Abstract
Although the adaptive mechanisms allowing the gastric pathogen Helicobacter pylori to survive acid shocks have been well documented, the mechanisms allowing growth at mildly acidic conditions (pH ∼5.5) are still poorly understood. Here we demonstrate that H. pylori strain 26695 increases the transcription and activity of its urease, amidase, and formamidase enzymes four- to ninefold in response to growth at pH 5.5. Supplementation of growth medium with NiCl2 resulted in a similar induction of urease activity (at low NiCl2 concentration) and amidase activity (at ≥500 μM NiCl2) but did not affect formamidase activity. Mutation of the fur gene, which encodes an iron-responsive repressor of both amidases, resulted in a constitutively high level of amidase and formamidase activity at either pH but did not affect urease activity at pH 7.0 or pH 5.5. In contrast, mutation of the nikR gene, encoding the nickel-responsive activator of urease expression, resulted in a significant reduction of acid-responsive induction of amidase and formamidase activity. Finally, acid-responsive repression of fur transcription was absent in the H. pylori nikR mutant, whereas transcription of the nikR gene itself was increased at pH 5.5 in wild-type H. pylori. We hypothesize that H. pylori uses a repressor cascade to respond to low pH, with NikR initiating the response directly via the urease operon and indirectly via the members of the Fur regulon.
Helicobacter pylori is an important human pathogen, which colonizes the mucus layer overlaying the gastric epithelium. Colonization with H. pylori results in chronic gastric mucosal inflammation, which can progress to peptic ulcer disease and gastric carcinomas (16). The pH in the gastric mucus layer is thought to vary between 4 and 6.5, with occasional acid shocks of pH <2 occurring when the mucus layer is damaged. Since H. pylori demonstrates optimal growth at neutral pH, survival and growth of H. pylori in the hostile environment of the stomach require mechanisms to survive acid shocks and allow growth at mildly acidic pH (31, 35). Resistance of H. pylori to acid shocks requires production of ammonia by urease-mediated degradation of urea (summarized in reference 35). H. pylori produces large amounts of urease, up to 10% of its total protein content, and expression of urease is controlled at the transcriptional, posttranscriptional, and posttranslational level (1, 31, 38, 39). Cytoplasmic urease activity is under strict pH-mediated control, since the intracellular urea concentration is controlled by the H+-gated urea transporter UreI (10, 30, 42).
In contrast to the mechanisms involved in the resistance to acid shocks, relatively little is known about the mechanisms, allowing growth at mildly acidic conditions (pH ∼5.5). The chronicity of colonization, however, suggests that H. pylori is able to grow at mildly acidic pH. Growth at acidic pH induces changes in lipopolysaccharide composition (23, 26, 36), increases expression of chaperone-like proteins (19), and affects expression of several genes at the transcriptional and protein expression level (4-6, 15, 24, 25, 34, 43). However, the exact role in acid resistance of many of these factors is largely unknown.
There is a close link between metal metabolism and acid resistance in H. pylori. Transcription of the urease structural genes, as well as the acid-activated enzyme activity of H. pylori urease, is dependent on the availability of the nickel cofactor (31, 38), probably via the NikR nickel-responsive regulatory protein (12, 39). In addition, the iron-responsive regulator Fur is required for growth at pH 5.5 (8). The relationship between the NikR and Fur metal-regulatory systems and acid resistance may not solely depend on a disturbed metal homeostasis, since we demonstrated recently that Fur mediates iron-responsive regulation of the H. pylori paralogous amidases AmiE and AmiF (40). Both amidases degrade amide substrates to ammonia and the corresponding carboxylic acid and, together with the amino acid deaminases, they probably represent alternative systems for ammonia production in times of urea shortage (9, 22, 25, 32, 33, 40). Recent studies using transcriptome analyses have provided evidence for acid-responsive regulation of diverse ammonia-generating pathways (25, 43), but thus far the responsible regulatory systems mediating this acid-responsive regulation were not described.
Based on our previous observations about the roles of the NikR and Fur proteins in the regulation of ammonia-producing enzymes (12, 38-40), we hypothesized that NikR and Fur may be two important mediators of acid-responsive gene regulation in H. pylori. Therefore, the effect of growth in mild acidic conditions on transcription and activity of the three main H. pylori enzymes involved in ammonia production was analyzed, and it was observed that transcription and the activity of all three enzymes is significantly induced upon acidification. Subsequently, it was shown that both NikR and Fur are responsible for the observed acid-responsive regulation of urease, amidase, and formamidase activity, and we present here a model wherein a cascade of two or more regulatory proteins is enough to mediate adaptation to acidic conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
H. pylori wild-type strain 26695 (37) and its isogenic fur (8), nikR (39), and nikR fur (the present study) mutants were routinely cultured on Dent agar, consisting of Columbia agar supplemented with 7% saponin-lysed horse blood, 0.004% triphenyltetrazolium chloride (Sigma, St. Louis, Mo.), and Dent selective supplement (Oxoid, Basingstoke, United Kingdom) at 37°C under microaerophilic conditions (10% CO2, 5% O2, and 85% N2). Broth cultures were grown in brucella broth (Difco, Sparks, Md.) supplemented with either 0.2% β-cyclodextrins (Fluka, Buchs, Switzerland) (BBC) or 3% newborn calf serum (Gibco/Life Technologies, Breda, The Netherlands) (BBN). NiCl2 was purchased from Sigma, filter sterilized, and used at the indicated concentrations (38, 39). The pH of broth medium was adjusted to 5.5 by using HCl as described previously (8) and did not change by more than 0.5 pH during growth experiments or by NiCl2 supplementation. For antibiotic selection, growth media were supplemented with kanamycin or chloramphenicol to final concentrations of 20 or 10 μg/ml, respectively.
