the plasmids used in this study, are listed in Table 1. For transposon mutagenesis, the mini-transposon mini-Tn5 Sm/Sp (with resistance to streptomycin [STR] and spectinomycin [SPT]) (19) was delivered into A. hydrophila by the pUT plasmid, which is a derivative of the pGP704 suicide vector (38). The suicide vector pJQ200SK contained a P15A origin of replication (ori), a levansucrase gene (sacB) from Bacillus subtilis, and a gentamicin resistance (Gmr) gene (46). The pJQactphoA plasmid contained an in-frame fusion of the act gene with the reporter alkaline phosphatase gene (phoA) in suicide vector pJQ200SK. This plasmid was used to generate reporter A. hydrophila SSU strains by single-crossover recombination in the chromosome for monitoring the act gene expression level by measuring phosphatase activity (Table 1) (53).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| A. hydrophila | ||
| SSU | CDCa | |
| SSU-R | Rifr strain of A. hydrophila SSU | Laboratory stock |
| M1 | gidA transposon mutant of A. hydrophila SSU-R; Rifr Sp/Smr | This study |
| M19 | gidA transposon mutant of A. hydrophila SSU-R; Rifr Sp/Smr | This study |
| MgidA | gidA isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr, sucrose resistance | This study |
| SSU-R/pBR322 | SSU-R containing pBR322 vector; Rifr Tcr | This study |
| M19/pBR322 | gidA transposon mutant M19 containing pBR322 vector; Rifr Sp/Smr Tcr | This study |
| MgidA/pBR322 | gidA isogenic mutant containing pBR322 vector; Rifr Kmr Tcr | This study |
| M19/pBRgidA | gidA transposon mutant M19 complemented with the gidA gene via pBR322 vector; Rifr Sp/Smr Tcr | This study |
| MgidA/pBRgidA | gidA isogenic mutant complemented with the gidA gene via pBR322 vector; Rifr Kmr Tcr | This study |
| SSU66 | A. hydrophila SSU-R with chromosomally integrated act::phoA fusion; Rifr Gmr sucrose sensitive | This study |
| M19phoA | Transposon mutant M19 with chromosomally integrated act::phoA fusion; Rifr Gmr Sp/Smr, sucrose sensitive | This study |
| MgidAphoA | gidA isogenic mutant with chromosomally integrated act::phoA fusion; Rifr Gmr Kmr, sucrose sensitive | This study |
| SSU66/pBR322 | SSU66 containing pBR322 vector; Tcr Rifr Gmr, sucrose sensitive | This study |
| M19phoA/pBR322 | M19phoA containing pBR322 vector; Tcr Rifr Gmr Sp/Smr, sucrose sensitive | This study |
| MgidAphoA/pBR322 | MgidAphoA containing pBR322 vector; Tcr Rifr Gmr Kmr, sucrose sensitive | This study |
| M19phoA/pBRgidA | M19phoA complemented with the gidA gene via pBR322; Tcr Rifr Gmr Sp/Smr, sucrose sensitive | This study |
| MgidAphoA/pBRgidA | MgidAphoA complemented with the gidA gene via pBR322; Tcr Rifr Gmr Kmr, sucrose sensitive | This study |
| E. coli | ||
| DH5α | recA gyrA | Laboratory stock |
| S17-1 | Streptomycin and trimethoprim resistance, λpir | Laboratory stock |
| Plasmids | ||
| pRK2013 | Helper plasmid, Kmr | ATCCb |
| pBR322 | Apr Tcr | Amersham |
| pBluescript SK | Apr | Stratagene |
| pUC-4K | Contains a 1.2-kb kanamycinr gene cassette | Amersham |
| pHP45Ω | Contains a 2.0-kb Sm/Spr gene cassette | 45 |
| pJQ200SK | Suicide vector; P15A, sacB Gmr | 46 |
| Mini-Tn5 Sm/Sp | Minitransposon containing Sp/Smr gene cassette delivered by the pUT plasmid | 19 |
| pJQ200actphoA | Vector pJQ200SK containing act::phoA fusion; Gmr | 53 |
| pBlueM1 | pBluescript vector containing a 6.6-kb PstI DNA fragment from M1, which harbored the interrupted gidA gene generated by mini-Tn5 transposition | This study |
| pBlueM19 | pBluescript vector containing a 6.6-kb PstI DNA fragment from M19, which harbored the interrupted gidA gene generated by mini-Tn5 transposition | This study |
| pBlgidA | pBluescript vector containing coding region of gidA gene of A. hydrophila | This study |
| pBlgidA-Km | gidA gene in plasmid pBluegidA interrupted at the BglII site with a Kmr gene cassette | This study |
| pJQgidA-Km | Vector pJQ200SK containing a Kmr gene cassette interrupting the gidA gene for generating a gidA isogenic mutant, MgidA | This study |
| pBRgidA | A. hydrophila gidA gene with its putative promoter region cloned in pBR322 at the EcoRI/PstI sites | This study |
CDC, Centers for Disease Control and Prevention, Atlanta, Ga.
ATCC, American Type Culture Collection.
Enzymes, chemicals, and recombinant DNA techniques.
The antibiotics ampicillin, gentamicin, tetracycline (TET), kanamycin (KAN), SPT, and STR were used at concentrations of 100, 15, 30, 50, 50, and 50 μg/ml, respectively. Unless otherwise stated, rifampin (RIF) was used at a concentration of 100 μg/ml for bacterial growth and 300 μg/ml during conjugation experiments. All of the antibiotics used were obtained from Sigma (St. Louis, Mo.). Restriction endonucleases and T4 DNA ligase were obtained from Promega (Madison, Wis.) and New England BioLabs (Beverly, Mass.). The Advantage cDNA PCR kit was purchased from BD Bioscience Clontech (Palo Alto, Calif.). Chromosomal DNA from various A. hydrophila mutants was isolated using a QIAamp DNA Mini kit (Qiagen, Inc., Valencia, Calif.). The digested plasmid DNA and the DNA fragments from the agarose gel were prepared and purified using a QIAprep Miniprep kit (Qiagen). All of the basic molecular biology techniques used in this study were previously described (5, 50, 57).
Bacterial growth curves and Act production.
The growth rate of WT and various A. hydrophila mutants over a period of the first 24 h was determined by plating (100 μl) of various dilutions (10−2 to 10−10) of the cultures on Luria-Bertani (LB) agar plates containing appropriate antibiotics. An aliquot of the sample was taken every hour for 14 h and then every 2 h up to 24 h. After overnight incubation of the plates at 37°C, the number of CFU per milliliter of sample was calculated. The CFU count was also correlated with the optical density of the culture by measuring the A600. The culture supernatants at various time points were used for examining the hemolytic activity as described later.
Transposon mutagenesis.
The mini-transposon mini-Tn5 Sm/Sp, from plasmid pUT in E. coli, was delivered to A. hydrophila by conjugation as previously described (27, 48). Briefly, both E. coli and Rifr A. hydrophila SSU were grown under static conditions at 37°C overnight. The cultures were mixed in different ratios and centrifuged at 5,000 rpm (Sorvall RC 5B plus centrifuge; Kendo Laboratory Products, Newtown, Conn.) for 10 min. The cells were resuspended in 200 μl of LB medium and transferred onto LB plates without any antibiotic pressure. After 4 h of incubation at 37°C, the cultures were removed from the plates and various dilutions (10−4 to 10−9) of the sample were transferred to LB agar plates with RIF, SPT, and STR antibiotics. Subsequently, the transconjugants were spotted onto 5% sheep blood agar plates (Fisher Scientific, Pittsburgh, Pa.) for examination of hemolytic activity. The cultures were identified as Aeromonas by using oxidase test reagent (Difco, Sparks, Md.) to differentiate them from E. coli (52).
Northern blot analysis to depict act gene transcription in the gidA transposon mutants.
By using an RNAqueous kit (Ambion, Inc., Austin, Tex.), we isolated RNA from WT A. hydrophila and its gidA transposon and isogenic mutants. The isolated RNA samples (10 μg) were then subjected to Northern blot analysis as described by us previously (52). A 1.4-kb [α-32P]dCTP-labeled act gene that was PCR amplified from the plasmid pXHC95 (57) was used as a probe to detect the act transcript. The amount of RNA in each lane was quantitated by scanning 23S or 16S rRNA bands after ethidium bromide staining of the gel, using the Gel Doc 2000 System (Bio-Rad Laboratories, Hercules, Calif.).
