Summary
The oral microbial community represents the best characterized consortium associated with the human host. There are strong correlations between the qualitative composition of the oral microbiota and clinically healthy or diseased states. However, additional studies are needed to elucidate the mechanisms that define these microbial/host relationships.
History of oral microbiology
One milliliter of human saliva from a healthy adult contains approximately 100 million bacterial cells. Given a normal salivary flow rate of 750 ml per day, approximately 8×1010 bacteria are shed from the surfaces of the mouth every 24 hours, equivalent to 5–10g of wet weight of bacterial cells. These bacteria originate from the highly specialized and distinctive communities of organisms that reside on a variety of different environmental niches in the human mouth. Hence, the human oral microbiome can be viewed as a summation of discrete microbial communities drawn from, for example, the mucosal surfaces of the tongue, cheeks, palate and tonsils and the microbial biofilms that accumulate on the hard, non-shedding surfaces of the teeth.
The ease of accessing and sampling the mouth and the long acknowledged role of bacteria in dental caries and periodontal disease, two of the most common diseases of humans, have driven extensive investigations on the microbial communities on the tooth surface. As a result, oral bacteria are now the most well characterized microbiota of the human microbiome. These studies extend back to over 3 centuries to the very first description of bacterial cells by Antonie van Leeuwenhoek, who in 1676 used his newly manufactured microscopes to describe the “animacules” in biofilms from human teeth. In the intervening period, our understanding of the complexity, site specificity and environmentally driven nature of these microbial communities has expanded with each technological advance in microbial identification and classification. These advances include the introduction of standardized culture techniques on solid media, the development of anaerobic culture systems, the introduction of non-culture techniques based on molecular phylogeny through nucleic acid analyses using DNA:DNA hybridization, PCR, Sanger sequencing and the more recent developments in high throughput pyro-sequencing-based analyses and metagenomics (Wade, 2011). These culture- and non-culture-based investigations have culminated in the development of the Human Oral Microbiome Database (http://www.homd.org), which lists all bacterial species found in the human mouth (Dewhirst et al., 2010) and more recently CORE (http://microbiome.osu.edu) (Griffen et al., 2011), a phylogenetically curated 16S rDNA database of the core oral microbiome that is representative of the bacteria that regularly reside in the human oral cavity.
Current understanding of the oral microbiome
As in other environments, a significant proportion of the total oral microbiota remains unculturable and hence non-culture methods are required to describe the overall species richness of the oral microbiome. Sequence analysis of 16S ribosomal RNA has been the method of choice because of its universal presence in all organisms and because, through PCR primer design, it is possible to describe either all the species present in a given sample or to target specific genera. The application of this approach has led to the description of 11 phyla in the domain Bacteria in the oral microbiome in addition to methanogenic species of the Methanobrevibacter genus from the domain Archaea. The phyla of the domain Bacteria that are reliably present include Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Tenericutes, Fusobacteria, Proteobacteria, Spirochaetes, Synergistetes and two currently un-named phyla, SR1 and TM7. Several hundred distinct species are contained within these divisions representing the highly diverse microbial communities of the mouth. The periodontal microbiota is particularly heterogeneous and over 400 species have been described in this habitat alone using a 16S rRNA amplification, cloning and Sanger sequencing approach (Dewhirst et al., 2010).