RNA hybridization.
RNA was isolated by using TRIzol (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions, separated on 2% formaldehyde-1.5% agarose gels in 20 mM sodium phosphate buffer, and subsequently transferred to nylon membranes (Roche, Basel, Switzerland) by using standard protocols (38). After transfer, RNA was covalently bound to the membrane by cross-linking with 0.120 J of UV light (254-nm wavelength)/cm2. Transferred RNA was visualized by methylene blue staining, and RNA samples were normalized based on 16S and 23S rRNA band intensities as described previously (38, 41).
Internal fragments of the ureA, amiE, amiF, hspA, fur, and nikR genes were PCR amplified from genomic DNA of H. pylori 26695 with the primers listed in Table 1. The resulting PCR fragments contained a T7 promoter sequence on the noncoding strand and were used for the production of antisense RNA probes labeled with digoxigenin (DIG) by in vitro transcription with T7 RNA polymerase (Roche). Northern hybridization and stringency washes were performed at 68°C, and bound probe was visualized with the DIG detection kit (Roche) and the chemiluminescent substrate CPD-Star (Amersham Pharmacia) (38). The sizes of hybridizing mRNA species were determined by comparison with the DIG-labeled RNA Molecular Weight Marker I (Roche).
TABLE 1.
Oligonucleotide primers used in this study
| Gene | Primer | Sequence (5′→3′)a |
|---|---|---|
| ureA | UreA-F1 | ATGAAACTCACCCCAAAAGA |
| UreA-R-T7b | ctaatacgactcactatagggagaGGAAGTGTGAGCCGATTTGA | |
| amiE | Amid-F1 | AGTAGCAGCCCAGATACTGT |
| Amid-R-T7b | ctaatacgactcactatagggagaGCACGATCTCACCCTTATCA | |
| amiF | Form-F1 | TCAGTTTCCTGTGCCAATTGTCA |
| Form-R-T7b | ctaatacgactcactatagggagaCTCAATGGGATTCCATGGGAATA | |
| hspA | HspA-F1 | CCAGTTCAGGCATCATCATC |
| HspA-R-T7b | ctaatacgactcactatagggagaCAAGAGCCTGAGCCCACAAT | |
| fur | Fur-F1 | TCCATTTTAGAGCGCTTGAG |
| Fur-R-T7b | ctaatacgactcactatagggagaTTAACGACTTCATTCTGGCGGTT | |
| nikR | NikR-F1 | CACCCAATAAAGACGATTCA |
| NikR-R-T7b | ctaatacgactcactatagggagaAGCTAGACGCCTTAGTCAAT |
Urease, amidase, and formamidase enzyme assays.
The enzymatic activity of urease, amidase, and formamidase were determined in fresh H. pylori lysates by measuring ammonia production from hydrolysis of urea, acrylamide, or formamide, respectively, by using the Berthelot reaction as described previously (40). The concentration of ammonia in the samples was inferred from a standard NH4Cl concentration curve. Enzyme activity was expressed as micromoles of substrate hydrolyzed per minute per milligram of protein.
Recombinant DNA techniques.
Restriction enzymes and DNA-modifying enzymes were purchased from Promega (Madison, Wis.), and standard protocols were used for the manipulation of DNA and the transformation of H. pylori (7). PCR was carried out with Taq polymerase (Promega). The H. pylori 26695 nikR fur double mutant was constructed by natural transformation of the H. pylori 26695 fur mutant with plasmid pAV364 as described previously (39). Correct replacement of the nikR gene by the interrupted version was confirmed by using PCR (39).
RESULTS
Transcription and activity of H. pylori ammonia-producing enzymes is induced at pH 5.5.