Southern blot analysis on the chromosomal DNA of transposon mutants of A. hydrophila SSU with act gene and Sp/Smr gene cassette probes.
An aliquot (10 μg) of the chromosomal DNA from the transposon mutants M1 and M19 (Table 1), as well as WT A. hydrophila, was digested with suitable enzymes and subjected to 0.8% agarose gel electrophoresis (52). Southern blot analysis was performed with the act and Sp/Smr gene probes as described in our previous studies (52, 53, 57).
Localization of the transposon insertion sites in the mutants of A. hydrophila SSU.
Based on Southern blot analysis data obtained with the Sp/Smr gene probe, the PstI-digested chromosomal DNA fragments in the size range of 6.4 to 6.7 kb from the transposon mutants were recovered and ligated to the pBluescript vector (Table 1). The correct transformants were screened on LB agar plates containing SPT and STR, and their identities were confirmed again by Southern blot analysis using an Sp/Smr gene probe. The cloned DNA fragments were further analyzed by automated DNA sequencing. The automated DNA sequencing was performed in the Protein Chemistry Core Facility at The University of Texas Medical Branch, Galveston. Based on the Blast search with the GenBank database, the interrupted open reading frames (ORFs) on the chromosome of transposon mutants M1 and M19 were located. The precise transposon insertion sites within the ORFs were determined by DNA sequence comparison between the transposon mutants and WT A. hydrophila.
Generation of a mutated gidA gene (MgidA) of A. hydrophila SSU by marker exchange mutagenesis.
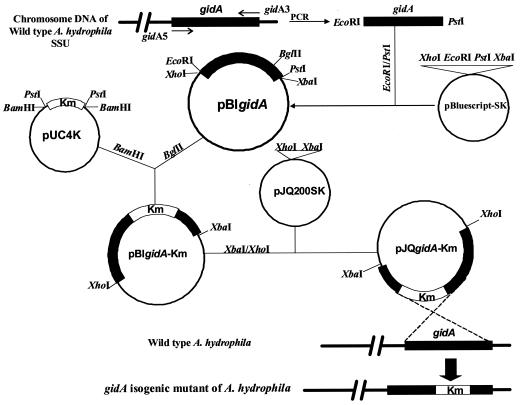
As shown in Fig. 1, based on the DNA sequence obtained from M1 and M19 transposon mutants, two primers (gidA-5 with an EcoRI restriction site [underlined], 5′ GTGAATTCATGCAATACCATGAACAATTTG 3′ [forward], and gidA-3 with a PstI restriction site, [underlined] 5′ GTCTGCAGGTAGGATCCTCAGGCGGTT 3′ [reverse]) were synthesized to depict the coding region of the gidA gene. The gidA gene (1,890 bp) was PCR amplified from the chromosomal DNA of WT A. hydrophila and subsequently cloned into pBluescript vector at the EcoRI/PstI sites, forming a recombinant plasmid, pBlgidA (Fig. 1). In the gidA gene coding region, there was a unique BglII restriction site; the plasmid pBlgidA was thus linearized with BglII digestion (Fig. 1). A 1.2-kb Kmr gene cartridge was isolated from plasmid pUC4K (Amersham) by using restriction enzyme BamHI, which bordered the Kmr gene cassette. This Kmr gene cassette was ligated to plasmid pBlgidA at the BamHI-compatible BglII site to interrupt the gidA gene, which generated a new recombinant plasmid, pBlgidA-Km (Fig. 1). Subsequently, the DNA fragment, which was now 3.2 kb, was removed from the plasmid pBlgidA-Km by XbaI/XhoI digestion and ligated to a suicide vector, pJQ200SK, at the XbaI/XhoI sites, forming a new recombinant plasmid, pJQgidA-Km, in E. coli strain S17-1 (Fig. 1; Table 1). This strategy to prepare an isogenic mutant provided 358 bp and 1.5 kb of the flanking 5′ and 3′ DNA sequences to the mutated gidA gene, respectively, to permit double-crossover homologous recombination.
FIG. 1.
Flow diagram showing the strategy used for preparation of the gidA isogenic mutant of A. hydrophila SSU. By using PCR, the coding region of the gidA gene was amplified from the chromosomal DNA of WT A. hydrophila and was subsequently cloned into pBluescript vector at the EcoRI/PstI sites to generate a recombinant plasmid, pBlgidA. A Kmr gene cassette from plasmid pUC4K interrupted the gidA gene at the BglII restriction site and generated the recombinant plasmid pBlgidA-Km. The mutated gidA gene was removed from the pBlgidA-Km plasmid by XhoI/XbaI digestion and cloned into a suicide vector, pJQ200SK, at the compatible sites, forming a recombinant plasmid, pJQgidA-Km, for the generation of the gidA mutant of A. hydrophila. The solid bar represents the gidA gene, while the open bar represents the Kmr gene cassette. These plasmids are not drawn to scale. The sequences of the primers (gidA5 and gidA3) used to amplify the gidA gene are described in Materials and Methods.
Complementation of the transposon mutant (M19) and gidA isogenic mutant (MgidA) of A. hydrophila SSU.
By using specific primers (gidAC5 with an EcoRI site [underlined], 5′ GAGAATTCGTGCTTATCAACAGAAGCGG 3′ [forward], and gidA-3 with a PstI site [underlined], 5′ GTCTGCAGGTAGGATCCTCAGGCGGTT 3′ [reverse]), a 2.1-kb DNA fragment containing the gidA gene and its putative promoter region was amplified from the chromosomal DNA of A. hydrophila. It was then ligated to vector pBR322 at the EcoRI/PstI restriction sites to generate a recombinant plasmid pBRgidA (Table 1), which was first transformed into E. coli HB101 that carried a helper plasmid, pRK2013 (with Kmr gene) (52, 53). Subsequently via conjugation, the recombinant pBRgidA plasmid was transferred into the gidA mutants of A. hydrophila (M19 and MgidA) (Table 1). The transconjugants were screened on LB agar plates containing TET, RIF, and SPT-STR (for M19) or TET, RIF, and KAN (for MgidA). The presence of pBRgidA recombinant plasmid in M19 and MgidA mutants was confirmed by plasmid isolation and restriction enzyme analysis. By using the same method, pBR322 vector alone was introduced into M19 and MgidA mutants, along with WT A. hydrophila to be used as negative controls (Table 1).
Reverse transcriptase PCR (RT-PCR).
Total RNA from WT A. hydrophila SSU, different gidA mutants, and their complemented strains was isolated, and an aliquot (1 μg) of the RNA was subjected to reverse transcription by using the Advantage RT for PCR kit. A negative control was set for each reverse transcription reaction by omitting RT from the reaction. A 5-μl aliquot from each of the reverse transcription reaction mixtures was used as a template for further PCRs by using the primers that were within the gidA gene: 5′ GCCTGATCCACATCGGCATG 3′ (forward) and 5′TGCCGATGCACTTGCTCTCC3′ (reverse). The PCR program used was described by us previously (52).
Integration of act::phoA reporter gene in the chromosome of A. hydrophila SSU mutants.
The strategy used to integrate an act::phoA reporter gene into the chromosome of A. hydrophila was described previously (Table 1) (53). The A. hydrophila mutants with integration of act::phoA in the chromosome exhibited diffused blue color around the colonies as a result of the secretion of PhoA, which reacted with the substrate BCIP (5-bromo-4-chloro-3-indolylphosphate) (53). The identity of the genuine single-crossover mutants (e.g., MgidAphoA and M19phoA) (Table 1) was confirmed by Southern blot analysis using the act gene probe (53, 57). A strain that was designated SSU66 represented WT A. hydrophila containing the fusion reporter gene cassette in the chromosome and was generated in our previous study (53). The complementation of these reporter strains was performed by delivering the gidA gene in trans using a pBR322 vector (52, 53).
Alkaline phosphatase assay to quantitate the act::phoA fusion gene expression.
The PhoA activity in the culture supernatants of the act::phoA reporter gene strains of A. hydrophila was measured as previously described (53). The density of the blue color was measured at 630 nm. The PhoA activity was calculated per milliliter of the culture supernatant per 108 CFU.
Measurement of biological activities.
The biological activities associated with Act in the culture filtrates of various mutants, WT, and complemented strains of A. hydrophila were determined by hemolytic and cytotoxic assays.
(i) Hemolytic assay.
The culture filtrates from various Aeromonas cultures (grown for 18 h in LB medium at 37°C with shaking [180 rpm]) were first treated with trypsin (final concentration, 0.05%) at 37°C for 1 h and then subjected to hemolytic assay as follows. A 100-μl aliquot of phosphate-buffered saline (PBS) was added to each of the wells of a 96-well microtiter plate. Next, 100-μl aliquots of culture filtrates were added, followed by a serial twofold dilution, with subsequent addition of 100 μl of 2.5% rabbit erythrocytes (Colorado Serum Company, Denver, Colo.). The plate was incubated at 37°C for 1 h and observed for the lysis of red blood cells (RBCs). The supernatant was taken from those wells that showed partial lysis of rabbit erythrocytes, and the hemoglobin release was recorded at 540 nm (21). The hemolytic titers were calculated as the value of the hemoglobin release multiplied by the dilution of the culture supernatant. The hemolytic units were reported per milliliter of the culture filtrate per 108 CFU. As RBCs do not produce any protease, the culture filtrates were treated with trypsin to convert all of the precursor form of Act to a mature form of the toxin. RBCs treated with trypsin alone served as a negative control, and it had no effect on the lysis of RBCs.
(ii) Cytotoxicity assay.
The RAW264.7 murine macrophage cell line (American Type Culture Collection, Manassas, Va.) was used for the cytotoxicity assay. The cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum supplemented with penicillin-STR (Invitrogen) and were incubated at 37°C with 5% CO2. As stated above, the culture filtrates from various Aeromonas cultures were serially diluted and added to the macrophages. After 1 h of incubation, the lactate dehydrogenase release was measured at 490 nm by using the CytoTox96 nonradioactive cytotoxicity assay kit from Promega. The cytotoxic activity was calculated as the value of the lactate dehydrogenase release multiplied by the dilution of the culture supernatant. The cytotoxicity units were measured per milliliter of the culture filtrate per 108 CFU. For cytotoxicity assay, culture filtrates were not treated with trypsin, since furin-like protease produced by host cells could activate Act (2).
Lethality associated with gidA mutants of A. hydrophila SSU in mice and histopathology.
WT A. hydrophila, its gidA transposon and isogenic mutants, and the complemented strains were grown in LB medium overnight. The cells were harvested by centrifugation and washed twice with PBS. The bacterial cells were resuspended in PBS and injected intraperitoneally into female Swiss-Webster mice weighing 20 to 25 g (Taconic Farms, Inc., Germantown, N.Y.) (five mice per group in triplicate) at a dose that was 3 logs higher (5.0 × 108 CFU) than the 50% lethal dose, and the mouse deaths were recorded for 2 weeks after challenge (57). A portion of the tissues, from, for example, the lungs, liver, and spleen was excised from mice, and the tissues were fixed in 10× buffered formalin and stained with hematoxylin-eosin in the Histology Core Facility, The University of Texas Medical Branch. The tissues were taken from mice infected with WT and complemented strains of A. hydrophila 24 h postchallenge, a time at which they died. For mice infected with the mutants, tissues were excised randomly from three mice at days 1, 3, and 7 postchallenge. The remaining six mice were kept under observation for 2 weeks. Histology data at 3 days after challenge were shown; however, the pathology was very similar, irrespective of the day the mice were sacrificed.
A three-tiered scoring system (0, 1+, and 2+) was developed to evaluate the histopathology observed in the lungs, spleen, and liver. Observed differences between the variously treated groups of animals were very clear based on histologic analysis. In addition, no variability within animals in the same group was observed. Measurements of capillary septa were obtained by using an Olympus 10× ocular equipped with a 1-cm linear reticule containing 1- and 0.1-mm subdivisions. Measurements were obtained perpendicularly to the alveolar septa. Cell densities were obtained by using the same ocular equipped with square chambers for cell counting. A total of 10 noncontiguous fields were evaluated with either 25 × 10−4-mm2 (spleen and lungs) or 318 × 10−4-mm2 square chambers (for liver).
Lung scoring was as follows: 0, normal histology; 1+, nondiscernible or barely discernible capillaries in alveolar septa and/or interstitium with one to three polymorphonuclear neutrophils (PMNs) per square chamber in alveolar spaces and/or interstitium and alveolar septal thickness ranging from 2.5 to 8.0 μm; 2+, prominent dilatation of capillaries in alveolar septa and/or interstitium with five to eight PMNs per square chamber in alveolar spaces and/or interstitium and alveolar septal thickness ranging from 7 to 13 μm.
Spleen scoring was as follows: 0, normal histology; 1+, small germinal centers in splenic follicles (10 to 40% of total diameter) with >50% immunoblasts per total number of mononuclear cells in splenic follicles and 0 to 2 apoptotic bodies per square chamber in splenic follicles; 2+, prominent germinal center formation (>40% of total diameter) with <20% immunoblasts per total number of mononuclear cells in splenic follicles and 3 to 10 apoptotic bodies per square chamber in splenic follicles.
Liver scoring was as follows: 0, normal histology; 1+, 0 to 3 inflammatory cells per square chamber in hepatic sinusoids and no sinusoidal congestion; 2+, 10 to 30 inflammatory cells per square chamber in hepatic sinusoids.
The liver and spleen tissue specimens were also homogenized and transferred to LB agar plates containing RIF (300 μg/ml) to monitor bacterial load in the tissues at days 1, 3, and 7 postinfection. With the WT and complemented strains of A. hydrophila, tissues were excised 1 day after infection (time of their death). Data for mutants were obtained 3 days postinfection; however, no bacteria were found in these tissues at days 1, 3, and 7 postinfection. The data were expressed as the number of bacteria recovered per 100 mg of liver and spleen tissues.
Statistical analysis.
Wherever appropriate, the data were analyzed using Student's t test, and P values of ≤0.05 were considered significant.
Nucleotide sequence accession number
The complete DNA sequence of the cloned chromosomal DNA fragment (4,574 bp) without the mini-Tn5 was submitted to GenBank under accession number of AY333759.
RESULTS
Transposon mutants of A. hydrophila SSU.
Approximately 5,000 transposon mutants of A. hydrophila were initially screened on 5% sheep blood agar plates for hemolytic activity, and a total of 24 colonies which exhibited reduced or no Act production were picked up. These 24 isolates were further tested for hemolytic activity in the culture filtrates, and a total of 20 mutants out of the 24 were confirmed to have no hemolytic activity. Southern blot analysis on the chromosomal DNA of these 20 transposon mutants with the act gene probe revealed that transposition occurred within the act structure gene. In the remaining four mutants, one contained free plasmid, while in another the transposon insertion occurred at more than one site on the chromosome, based on Southern blot analysis with the Sp/Smr gene cassette probe. Thus, these mutants were not the ideal candidates for further study.
Hemolytic activity was measured in culture filtrates, cell lysates, and membrane fractions of the remaining two transposon mutants (designated M1 and M19). Minimal or no hemolytic activity was detected in the cell lysates and membrane fractions (data not shown), and a 50 to 53% reduction in hemolytic activity and an 83 to 87% reduction in cytotoxic activity were observed in the culture filtrates of these mutants compared to those of the WT A. hydrophila (Table 2). These results indicated that transposition did not alter the export machinery in these two mutants. To rule out the possibility that transposition caused delayed toxin production, WT A. hydrophila and its transposon mutants M1 and M19 were grown for 96 h. At 4-h intervals, a sample was removed, and the CFU count (for measuring bacterial growth) and the hemolytic activity (by measuring hemoglobin release at 540 nm) in the culture filtrates were monitored. The hemolytic and cytotoxic activities were first detected at 6 to 8 h and reached a maximum at 18 h in both of the mutants, which was similar to those recorded for the parental A. hydrophila strain. It was noted that the growth of transposon mutants was 10% slower than that of WT A. hydrophila during the first 3 to 4 h; however, no difference in the CFU count was noted at the subsequent time points.
TABLE 2.
Hemolytic and cytotoxic activities of WT A. hydrophila SSU, gidA mutants, and their complemented strains
| Aeromonas strain tested | Mean activity ± SDa (U/ml/108 CFU)
|
|
|---|---|---|
| Hemolytic | Cytotoxic | |
| WT A. hydrophila | 83.8 ± 3.2 | 1226.7 ± 36.8 |
| M1 | 39.2 ± 4.0b | 214.4 ± 18.3b |
| M19 | 42.2 ± 3.1b | 154.9 ± 9.9b |
| MgidA | 45.6 ± 0.3b | 229.4 ± 33.8b |
| WT A. hydrophila/pBR322 | 82.3 ± 5.2 | 1015.4 ± 68.9 |
| M19/pBR322 | 42.1 ± 2.1b | 241.4 ± 12.2b |
| M19/pBRgidA | 81.8 ± 2.7 | 867.4 ± 35.4 |
| MgidA/pBR322 | 46.9 ± 2.3b | 326.5 ± 31.2b |
| MgidA/pBRgidA | 85.0 ± 3.5 | 1129.5 ± 49.1 |
Three independent experiments were performed.