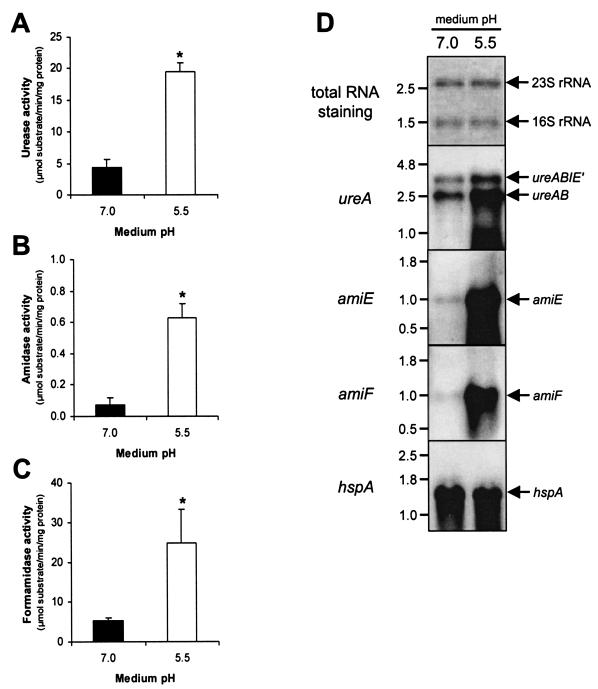
The effect of growth in acidic conditions on the activity of the three main ammonia-producing enzymes of the reference H. pylori strain 26695 was quantitated after overnight growth at pH 7.0 and pH 5.5. The latter pH was chosen to represent the average conditions thought to occur in the gastric mucus layer, and it also is the lower limit at which most H. pylori strains still grow in vitro (6, 8). When compared to enzyme activity at pH 7.0, growth at pH 5.5 resulted in a significant increase in urease, amidase, and formamidase activity (Fig. 1A to C). The increases were approximately fourfold for urease, ninefold for amidase, and fivefold for formamidase and were similar in β-cyclodextrin- and serum-supplemented media, thus excluding the effect of medium additives (not shown). There was no increase in urease, amidase, and formamidase activity when the medium was first adjusted to pH 5.5 and then subsequently readjusted to pH 7.0, excluding the possible artifact that the induction of enzyme activity was mediated by precipitation of medium components (not shown).
FIG. 1.
Acid-responsive induction of urease (A), amidase (B), and formamidase (C) in H. pylori is mediated at the transcriptional level (D). Graphs A, B and C represent enzyme activities, measured in wild-type H. pylori 26695, grown in pH 7.0 and pH 5.5 BBC medium. The activity of urease, amidase, and formamidase was determined by measuring ammonia production from urea, acrylamide, and formamide, respectively. The results shown are the averages of three independent growth experiments. Bars: ▪, pH 7.0□, pH 5.5. Errors bars denote the standard deviations. Asterisks indicate a significant difference in enzyme activity at pH 5.5 compared to pH 7.0 (P < 0.05, Mann-Whitney U test). (D) Northern hybridization with probes specific for the ureA, amiE, and amiF genes by using RNA isolated from cultures grown at pH 7.0 and pH 5.5. Staining of transferred RNA by methylene blue was included for comparison of the RNA amounts. A Northern hybridization with the pH-independently transcribed hspA gene is included as a control. The probes used are indicated on the far left, whereas the position of the specific mRNAs is indicated on the right; relevant RNA marker sizes (in kilobases) are indicated on the left.
The increase in enzyme activity was mediated by increased transcription of each of the genes, as tested by Northern hybridization (Fig. 1D). The increase in the two ureAB transcripts (1) was relatively minor and suggests that a major part of the induction in urease activity is mediated through activation of inactive urease apoenzyme (30, 31, 39). In contrast, the relative increase in the amiE and amiF transcripts after growth in acidic conditions was much more pronounced compared to ureAB transcription (Fig. 1D) and is likely to be mostly mediated via increased transcription of the amidase and formamidase genes. Transcription of the hspA (HP0011) chaperone-encoding gene was included as an example of a pH-independently transcribed gene and excludes the possibility that the observed acid-responsive changes in ureA, amiE, and amiF transcription are caused by global changes in transcriptional activity.
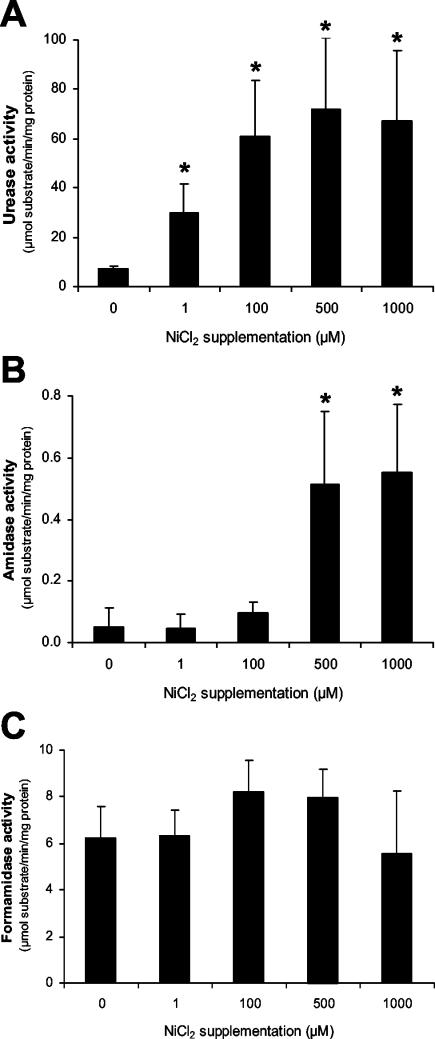
Nickel supplementation of growth media mimicks acidification.