Statistically significant value (P ≤ 0.05) compared to WT A. hydrophila SSU using Student's t test.
Cloning of the gidA gene from A. hydrophila SSU.
Based on Southern blot analysis, a 6.6-kb DNA fragment was detected in the PstI-digested chromosomal DNA of transposon mutant M19 when an Sp/Smr gene cassette was used as a probe (data not shown). This DNA fragment was cloned into pBluescript vector to generate a recombinant plasmid, pBlueM19 (Table 1). DNA sequence analysis revealed that the 6.6-kb DNA fragment contained a complete sequence of the gidA gene, which was interrupted by Tn5. Based on the entire gidA gene sequence, two primers, gidA-5 and gidA-3 (described in Materials and Methods), were used to amplify the native gidA gene from the chromosomal DNA of WT A. hydrophila. The native gidA gene was subsequently cloned into pBluescript vector at EcoRI/PstI sites.
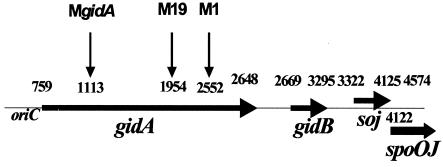
The DNA sequence of the gidA gene revealed that it contained 1,890 bp and had 68 and 74% homology at the DNA and amino acid levels, respectively, to the gidA gene recently isolated from the plant pathogen Pseudomonas syringae (29), while our studies were in progress. The homology of the gidA gene between A. hydrophila and E. coli was 68% at the DNA level and 71% at the amino acid level. By DNA sequence comparison between the native gidA gene with that of the interrupted version present on the 6.6-kb DNA fragment in pBlueM19 plasmid, the exact insertion site for transposition was determined to be at bp 1954 within the gidA gene ORF (bp 759 to 2648) (Fig. 2).
FIG. 2.
Schematic diagram showing the gidA region of A. hydrophila SSU and insertion sites in the gidA mutants. The gidA gene (long arrow) of A. hydrophila was located near the origin of the replication site (oriC), and three ORFs downstream of gidA included gidB, soj, and part of the spoOJ gene (short arrows). Based on the 4,574-bp DNA fragment that was sequenced, the transposition in mutants M1 and M19 occurred in the gidA gene at bp 2552 and 1954, respectively. The gidA isogenic mutant had an insertion, with the Kmr gene cassette at bp 1113 of the gidA gene. The figure is not drawn to scale.
By using a similar strategy, the transposon insertion in the M1 mutant was determined to be within the gidA gene. In the M1 mutant, transposition occurred at bp 2552 (Fig. 2). Other genes detected on the 6.6-kb PstI fragment of transposon mutants included gidB, soj, and spoOJ, based on sequence comparison with other bacterial genome sequences (Fig. 2). The function of these genes includes chromosome replication and partition and cell division (25, 40). Since transposition in both the mutants (M1 and M19) occurred within the gidA gene, subsequent detailed studies were performed only with the M19 mutant.
Characterization of gidA isogenic mutants (MgidA) of A. hydrophila SSU.
The strategy used to develop MgidA is depicted in Fig. 1. Conjugation of WT A. hydrophila with E. coli S17-1 harboring pJQgidA-Km plasmid (Table 1) with the interrupted gidA gene should result in transconjugants that were resistant to RIF, KAN, and 5% sucrose but sensitive to gentamicin. Such mutants should have undergone genuine, double-crossover homologous recombination, resulting in the replacement of the native gidA gene with an insertionally inactivated version of gidA, and with concomitant loss of the suicide vector harboring Gmr and sacB genes.
To confirm the correct genotype of the isogenic mutants, the chromosomal DNA from WT A. hydrophila and MgidA mutants was isolated and subjected to PCR and Southern blot analysis. For PCR analysis, the gidA gene primers gidA-5 and gidA-3 were used. These primers detected a 1.9-kb DNA fragment from the chromosome of WT A. hydrophila, which represented the correct size of the gidA gene. However, the size of the gidA gene was larger by 1.2 kb in the isogenic mutants, due to the insertion of the Kmr gene cassette. The PCR data were confirmed by performing Southern blot analysis.
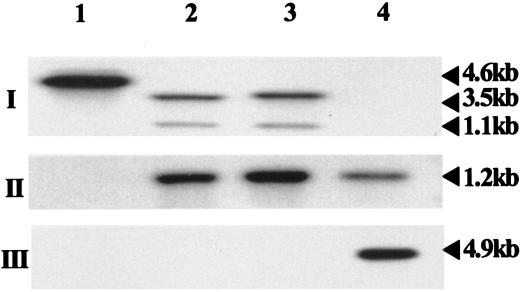
As depicted in Fig. 3, the size of the chromosomal DNA fragment from WT A. hydrophila digested with PstI restriction enzyme was 4.6 kb when a gidA-specific gene probe was used for Southern blot analysis (panel I, lane 1). However, in the MgidA mutants, the PstI restriction enzyme digestion divided this 4.6-kb DNA into two fragments of 1.1 and 3.5 kb (Fig. 3, panel I, lanes 2 and 3), due to the insertion of a Kmr gene cassette, which contained a PstI site at both ends (Fig. 1). A 1.2-kb DNA fragment was detected in the digested chromosomal DNA of the isogenic mutants when the Kmr gene cassette was used as a probe (Fig. 3, panel II, lanes 2 and 3). As expected, this probe did not react with the digested chromosomal DNA from the WT A. hydrophila (panel II, lane 1). Lane 4 in panel II contained a purified 1.2-kb Kmr gene cassette that was used as a probe and served as a positive control. No band was detected in the digested chromosomal DNA of both mutants (Fig. 3, panel III, lanes 2 and 3) and WT A. hydrophila (Fig. 3, panel III, lane 1) when suicide vector pJQ200SK was used as a probe. The probe reacted with the pJQ200SK plasmid digested with XbaI/XhoI restriction enzymes and served as a positive control (Fig. 3, panel III, lane 4). These data indicated that the MgidA mutants had completely lost the suicide vector sequences as a result of double-crossover homologous recombination. Compared with the WT A. hydrophila, the selected MgidA mutant exhibited 46 and 81% reduction in hemolytic and cytotoxic activities, respectively (Table 2). The growth rate of the MgidA was similar to that of transposon mutants, with no difference in the CFU count at 18 h, the time point that was used for measuring the biological activities. The growth of the mutant was slower in the first 3 to 4 h by 10% compared to the WT A. hydrophila; however, no toxin was synthesized during this time by both the mutant and the WT A. hydrophila. No difference in the CFU count was noted after the initial 3 to 4 h of growth.
FIG. 3.
Confirmation of the identity of the gidA isogenic mutants of A. hydrophila SSU based on Southern blot analysis. Chromosomal DNA from WT A. hydrophila (lane 1) and isogenic mutants (MgidA) (lanes 2 and 3) was digested with PstI restriction enzyme. As a positive control, a purified Kmr gene cassette was loaded in lane 4 of panel II, while an XbaI/XhoI-digested suicide vector, pJQ200SK, was used in lane 4 of panel III. Three different probes were used. In panel I, the probe was a gidA gene from A. hydrophila. In panel II, a 1.2-kb Kmr gene cassette was employed as the probe, while in panel III, plasmid pJQ200SK was used for hybridization. The two bands in panel I, lanes 2 and 3, reflect fragments containing the gidA gene. These fragments were generated by the PstI restriction enzyme digestion as a result of the insertion of a Kmr gene cassette within the gidA gene that introduced two additional PstI sites (Fig. 1).
As the gidA gene was shown to be involved in the cell division of E. coli (55), morphological examination of the MgidA mutant by Gram staining showed that the bacterial cells were filamentous (Fig. 4B) compared to the cocco-bacillus nature of parental A. hydrophila when grown in a medium containing 0.2% glucose (Fig. 4A). A similar pattern was noticed in the gidA transposon mutants (data not shown).
FIG. 4.
Morphology of A. hydrophila mutants as determined by Gram staining. (A) WT A. hydrophila. A typical cocco-bacillus nature of the cells was noted. (B) MgidA mutant; the cells were longer. (C) Complemented MgidA strain; a typical morphology of the cells was restored. The bacterial cells were grown in NY medium (N-Z amine [8 g], yeast extract [4 g], NaCl [5 g], distilled H2O to 1 liter) containing 0.2% glucose.
Restoration of biological activities of transposon mutant M19 and MgidA isogenic mutant by complementation.
The native gidA gene was supplied to the M19 and MgidA mutant via plasmid vector pBR322. As shown in Table 2, the hemolytic activity in transposon mutant M19 and isogenic mutant MgidA with pBR322 vector was 49 and 43% lower compared to that of WT A. hydrophila containing vector pBR322. However, the hemolytic activity was fully restored in the complemented strain M19, with a hemolytic titer of 81.8 ± 2.7 U/ml/108 CFU (results here and elsewhere are presented as arithmetic means ± standard deviations). Likewise, the complemented MgidA mutant restored the hemolytic titer to 85.0 ± 3.5 U/ml/108 CFU (Table 2). WT A. hydrophila with the pBR322 vector alone had a hemolytic titer of 82.3 ± 5.2 U/ml/108 CFU. A similar pattern was seen with the cytotoxic activity as well, with cytotoxicity titers of 1,015.4 ± 68.9, 241.4 ± 12.2, and 867.4 ± 35.4 U/ml/108 CFU in WT A. hydrophila, M19, and the complemented strain of the M19 transposon mutant. The cytotoxicity titers for WT A. hydrophila, MgidA, and its complemented strain were 1,015.4 ± 68.9, 326.5 ± 31.2, and 1,129.5 ± 49.1 U/ml/108 CFU (Table 2). Further, the cocco-bacillus nature of A. hydrophila was restored after complementation (Fig. 4C).
Detection of the gidA gene transcription in WT A. hydrophila SSU and its various mutants.
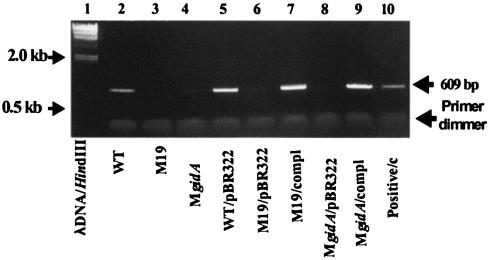
Total RNA from WT A. hydrophila, gidA mutants and their complemented strains was subjected to cDNA synthesis, and the synthesized cDNA was used for PCR using two specific primers (5′ GCCTGATCCACATCGGCATG 3′ [forward] and 5′ TGCCGATGCACTTGCTCTCC 3′ [reverse]) generated within the gidA gene of A. hydrophila. As shown in Fig. 5, a 609-bp DNA fragment, which represented a portion of the gidA gene transcript, was amplified from WT A. hydrophila (lanes 2 and 5) and the complemented mutants M19 and MgidA (lanes 7 and 9). The fragment was missing from the gidA mutants M19 (lanes 3 and 6) and MgidA (lanes 4 and 8). The same-size band was amplified from the chromosomal DNA of WT A. hydrophila and served as a positive control (lane 10). No band was amplified in any of these strains when RT was omitted from the reverse transcription reaction mixture. This control ensured that the PCR amplification was occurring only from the gidA gene mRNA, and not from the chromosomal DNA (data not shown).
FIG. 5.
RT-PCR depicting transcript of the gidA gene in various A. hydrophila mutants. Total RNA was isolated from different cultures of A. hydrophila. A hexamer primer was used for the cDNA synthesis. Subsequently, PCR was performed by using a pair of primers designed within the gidA gene, as stated in Materials and Methods. An aliquot (5 μl) of the PCR product from each of the cDNA reactions was loaded onto a 0.8% agarose gel for electrophoresis. Lane 1, λ DNA digested with HindIII marker; lanes 2 and 5, WT A. hydrophila; lanes 3 and 6, transposon mutant M19; lanes 4 and 8, MgidA mutant; lane 7, complemented M19 mutant; lane 9, complemented MgidA mutant. Chromosomal DNA from the WT of A. hydrophila was used as a template for the PCR and served as a positive control (lane 10). Abbreviations: compl, complementation; c, control.
Evaluation of act gene expression level in gidA mutants of A. hydrophila SSU.
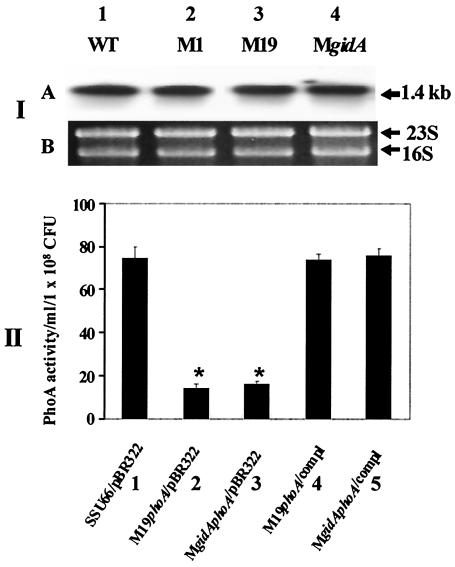
The isolated RNA from WT A. hydrophila, the M1, M19, and MgidA mutants, and their proper control strains was subjected to Northern blot analysis with the act gene-specific probe. We noticed no significant difference in the act gene transcription level between the gidA mutants and the WT of A. hydrophila (Fig. 6, panel I). To evaluate whether the effect of the gidA gene was at the translational level of Act, several reporter gene constructs with proper control strains were constructed and tested (Table 1). The amount of PhoA secreted from these strains reflected the level of Act produced (56). As shown in Fig. 6, panel II, PhoA activity was decreased in the gidA mutants compared to that of WT A. hydrophila. PhoA activity was restored after complementation. These data indicated that translation of the act gene was affected in the gidA mutants.
FIG. 6.
Translation of the act gene was affected in gidA-deficient mutants of A. hydrophila SSU based on Northern blot analysis and production of PhoA from the act::phoA reporter gene constructs. In panel I, total RNA from different cultures of A. hydrophila was isolated and subjected to Northern blot analysis. Lanes 1, RNA from WT A. hydrophila; lanes 2 and 3, RNA from gidA transposon mutants M1 and M19; lanes 4, RNA from the gidA isogenic mutant (MgidA). (A) The probe used was a 1.4-kb act gene. (B) The RNA loaded in each lane was quantitated by scanning 16S and 23S rRNA bands after ethidium bromide staining of the gel. In panel II, the PhoA activity was measured to represent the translation of the act mRNA in various A. hydrophila strains containing the act::phoA reporter gene construct. The arithmetic mean values and standard deviations (error bars) were plotted. The unit of measurement for the y axis is PhoA activity per milliliter per 108 CFU. Column 1, WT A. hydrophila; column 2, M19 transposon mutant; column 3, isogenic mutant MgidA; column 4, complemented (compl) M19 transposon mutant; column 5, complemented MgidA mutant. An asterisk denotes statistically significant difference (P ≤ 0.05) compared to WT A. hydrophila using Student's t test.
The gidA gene mutants of A. hydrophila SSU were avirulent in a mouse model.
WT A. hydrophila and the M19 and MgidA mutants with their complemented strains were tested for the ability to induce death in mice. In the WT A. hydrophila group, all of the animals died within 24 h. In the transposon mutant M19 and MgidA isogenic mutant group of mice, all of the mice survived up to a tested period of 2 weeks. Complementation restored the virulence, and all mice died within 24 h, as noted for WT A. hydrophila (data not shown). The lungs, liver, and spleen were excised from mice in each group for further analysis. Figure 7 shows histopathological findings in the lungs 3 days postinfection with the gidA mutants and 1 day after infection with the WT and complemented strains of A. hydrophila. However, the pathology was very similar on days 1 and 7 postinfection with the gidA mutants as seen on day 3.
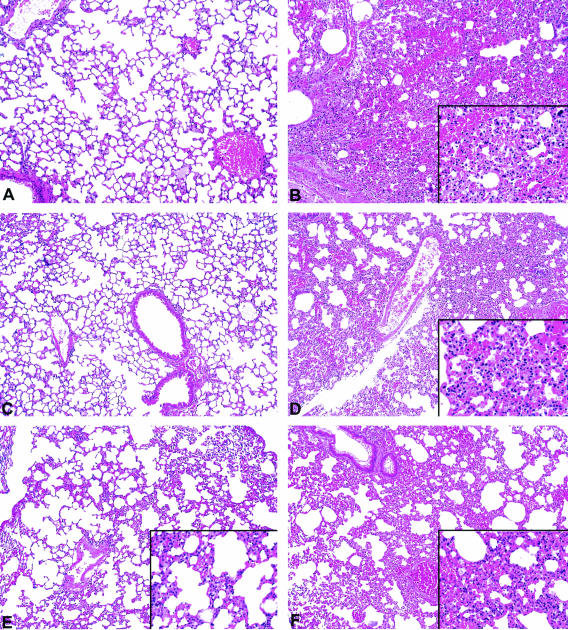
FIG. 7.