To further define the link between nickel and acid, we investigated the effect of nickel supplementation on urease, amidase, and formamidase activity. When H. pylori strain 26695 was grown in media of pH 7.0 supplemented with NiCl2 to final concentrations ranging from 1 to 1,000 μM, urease activity was already induced at low concentrations of NiCl2 to levels similar to activity at pH 5.5 (Fig. 2A), a finding consistent with earlier reports (12, 38, 39). In contrast, amidase and formamidase activity was not affected by low concentrations of NiCl2 (Fig. 2B,C). However, at NiCl2 concentrations of 500 μM and higher, amidase activity increased significantly (Fig. 2B), while formamidase activity was not affected (Fig. 2C). The level of amidase activity at NiCl2 concentrations of ≥500 μM was similar to that observed with unsupplemented growth media adjusted to pH 5.5 (Fig. 1B).
FIG. 2.
Nickel supplementation of H. pylori growth medium mimics acidification. Wild-type H. pylori 26695 was grown in unsupplemented BBC medium or in medium supplemented with NiCl2 to final concentrations of 1 to 1,000 μM (as indicated on the x axis). The activities of urease (A), amidase (B), and formamidase (C) were determined after ∼20 h of growth. The results shown are the averages of three independent growth experiments; errors bars denote the standard deviations. Asterisks indicate a significant change in enzyme activity of the respective enzymes upon NiCl2 supplementation compared to unsupplemented medium (P < 0.05; Mann-Whitney U test).
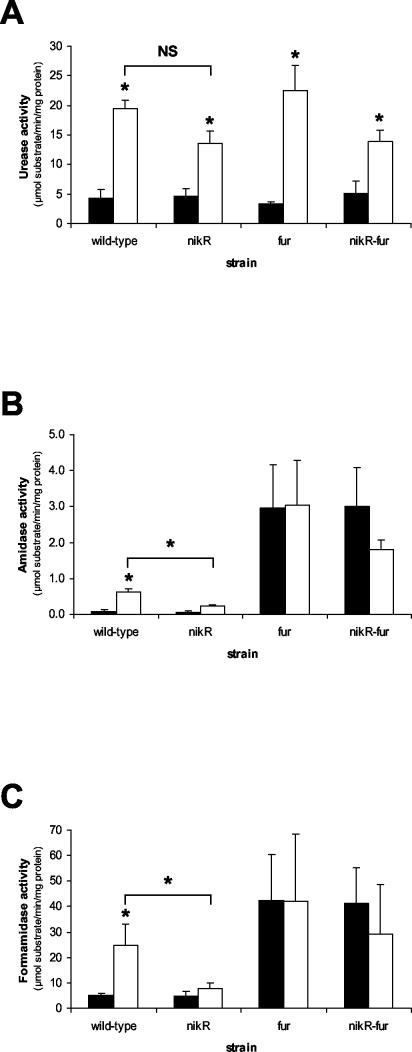
The nikR and fur gene products govern acid-responsive gene regulation in H. pylori.
The H. pylori genome contains two genes encoding regulatory proteins known to be involved in urease, amidase, and formamidase regulation: the HP1027 (fur) gene encoding the Fur regulator, which mediates iron-responsive regulation of both amidases (40), and the HP1338 (nikR) gene encoding the NikR regulator, which mediates nickel-responsive induction of urease (12, 39). Isogenic fur and nikR mutants, as well as a nikR fur double mutant, were used to investigate the role of the Fur and NikR proteins in acid-responsive induction of the urease, amidase, and formamidase enzymes (Fig. 3). Amidase and formamidase activity was constitutively high in the fur and nikR fur double mutants and was not further induced at acidic pH (Fig. 3B and C). In contrast, the fur mutation did not affect urease activity at either pH (Fig. 3A). Conversely, mutation of the nikR gene did not affect the activity of any of the three enzymes at pH 7.0 but had a significant effect on the acid-responsive induction of amidase and formamidase (Fig. 3B and C). The nikR mutant also displayed lowered acid-responsive induction of urease activity, albeit not statistically significant (Fig. 3A). This is similar to the effect of the nikR mutation on nickel-responsive induction of urease expression reported previously (39). The induction still present is probably due to increased activation of urease apoenzyme, as suggested previously (30, 31, 39). The effect of the nikR mutation on acid-responsive induction of amidase and formamidase activity was similar, since the nikR mutant was unable to induce enzyme activity at pH 5.5 to the levels found in the wild-type strain (Fig. 3B and C). The nikR fur double mutant displayed a phenotype similar to the nikR mutant with regard to urease activity and induction (Fig. 3A) but was virtually identical to the fur mutant with regard to amidase and formamidase activity and induction (Fig. 3B and C).
FIG. 3.
The H. pylori NikR and Fur regulators mediate acid-responsive induction of urease and both amidase enzymes. H. pylori 26695 wild-type and mutant derivatives were grown for 24 h in pH 7.0or pH 5.5 BBC medium, and the urease (A), amidase (B), and formamidase (C) enzyme activities were determined. The results shown are the averages of three independent growth experiments. Bars: ▪, pH 7.0; □, pH 5.5. Errors bars denote the standard deviations. Asterisks indicates a significant difference in enzyme activity at pH 5.5 compared to pH 7.0 (P < 0.05, Mann-Whitney U test), whereas the statistical evaluation of the differences between the wild-type and nikR mutant strains at pH 5.5 is given above the respective graphs. NS, not significant.
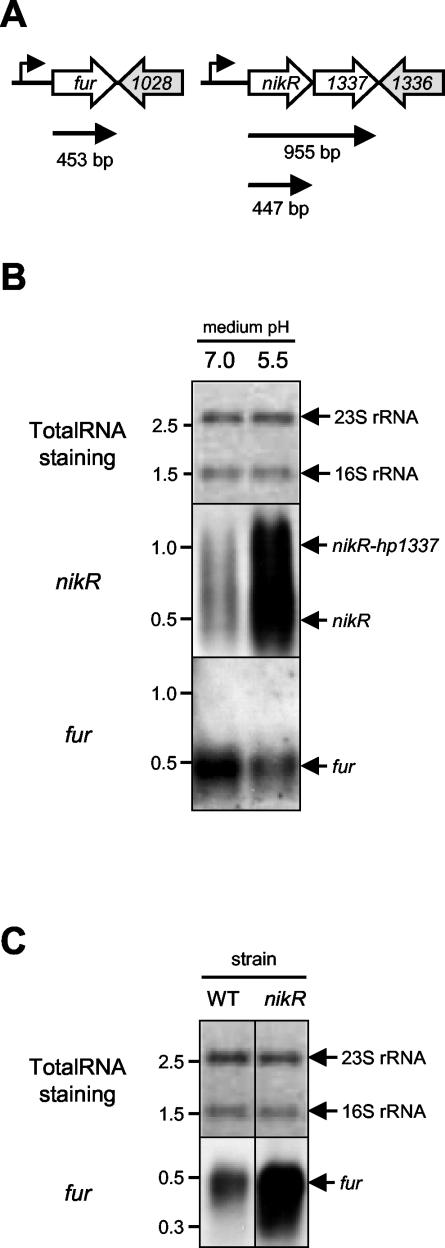
Acid-responsive transcriptional regulation is mediated by a repressor cascade.
The inverse effects of the nikR and fur mutations on expression of ammonia-producing enzymes indicated interaction or overlap between the two regulatory systems. Therefore, we investigated the effect of medium acidification on the transcription of fur and nikR genes themselves (Fig. 4). The fur gene displayed acid- responsive repression of transcription (Fig. 4B), as described previously (8). In contrast, transcription of the nikR gene was induced by medium acidification (Fig. 4B). The nikR transcript was relatively unstable, resulting in partial degradation of the nikR mRNA on Northern hybridization (Fig. 4B). Mutation of the nikR gene had a profound effect on the transcription of the fur gene, which was significantly increased in the nikR mutant, indicating that NikR represses fur transcription (Fig. 4C) (12). There was, however, no discernible effect of the fur mutation on nikR transcription (not shown), indicating that the NikR regulator is hierarchically higher ranked than the Fur regulator in the response to environmental acidification.
FIG. 4.
Acid-responsive regulation in H. pylori is mediated by a regulatory NikR-Fur cascade. (A) Putative transcriptional organization and predicted length of the nikR and fur mRNA species. (B) Transcription of the nikR gene is acid-induced, whereas fur transcription is acid repressed. Northern hybridization of H. pylori 26695 RNA isolated from cultures grown in BBC medium of pH 7.0 and pH 5.5, withprobes specific for the nikR and fur genes. (C) Transcription of the fur gene is derepressed in a H. pylori 26695 nikR mutant. Northern hybridization of RNA isolated from H. pylori 26695 (WT) and its isogenic nikR mutant (39) grown in BBC medium of pH 7.0 was carried out with a probe specific for the fur gene. Staining of transferred RNA by methylene blue is included for comparison of the RNA amounts. The probes used are indicated on the far left, whereas the position of the specific mRNAs is indicated on the right; relevant RNA marker sizes (in kilobases) are indicated on the left.
DISCUSSION
The ability to survive and adapt to the hostile environmental conditions within the mammalian host is a hallmark of successful pathogens. Adaptation to such conditions is mostly mediated at the transcriptional level via regulation of transcription, but alternative approaches such as differential mRNA processing, phase variation, posttranslational protein modification, and genetic reshuffling are all known to play important roles as adaptive response mechanisms in pathogenic microorganisms. The human gastric pathogen H. pylori is unique among pathogens, since it is known to utilize all of these methods (13). H. pylori is the sole colonizer of an ecological niche which is hostile with respect to environmental pH, inflammatory response, and availability of nutrients and is extraordinarily adapted to the conditions occurring in the human stomach. Here part of the regulatory mechanisms allowing H. pylori to respond to mildly acidic growth conditions by increasing the expression of three of its ammonia-producing enzymes are described.