Lung sections of mice infected with various mutants of A. hydrophila. All sections were stained with hematoxylin and eosin. (A) Uninfected mouse showing normal lung histology. (B) Mouse infected with WT A. hydrophila (24 h postinfection). Note marked infiltration of alveolar spaces and interstitium by PMNs (inset). Severe vascular congestion was also observed. (C) Mouse infected with the M19 transposon mutant (3 days postinfection). The histology was virtually identical to that of the uninfected mouse. (D) Mouse infected with the complemented M19 mutant. Note marked infiltration of interstitium and alveolar spaces by PMNs (inset) as shown in panel B. (E) Mouse infected with the MgidA mutant (3 days postinfection). Minimal to mild inflammatory infiltrates composed mostly of PMNs in the interstitial areas (inset) were noted. Alveolar spaces were free of infiltrates. (F) Mouse infected with the MgidA-complemented strain. Note marked infiltration of alveolar spaces and interstitium as depicted in panel B. All panels and insets are shown at magnifications of ×100 and ×400, respectively.
Severe vascular congestion and acute inflammatory infiltrates (score, 2+) composed of mostly PMNs in the alveolar spaces and interstitium were seen in the animals infected with WT and complemented strains (Fig. 7B, D, and F). The animals infected with the MgidA mutant exhibited mild histopathology that consisted of mostly mild inflammatory infiltrates (score, 1+) in the interstitium of the lungs (Fig. 7E). Lung histologic sections from mice infected with the M19 mutant (Fig. 7C) were virtually identical to those of the uninfected animals (score, 0) (Fig. 7A).
The histopathologic findings in the liver and spleen were also evaluated. In animals infected with the mutant strains, the liver did not show any histologic abnormalities except for the presence of scattered mononuclear inflammatory cells in the hepatic sinusoids (score, 1+). Spleen sections from these animals revealed follicular hyperplasia with germinal center formation and >50% immunoblasts; occasional apoptotic lymphocytes (tingible body macrophages) were also observed (score, 1+). In contrast, animals infected with the WT and complemented strains revealed prominent inflammatory infiltrates composed of mononuclear cells and neutrophils in the portal triads and sinusoidal spaces in the liver (score, 2+). Sections of spleen revealed marked follicular hyperplasia with large germinal centers formation, <20% of immunoblasts in the germinal centers, and abundant apoptotic bodies within the splenic follicles (score, 2+). Scattered neutrophils were seen in the splenic sinusoids. Overall, the differences were clearly evident between the animals infected with either the WT strain or the complemented strains versus the animals infected with the mutant strains.
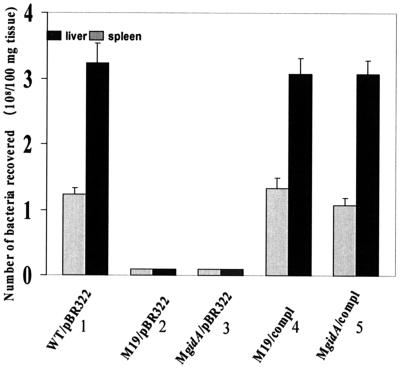
The number of bacteria recovered from the livers and spleens of the mice infected with WT A. hydrophila and the complemented strains at the time of death (24 h) was in the range of 107 to 108 CFU (Fig. 8). No bacteria were recovered from the tissues of animals infected with gidA mutants on day 1, 3, and 7 postinfection. These data indicated that systemic infection occurred in mice infected with WT A. hydrophila and the complemented strains. However, the gidA mutants failed in infected mice to cause systemic infection because of their rapid clearance from the host system.
FIG. 8.
Systemic infection by various strains of A. hydrophila in mice. As stated in Materials and Methods, the bacteria recovered from the liver and spleen tissue specimens (1 day for the WT and complemeneted strains and 3 days postinfection for the mutants) were plated and expressed as the number of recovered bacteria per 100 mg of tissue. Column 1, WT A. hydrophila; column 2, gidA transposon mutant M19; column 3, isogenic mutant MgidA; column 4, complemented (compl) M19 mutant; column 5, complemented MgidA mutant. The gray bar represents the number of bacteria recovered from the spleen, while the black bar represents bacteria recovered from the liver. The arithmetic mean ± standard deviation (error bar) was plotted.
DISCUSSION
In this study, a Tn5-derived mini-transposon was used to generate insertion mutants in A. hydrophila (19). Among the 24 genuine transposon mutants obtained, 2 mutants (M1 and M19) had reduced hemolytic (50 to 53%) and cytotoxic (83 to 87%) activities with an intact act gene in their genomes (Table 2). Transposition in both of these mutants occurred in the same ORF that encoded the GidA protein consisting of 630 amino acid residues (Fig. 2).
Since transposon mutants could lead to polar effects, a gidA isogenic mutant was prepared, which similarly exhibited significantly reduced hemolytic (46%) and cytotoxic (81%) activities, and these effects could be fully restored in both the isogenic and transposon mutants after complementation (Table 2). It was noticed that reduction in cytotoxic activity in the mutants was much more pronounced than the effect on hemolytic activity compared to WT A. hydrophila. This possibly could be due to the sensitivity of the assays used.
As shown in Fig. 4, the gidA isogenic mutant of A. hydrophila showed filamentous morphology (Fig. 4B) compared to WT A. hydrophila (Fig. 4A). We noticed that this observed phenotype caused the optical density of the mutant culture at 600 nm to appear less compared to that of WT A. hydrophila due to differences in the light-scattering properties of normal and filamentous cells.
We therefore determined the growth rates of the WT and the mutant cultures by counting CFU rather than measuring the absorbance of the culture. We noted no difference in the CFU count between the gidA mutants and WT A. hydrophila after 3 to 4 h of growth. During this early growth period, the mutants grew 10% more slowly and no toxin production occurred. Toxin in both the WT and the mutants was first detected at around 6 to 8 h, and became maximal at 18 h, with a parallel increase in the bacterial number for both the mutants and WT A. hydrophila. Taken together, these data indicated a direct role of GidA in regulating Act production and not a pleiotropic effect of reduced bacterial growth leading to low toxin production.
The function of GidA was initially thought to be involved in chromosome replication and cell division. This was based on several pieces of evidence. First, disruption of the gidA gene in E. coli affected cell division when cells were grown in a medium containing glucose (55). Second, the gidA gene is located in close proximity to the oriC (26, 30, 41, 42, 58), and initiation of chromosomal replication is still inhibited by RIF after a point in the replication process that requires protein synthesis, indicating that a transcriptional event might be involved (29, 31, 36). Third, in the synchronous cell studies, the activity of the gidA promoter was turned off after the chromosomal replication initiation events were accomplished (8, 43, 54). However, recent studies implicated GidA in a number of biological and pathogenic processes, as it is a widely distributed and conserved protein with multiple functions (56).
We previously showed that addition of glucose in the medium repressed Act production by A. hydrophila (53). Since in the gidA mutants of Aeromonas Act production was reduced, we performed an experiment in which Act production by the gidA mutants was evaluated in the presence and absence of glucose. The premise of our study was whether there is a link between regulation of Act production by glucose and GidA. Our initial study indicated that the addition of glucose (0.1 to 0.2%) in the medium still suppressed Act production, as measured by the hemolytic activity assay, in the GidA mutants of A. hydrophila. These data suggested that a mechanisms(s) in addition to GidA was responsible for reduced Act production in the presence of glucose. These studies will be pursued in the future.
GidA-like protein is widely distributed in nature (29, 56). It is conserved among eubacteria and has been classified into two groups, based on the size of the protein. The larger protein (GidAL) has 611 to 679 amino acid residues, and the smaller protein (GidAS), which is truncated at the carboxyl-terminal end compared to GidAL, contains 435 to 482 amino acid residues (56). The GidA of A. hydrophila belongs to the longer version (GidAL) of the GidA family of proteins, which is usually located near the replication origin (oriC) of the chromosome (Fig. 2), whereas the chromosomal positioning of the gene encoding GidAS is variable (56). Some eubacteria, such as Myxococcus xanthus, B. subtilis, Aquifex aeolicus, and Thermotoga maritima carry both versions of GidA (56). The GidAS shares 24% identity with the GidAL in M. xanthus (56).