Acidic pH is probably the most important environmental stress for H. pylori, which it encounters both during the initial and the chronic phase of colonization (35). Initial colonization requires protection against short-term acid exposure in the lumen of the stomach, where the pH is usually <2. Once established, H. pylori is present in the gastric mucus layer, where the average pH is ≥5.5 (27, 29). Despite its small genome size of 1.7 Mbp (3, 37), H. pylori is able to survive both these short-term low pH shocks and long-term exposure to mildly acidic conditions without being a truely acidophilic organism.
The urease enzyme mediates resistance of H. pylori to acid shocks via hydrolysis of urea to form ammonia, with subsequent buffering of either the periplasm or cytoplasm of H. pylori (28, 35). This short-term response to acid also requires acid-activated urea import via UreI and acid activation of urease apoenzyme via increased nickel availability (31, 42). In contrast, the long-term H. pylori response to acidic conditions is multifactorial, leading to modifications in membrane composition and H. pylori metabolism (4-6, 15, 19, 23-26, 34, 36, 43). Here it is demonstrated that growth at acidic conditions results in increased transcription and activity of the ammonia-producing enzymes urease, amidase, and formamidase and that part of this response is mediated via a regulatory cascade consisting of the NikR and Fur metal-regulatory proteins of H. pylori. In two recent studies on acid-responsive transcriptome analyses, it was also reported that several genes encoding for ammonia-producing enzymes were induced by medium acidification, and these included the ureAB, amiE, and amiF genes (25, 43). However, these findings were not further confirmed on the enzyme activity level, nor were the regulatory systems mediating the response identified. Many of the genes identified as acid regulated in these recent studies may well be regulated via either the NikR or Fur regulators but, again, this requires experimental validation.
The connection between pH and metal bioavailability in bacterial pathogens has been well established for iron, since a low pH stabilizes the readily oxidated ferrous form of iron. However, whereas iron plays an important role in H. pylori gene regulation, its role in acid resistance has been less clear. The iron-responsive regulator Fur is required for acid resistance, but this phenotype was reported to be independent of its role in iron regulation (8). Together with the previously reported role of Fur in regulation of the H. pylori amidases (40), this suggests that mutation of the fur gene disturbs the response to acidification either by constitutive overexpression of amidase or via another regulatory function not yet identified.
In H. pylori, the main link between metals and acid resistance is nickel. Nickel is cofactor of the urease enzyme, which is absolutely essential for acid survival of H. pylori. Urease expression and activity is regulated via nickel availability by the NikR regulatory protein (12, 31, 38, 39). The H. pylori NikR protein is a homolog of the Escherichia coli NikR protein, which regulates nickel acquisition via the Nik ABC transporter (11, 14). In H. pylori, the acquisition of nickel is required in the induction of urease activity (but not transcription) upon acid shock (31) and is likely to play a role in the transcriptional induction of urease upon growth in acidic conditions described here. Interestingly, mutation of the nikR gene resulted only in a small decrease in urease activity, which was not statistically significant (Fig. 3A). This is probably due to the NikR-independent activation of inactive urease apoenzyme previously described (31, 39), which masks the effect of the nikR mutation as shown previously for the effect of low concentrations of NiCl2 (39).
NikR also plays a role in regulation of nickel uptake in H. pylori (12) and, as such, the NikR protein can mediate regulation of several necessary processes for ammonia production in H. pylori: (i) induction of urease expression in acidic conditions (the present study) (30, 31) or nickel-rich (12, 38, 39) conditions and (ii) indirect regulation of amidase via regulation of the fur gene (the present study). Identification of additional targets for the NikR regulatory system via transcriptional profiling should allow for the identification of alternative or additional roles of NikR in acid resistance or ammonia production by H. pylori, as has been done recently for the identification of acid-responsive genes in H. pylori (2, 4, 15, 25, 43).
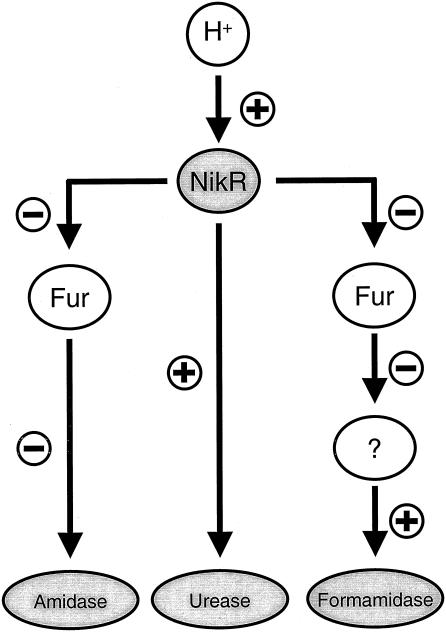
The NikR and Fur proteins generally function as transcriptional repressor proteins through binding of operators in their target promoters, blocking access of RNA polymerase, and subsequent transcription (11, 17). The repressor cascade of NikR and Fur described in the present study adds another level of complexity to this model. We hypothesize that the NikR protein ranks higher in the hierarchy, since NikR regulates fur transcription (Fig. 5) (12), whereas the reverse regulation does not occur (not shown). This view is also supported by the phenotypes of a H. pylori nikR fur double mutant (Fig. 3), which resembled the nikR mutant with regard to urease activity but resembled the fur mutant with regard to amidase and formamidase activity. A putative role for NikR as master regulator for acid-responsive regulation is also supported by the role of nickel in acid resistance of H. pylori (12, 31, 38, 39). The regulatory cascade proposed here utilizes two repressors each only allowing on/off regulation but, by varying the levels of Fur protein in cells, NikR not only regulates expression of its own regulon but also mediates changes in all members of the Fur regulon indirectly via regulation of fur transcription (Fig. 5). Since the effect of Fur on formamidase is indirect (40), it is likely that there is a third, as-yet-unknown regulator involved that ranks hierarchically below Fur (Fig. 5). These added levels of complexity should allow for the fine-tuning of the response to acid and possibly other relevant stresses. Other regulatory systems may also communicate or interact with the NikR or Fur regulons (12, 18, 20, 21, 25), since such intensive regulatory cross talk may allow H. pylori to mount protective responses to a multitude of different stresses despite a paucity of regulatory systems. In conclusion, H. pylori displays acid-responsive induction of its ammonia-producing enzymes at the transcriptional level via a regulatory cascade which contains the NikR and Fur repressor proteins (Fig. 5). The characterization of the Fur and NikR regulons may provide further insight in the mechanisms allowing H. pylori to persistently colonize the gastric environment.
FIG. 5.
Model of the H. pylori regulatory cascade mediating the acid-responsive transcriptional induction of ammonia-producing enzymes as described in the present study. Acidification of the environment results in increased transcription of the nikR gene; the corresponding increase in NikR protein directly affects the urease system and indirectly affects expression of the amidase system via repression of transcription of the fur gene. The decrease in Fur repressor subsequently leads to increased activity of the amidase enzyme. Since the Fur protein does not directly regulate formamidase expression (40), one or more additional regulatory systems are putatively involved, and this is represented by the circled question mark.
Acknowledgments
We thank Angela Heijens for technical assistance and Florian D. Ernst for helpful discussions.
Editor: V. J. DiRita
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 2.Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauerfeind, P., R. Garner, B. E. Dunn, and H. L. Mobley. 1997. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 40:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bijlsma, J. J. E., M. Lie-a-Ling, I. C. Nootenboom, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566-1569. [DOI] [PubMed] [Google Scholar]
- 7.Bijlsma, J. J. E., C. M. J. E. Vandenbroucke-Grauls, S. H. Phadnis, and J. G. Kusters. 1999. Identification of virulence genes of Helicobacter pylori by random insertion mutagenesis. Infect. Immun. 67:2433-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bijlsma, J. J. E., B. Waidner, A. H. M. van Vliet, N. J. Hughes, S. Hag, S. Bereswill, D. J. Kelly, C. M. J. E. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The ferric uptake regulator (Fur) homologue of Helicobacter pylori is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bury-Mone, S., S. Skouloubris, C. Dauga, J. M. Thiberge, D. Dailidiene, D. E. Berg, A. Labigne, and H. De Reuse. 2003. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect. Immun. 71:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bury-Mone, S., S. Skouloubris, A. Labigne, and H. De Reuse. 2001. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol. Microbiol. 42:1021-1034. [DOI] [PubMed] [Google Scholar]
- 11.Chivers, P. T., and R. T. Sauer. 2000. Regulation of high-affinity nickel uptake in bacteria: Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 275:19735-19741. [DOI] [PubMed] [Google Scholar]
- 12.Contreras, M., J. M. Thiberge, M. A. Mandrand-Berthelot, and A. Labigne. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49:947-963. [DOI] [PubMed] [Google Scholar]
- 13.de Vries, N., A. H. M. van Vliet, and J. G. Kusters. 2001. Gene Regulation, p. 321-334. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 14.De Pina, K., V. Desjardin, M. A. Mandrand-Berthelot, G. Giordano, and L. F. Wu. 1999. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J. Bacteriol. 181:670-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, Q., D. Hyde, C. Herra, C. Kean, P. Murphy, C. A. O'Morain, and M. Buckley. 2001. Identification of genes regulated by prolonged acid exposure in Helicobacter pylori. FEMS Microbiol. Lett. 196:245-249. [DOI] [PubMed] [Google Scholar]