White et al. (56) reported that GidA is a flavin adenine dinucleotide (FAD)-binding protein involved in development of M. xanthus. The gidA knockout mutant of M. xanthus was unstable, and after a few generations, was converted into a strain that was no longer able to form fruiting bodies (56). Although these investigators focused on the smaller version of GidA in M. xanthus, both larger and smaller forms of GidA from different bacteria revealed a conserved dinucleotide-binding motif at the N terminus. The distribution of GidAS in M. xanthus varied, based on different growth conditions (56). GidAS was found in both cytoplasm and periplasm when M. xanthus was vegetatively grown, while starved cells contained only the cytoplasmic GidAS. Further studies revealed that the cytoplasmic GidAS was associated with the inner membrane and was transported to the periplasm (56). FAD proteins involved in oxidation-reduction reactions required for energy production are associated with the membrane or located in the periplasm. The FAD moiety of GidA and conservation of the FAD-binding site among all GidA proteins suggest that GidA might act as a sensor for the redox state of cells similar to the flavoproteins, e.g., aerotaxis signal transducer in E. coli (56).
In another study, Karita et al. (28) noted that the gidA gene was closely located near the dapE gene (which encodes succinyl-diaminopimelate desuccinylase) and that the gidA gene was essential for the cell viability of Helicobacter pylori. These investigators were unable to obtain a gidA-deficient mutant while providing evidence that insertion mutagenesis indeed occurred within the target gene (28). By using transposon mutagenesis, Duwat et al. (20) generated transposon mutants of Lactococcus lactis, which were sensitive to mitomycin and UV light. In one of these mutants, the transposition occurred in the gidA gene, and the capacity of this mutant to mediate homologous recombination was affected (20). Lidholm and Gustafsson (32) reported that the presence of the gidA gene on the chloroplast genomes of conifers was important for the ability of these plants to synthesize chlorophyll in the dark (23, 32). GidA was also found as a mitochondrial protein in Saccharomyces cerevisiae. Mutations in an MTO1 gene (which codes for a GidA homolog) were pleiotropic on the expression of several mitochondrial genes in yeast (18). Most recently, while our studies were in progress, Kinscherf and Willis (29) reported GidA as a global regulator in the plant pathogen P. syringae. Mutation in the gidA gene affected diverse phenotypic traits, such as lipodepsipeptide antibiotic production, swarming, presence of fluorescent pigment, and virulence. In A. hydrophila, we have now shown that disruption in the gidA gene reduced the hemolytic and cytotoxic activities associated with Act. Further, the gidA mutants were avirulent in mice (Table 2; Fig. 8). However, unlike in H. pylori, mutation in the gidA gene was not lethal to P. syringae and A. hydrophila.
The mechanism of how GidA regulates gene expression is not clear. One study demonstrated that the gidA gene has a function similar to that of miaA (which encodes aminoacyl tRNA synthetase), a gene involved in tRNA modification that stabilizes codon-anticodon interactions (39). In another study, inactivation of either the gidA or the mnmE gene (allele of trmE gene that encodes GTP binding protein) in E. coli greatly increased the occurrence of a two-base frameshift during the translation of particular sites in mRNA (10). Since the mnmE gene product is known to be involved in the modification of some tRNAs (11, 29), a similar role was proposed for the gidA gene. Detailed studies showed that GidA up regulated the expression of salA (which codes for a putative DNA-binding protein) and that of syrB (which encodes syringomycin) genes at the translational level in P. syringae (29). In our study, we also noted that GidA affected the translation of Act, as the transcription of the act gene remained unchanged in the gidA mutants (Fig. 6). A role has been suggested for gidA in moderating translation fidelity, and this may explain the striking absence of gidA analogs in Archaea, since it had different ribosomes and tRNAs (7, 29).
Despite the conservation of the gidA gene among different bacteria, the phenotypes of different gidA mutants do not point to a common mechanism for GidA function in these organisms. Moreover, a detailed understanding of GidA is complicated by the fact that homologs of GidA exist in two sizes and are transcribed from genes that are not linked genetically (56). In addition to size, other differences have also been noticed between these two versions of GidA, such as distribution in the cells and the export machinery. GidAL was found only in the periplasm fraction of M. xanthus cells, while GidAS was found in cytoplasm as well as in the periplasm. An SRR motif was found in the N terminus of the small GidA protein of M. xanthus, a twin-arginine translocation-like (Tat) system (6) that was suggested for GidAS export (56). While the GidAL lacks the (S/T)RR motif, it may be exported through a different pathway. These differences might reflect various roles of different versions of GidA.
Microbial infection represents interactions between the host and pathogen, in which the microbe uses its strategies for survival and multiplication by combating defenses of the immune system of the host (33). One such microbial strategy is to use the regulatory system by which bacteria modulate their genes, as well as genes of the host, during infection. There are reports of such regulators in the literature, e.g., the phoP-phoQ regulatory system in Salmonella enterica serovar Typhimurium. This system controls expression of several bacterial genes, such as genes involved in macrophage survival (22, 37). Likewise, the accessory gene regulator (agr) in Staphylococcus aureus controls expression of alpha-hemolysin (hla) and staphylococcal enterotoxin C (s+) genes (47).
In this study, we have identified for the first time a regulatory gene (gidA) from an enteric pathogen, A. hydrophila, that regulates expression of a crucial virulence gene act. The biological activity of Act was decreased in both the gidA transposon and isogenic mutants and was restored after complementation. Most importantly, these gidA mutants were avirulent in the mice. Our future studies will be targeted at defining whether gidA regulates other virulence factors produced by A. hydrophila and the specific interaction that occurs between Act and GidA. GidA could be an important regulatory molecule, controlling virulence-associated factors of other enteric pathogens.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (AI41611). Jian Sha was partly supported by the J. W. McLaughlin Postdoctoral Fellowship.
The editorial assistance of Mardelle Susman is greatly appreciated.
Editor: J. T. Barbieri
REFERENCES
- 1.Abbott, S. L., S. Seli, M. Catino, Jr., M. A. Hartley, and J. M. Janda. 1998. Misidentification of unusual Aeromonas species as members of the genus Vibrio: a continuing problem. J. Clin. Microbiol. 36:1103-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrami, L., M. Fivaz, E. Decroly, N. G. Seidah, F. Jean, G. Thomas, S. H. Leppla, J. T. Buckley, and F. G. van der Goot. 1998. The pore-forming toxin proaerolysin is activated by furin. J. Biol. Chem. 273:32656-32661. [DOI] [PubMed] [Google Scholar]
- 3.Alavandi, S. V., M. S. Subashmi, and S. Ananthan. 1999. Occurrence of haemolytic and cytotoxic Aeromonas species in domestic water supplies in Chennai. Indian J. Med. Res. 110:50-55. [PubMed] [Google Scholar]
- 4.Annapurna, E., and S. C. Sanyal. 1977. Enterotoxicity of Aeromonas hydrophila. J. Med. Microbiol. 10:317-323. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 6.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 7.Bernander, R. 1998. Archaea and the cell cycle. Mol. Microbiol. 29:955-961. [DOI] [PubMed] [Google Scholar]
- 8.Bogan, J. A., and C. E. Helmstetter. 1997. DNA sequestration and transcription in the oriC region of Escherichia coli. Mol. Microbiol. 26:889-896. [DOI] [PubMed] [Google Scholar]
- 9.Brandi, G. M., F. Sisti, F. Giardini, G. F. Schiavano, and A. Albano. 1999. Survival ability of cytotoxic strains of motile Aeromonas spp. in different types of water. Lett. Appl. Microbiol. 29:211-215. [DOI] [PubMed] [Google Scholar]
- 10.Bregeon, D., V. Colot, M. Radman, and F. Taddei. 2001. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 15:2295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabedo, H., F. Macian, M. Villarroya, J. C. Escudero, M. Martinez-Vicente, E. Knecht, and M. E. Armengod. 1999. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 18:7063-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chopra, A. K., and C. W. Houston. 1989. Purification and partial characterization of a cytotonic enterotoxin produced by Aeromonas hydrophila. Can. J. Microbiol. 35:719-727. [DOI] [PubMed] [Google Scholar]
- 13.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1:1129-1137. [DOI] [PubMed] [Google Scholar]
- 14.Chopra, A. K., C. W. Houston, J. W. Peterson, and G.-F. Jin. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can. J. Microbiol. 39:513-523. [DOI] [PubMed] [Google Scholar]
- 15.Chopra, A. K., J. W. Peterson, X.-J. Xu, D. H. Coppenhaver, and C. W. Houston. 1996. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb. Pathog. 21:357-377. [DOI] [PubMed] [Google Scholar]
- 16.Chopra, A. K., R. Pham, and C. W. Houston. 1994. Cloning and expression of putative cytotonic enterotoxin-encoding genes from Aeromonas hydrophila. Gene 139:87-91. [DOI] [PubMed] [Google Scholar]
- 17.Chopra, A. K., X.-J. Xu, D. Ribardo, M. Gonzalez, K. Kuhl, J. W. Peterson, and C. W. Houston. 2000. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 68:2808-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colby, G., M. Wu, and A. Tzagoloff. 1998. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 273:27945-27952. [DOI] [PubMed] [Google Scholar]
- 19.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson, M. R., X.-J. Xu, C. W. Houston, J. W. Peterson, D. H. Coppenhaver, V. L. Popov, and A. K. Chopra. 1997. Hyperproduction, purification, and mechanism of action of the cytotoxic enterotoxin produced by Aeromonas hydrophila. Infect. Immun. 65:4299-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 23.Fujita, Y., H. Matsumoto, Y. Takahashi, and H. Matsubara. 1993. Identification of a nifDK-like gene (ORF467) involved in the biosynthesis of chlorophyll in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 34:305-314. [PubMed] [Google Scholar]
- 24.Galindo, C. L., J. Sha, D. A. Ribardo, A. A. Fadl, L. Pillai, and A. K. Chopra. 2003. Identification of Aeromonas hydrophila cytotoxic enterotoxin-induced genes in macrophages using microarrays. J. Biochem. 287:40198-40212. [DOI] [PubMed] [Google Scholar]
- 25.Gal-Mor, O., I. Borovok, Y. Av-Gay, G. Cohen, and Y. Aharonowitz. 1998. Gene organization in the trxA/B-oriC region of the Streptomyces coelicolor chromosome and comparison with other eubacteria. Gene 217:83-90. [DOI] [PubMed] [Google Scholar]
- 26.Gielow, A., C. Kucherer, R. Kolling, and W. Messer. 1988. Transcription in the region of the replication origin, oriC, of Escherichia coli: termination of asnC transcripts. Mol. Gen. Genet. 214:474-481. [DOI] [PubMed] [Google Scholar]
- 27.Harayama, S., M. Tsuda, and T. Lino. 1980. High frequency mobilization of the chromosome of Escherichia coli by a mutant of plasmid RP4 temperature sensitive for maintenance. Mol. Gen. Genet. 180:47-56. [DOI] [PubMed] [Google Scholar]
- 28.Karita, M., M. L. Etterbeek, M. H. Forsyth, M. K. Tummuru, and M. J. Blaser. 1997. Characterization of Helicobacter pylori dapE and construction of a conditionally lethal dapE mutant. Infect. Immun. 65:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinscherf, T. G., and D. K. Willis. 2002. Global regulation by gidA in Pseudomonas syringae. J. Bacteriol. 184:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolling, R., A. Gielow, W. Seufert, C. Kucherer, and W. Messer. 1988. asnC, a multifunctional regulator of genes located around the replication origin of Escherichia coli, oriC. Mol. Gen. Genet. 212:99-104. [DOI] [PubMed] [Google Scholar]
- 31.Lark, K. G. 1972. Evidence for the direct involvement of RNA in the initiation of DNA replication in Escherichia coli 15T. J. Mol. Biol. 64:47-60. [DOI] [PubMed] [Google Scholar]
- 32.Lidholm, J., and P. Gustafsson. 1991. Homologues of the green algal gidA gene and the liverwort frxC gene are present on the chloroplast genomes of conifers. Plant Mol. Biol. 17:787-798. [DOI] [PubMed] [Google Scholar]
- 33.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merino, S., M. M. Nogueras, A. Aguilar, X. Rubires, S. Alberti, V. J. Benedi, and J. M. Tomas. 1998. Activation of the complement classical pathway (C1q binding) by mesophilic Aeromonas hydrophila outer membrane protein. Infect. Immun. 66:3825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merino, S., X. Rubires, S. Knochel, and J. M. Tomas. 1995. Aeromonas spp. Int. J. Food Microbiol. 28:79-86. [DOI] [PubMed] [Google Scholar]
- 36.Messer, W. 1972. Initiation of deoxyribonucleic acid replication in Escherichia coli B/r: chronology of events and transcriptional control of initiation. J. Bacteriol. 112:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, S. I., A. M. Kukra, and J. J. Mekalanos. 1991. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayashiki, T., and H. Inokuchi. 1998. Novel temperature-sensitive mutants of Escherichia coli that are unable to grow in the absence of wild-type tRNA6Leu. J. Bacteriol. 180:2931-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nardmann, J., and W. Messer. 2000. Identification and characterization of the dnaA upstream region of Thermus thermophilus. Gene 261:299-303. [DOI] [PubMed] [Google Scholar]
- 41.Ogasawara, N., and H. Yoshikawa. 1992. Genes and their organization in the replication origin region of the bacterial chromosome. Mol. Microbiol. 6:629-634. [DOI] [PubMed] [Google Scholar]
- 42.Ogawa, T., and T. Okazaki. 1991. Concurrent transcription from the gid and mioC promoters activates replication of an Escherichia coli minichromosome. Mol. Gen. Genet. 230:193-200. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa, T., and T. Okazaki. 1994. Cell cycle-dependent transcription from the gid and mioC promoters of Escherichia coli. J. Bacteriol. 176:1609-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pemberton, J. M., S. P. Kidd, and R. Schmidt. 1997. Secreted enzymes of Aeromonas. FEMS Microbiol. Lett. 152:1-10. [DOI] [PubMed] [Google Scholar]
- 45.Prentki, P., and H. M. Krish. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 46.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 47.Regassa, L. B., R. P. Novick, and M. J. Betley. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (arg) in Staphylococcus aureus. Infect. Immun. 60:3381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rella, M., A. Mercenier, and D. Haas. 1985. Transposon insertion mutagenesis of Pseudomonas aeruginosa with a Tn5 derivative: application to physical mapping of the arc gene cluster. Gene 33:293-303. [DOI] [PubMed] [Google Scholar]
- 49.Ribardo D. A., K. R. Kuhl, I. Boldogh, J. W. Peterson, C. W. Houston, and A. K. Chopra. 2002. Early cell signaling by the cytotoxic enterotoxin of Aeromonas hydrophila in macrophages. Microb. Pathog. 32:149-163. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Sha, J., C. L. Galindo, V. Pancholi, V. L. Popov, Y. Zhao, C. W. Houston, and A. K. Chopra. 2003. Differential expression of the enolase gene under in vivo versus in vitro growth conditions of Aeromonas hydrophila. Microb. Pathog. 34:195-204. [DOI] [PubMed] [Google Scholar]
- 52.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sha, J., M. Lu, and A. K. Chopra. 2001. Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: characterization of an iron uptake regulator. Infect. Immun. 69:6370-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theisen, P. W., J. E. Grimwade, A. C. Leonard, J. A. Bogan, and C. E. Helmstetter. 1993. Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 10:575-584. [DOI] [PubMed] [Google Scholar]
- 55.von Meyenburg, K., B. B. Jorgensen, J. Nielsen, and F. G. Hansen. 1982. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol. Gen. Genet. 188:240-248. [DOI] [PubMed] [Google Scholar]
- 56.White, D. J., R. Merod, B. Thomasson, and P. L. Hartzell. 2001. GidA is an FAD-binding protein involved in development of Myxococcus xanthus. Mol. Microbiol. 42:503-517. [DOI] [PubMed] [Google Scholar]
- 57.Xu, X.-J., M. R. Ferguson, V. L. Popov, C. W. Houston, J. W. Peterson, and A. K. Chopra. 1998. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect. Immun. 66:3501-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, P., J. A. Bogan, K. Welch, S. R. Pickett, H. J. Wang, A. Zaritsky, and C. E. Helmstetter. Gene transcription and chromosome replication in Escherichia coli. J. Bacteriol. 179:163-169. [DOI] [PMC free article] [PubMed]