- 16.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron-box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsyth, M. H., P. Cao, P. P. Garcia, J. D. Hall, and T. L. Cover. 2002. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J. Bacteriol. 184:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huesca, M., A. Goodwin, A. Bhagwansingh, P. Hoffman, and C. A. Lingwood. 1998. Characterization of an acidic-pH-inducible stress protein (Hsp70), a putative sulfatide binding adhesin, from Helicobacter pylori. Infect. Immun. 66:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGee, D. J., C. Coker, T. L. Testerman, J. M. Harro, S. V. Gibson, and H. L. T. Mobley. 2002. The Helicobacter pylori flbA flagellar biosynthesis and regulatory gene is required for motility and virulence and modulates urease of H. pylori and Proteus mirabilis. J. Med. Microbiol. 51:958-970. [DOI] [PubMed] [Google Scholar]
- 21.McGee, D. J., C. A. May, R. M. Garner, J. M. Himpsl, and H. L. Mobley. 1999. Isolation of Helicobacter pylori genes that modulate urease activity. J. Bacteriol. 181:2477-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGee, D. J., F. J. Radcliff, G. L. Mendz, R. L. Ferrero, and H. L. Mobley. 1999. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 181:7314-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGowan, C. C., A. Necheva, S. A. Thompson, T. L. Cover, and M. J. Blaser. 1998. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol. Microbiol. 30:19-31. [DOI] [PubMed] [Google Scholar]
- 24.McGowan, C. C., A. S. Necheva, M. H. Forsyth, T. L. Cover, and M. J. Blaser. 2003. Promoter analysis of Helicobacter pylori genes with enhanced expression at low pH. Mol. Microbiol. 48:1225-1239. [DOI] [PubMed] [Google Scholar]
- 25.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran, A. P., Y. A. Knirel, S. N. Senchenkova, G. Widmalm, S. O. Hynes, and P. E. Jansson. 2002. Phenotypic variation in molecular mimicry between Helicobacter pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. Acid-induced phase variation in Lewisx and Lewisy expression by H. pylori lipopolysaccharides. J. Biol. Chem. 277:5785-5795. [DOI] [PubMed] [Google Scholar]
- 27.Quigley, E. M., and L. A. Turnberg. 1987. pH of the microclimate lining human gastric and duodenal mucosa in vivo. Studies in control subjects and in duodenal ulcer patients. Gastroenterology 92:1876-1884. [DOI] [PubMed] [Google Scholar]
- 28.Sachs, G., D. Scott, D. Weeks, and K. Melchers. 2002. The compartment buffered by the urease of Helicobacter pylori: cytoplasm or periplasm? Trends Microbiol. 10:217-218. [DOI] [PubMed] [Google Scholar]
- 29.Schade, C., G. Flemstrom, and L. Holm. 1994. Hydrogen ion concentration in the mucus layer on top of acid-stimulated and -inhibited rat gastric mucosa. Gastroenterology 107:180-188. [DOI] [PubMed] [Google Scholar]
- 30.Scott, D. R., E. A. Marcus, D. L. Weeks, A. Lee, K. Melchers, and G. Sachs. 2000. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infect. Immun. 68:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott, D. R., E. A. Marcus, D. L. Weeks, and G. Sachs. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187-195. [DOI] [PubMed] [Google Scholar]
- 32.Skouloubris, S., A. Labigne, and H. De Reuse. 2001. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: natural evolution of two enzyme paralogues. Mol. Microbiol. 40:596-609. [DOI] [PubMed] [Google Scholar]
- 33.Skouloubris, S., A. Labigne, and H. De Reuse. 1997. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol. Microbiol. 25:989-998. [DOI] [PubMed] [Google Scholar]
- 34.Slonczewski, J. L., D. J. McGee, J. Phillips, C. Kirkpatrick, and H. L. Mobley. 2000. pH-dependent protein profiles of Helicobacter pylori analyzed by two-dimensional gels. Helicobacter 5:240-247. [DOI] [PubMed] [Google Scholar]
- 35.Stingl, K., K. Altendorf, and E. P. Bakker. 2002. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 10:70-74. [DOI] [PubMed] [Google Scholar]
- 36.Tannaes, T., N. Dekker, G. Bukholm, J. J. Bijlsma, and B. J. Appelmelk. 2001. Phase variation in the Helicobacter pylori phospholipase A gene and its role in acid adaptation. Infect. Immun. 69:7334-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karpk, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 38.van Vliet, A. H. M., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. J. E. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Vliet, A. H. M., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, E. J. Kuipers, C. W. Penn, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Vliet, A. H. M., J. Stoof, S. W. Poppelaars, S. Bereswill, G. Homuth, M. Kist, E. J. Kuipers, and J. G. Kusters. 2003. Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori Fur repressor. J. Biol. Chem. 278:9052-9057. [DOI] [PubMed] [Google Scholar]
- 41.van Vliet, A. H. M., J. Stoof, R. Vlasblom, S. A. Wainwright, N. J. Hughes, D. J. Kelly, S. Bereswill, J. J. E. Bijlsma, T. Hoogenboezem, C. M. J. E. Vandenbroucke-Grauls, M. Kist, E. J. Kuipers, and J. G. Kusters. 2002. The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter 7:237-244. [DOI] [PubMed] [Google Scholar]
- 42.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 43.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]