While these findings have significantly enhanced our understanding of the oral microbiome, they have also highlighted the likelihood there may be an additional large number of low abundance species that have remained undetected using this standard methodological approach, largely because of the relatively time consuming and laborious nature of the techniques. This issue is now being addressed through the application of deep sequencing methods in particular pyro-sequencing technologies, which enable a more comprehensive coverage of the 16S rRNA sequences in large numbers of samples. Although relatively few large scale studies have been undertaken, there are indications that it may be necessary to revise our estimates of the species richness of the oral microbiome, perhaps by a factor of 10. For example, in a study of the microbiota of saliva and supragingival plaque from 71 and 98 healthy adults, respectively, amplicons from the V6 hypervariable region of the small-sub unit ribosomal RNA gene were generated by PCR and sequenced by 454 technology. 197,600 sequences were analyzed and suggested to represent 22 phyla comprising 3621 and 6888 species-level phylotypes in the saliva and plaque, respectively (Keijser et al., 2008). However, these early data need to be viewed with some caution as it is well recognized that the errors inherent in pyrosequencing, particularly of homopolymeric tracts, may lead to an over estimation of the total number of unique sequences in a given sample. The development of increasingly sophisticated software to minimize these problems should lead to increasingly accurate estimates of the species diversity of the oral microbiome and indeed more recent investigations have tended to be more conservative (Zaura et al., 2009). Nonetheless, the application of high throughout sequencing approaches, particularly to comparative analyses of health and disease, are likely to lead to increasing insights into the range of bacterial species associated with the development of pathology. A significant challenge in this area will be the analysis and interpretation of these high volumes of data. While the frequent condensation of high granularity phylotype information to the phylum or genus level, which is evident in much of the published literature on human microbiomes, enables ready comparison of different datasets, it is inadequate in terms of maximizing the value of these high throughput approaches.
A further issue to consider is the value of non-culture approaches to our understanding of the microbial pathogenesis of oral disease. Strategies based upon the sequence analysis of 16S rRNA limit the description of the microbiome to species level identifications. However, it is well established that there can be significant genomic variation within a species such that whilst a core set of genes may be common to all strains, there are others with a more restricted distribution. Along with other genetic modifications, including mutations, deletions and inversions, variability in the gene content contributes to the population diversity of that species – a heterogeneity that is not detected based on 16S rRNA analysis. As we will describe later, in one periodontal organism, this within species genetic variation leads to a clonal population structure where one particular clone, JP2 of Aggregatibacter actinomycetemcomitans, appears to have a much higher virulence potential in periodontitis than other clones of this species.
Formation of the dental plaque biofilm
The formation of the dental plaque biofilm has been well studied both in vitro and in vivo (Kolenbrander et al. 2010). It is clear that there is an orderly succession of early, intermediate, and late colonizing species. Colonization of the tooth surface begins with highly specific interactions between oral bacteria, mostly Streptococcus species, and the tooth pellicle. The pellicle is a thin layer of both saliva and gingival crevicular fluid that coats the dentin surface of the tooth. Oral bacteria have evolved highly specific adhesions to pellicle proteins and carbohydrates, not unlike specific adhesins found in other commensal and pathogenic bacteria that display highly specific tissue tropisms. After early colonizers have established themselves on the tooth surface through host derived pellicle interactions, these bacteria themselves then serve as additional binding sites for intermediate and late colonizers. This process, which has been eloquently studied and described by Kolenbrander’s group, reveals that each step in dental plaque biofilm formation is highly specific and represents co-evolution between different oral bacterial species as well as the host (Socransky and Haffajee, 2005). Recently, with the use of 16S or 18SrRNA probes the microbial spatial distribution of in vivo dental plaque biofilm was examined by fluorescent in situ hybridization (FISH) (Zijnge et al., 2010). This study confirmed many early observations yet also provided microbial definition to the species composition in different layers of the dental plaque biofilm. In addition, the demonstration that P. gingivalis directly binds Streptococcus gordonii, an early colonizer, provides evidence that potential perio-pathogens can colonize the biofilm early and this may represent a reservoir for the organism (Nobbs, et al., 2009).
Clinically Healthy Periodontal Tissue/Bacterial Interactions
The presence of a large and diverse microbial load on the tooth surface places a polymicrobial consortium in juxtaposition to host periodontal tissue. Nevertheless, normally, the periodontium remains healthy largely due to the numerous host protection mechanisms that operate in the oral cavity (Darveau, 2010).
Similar to the intestinal approach of handling a large microbial load, the oral cavity employs the tactic of first limiting exposure to host tissues. Perhaps the single most important component that limits the numbers of bacteria that can accumulate on the tooth surface is saliva. Saliva contains numerous components that contribute to either limiting bacterial accumulation or directing killing bacteria in the oral cavity. For example, similar to the intestine, saliva contains mucin proteins; however, in contrast to their function in the intestine, the mucins in the oral cavity do not form a thick layer that bacteria may need to penetrate through in order to approach host tissue. Rather, oral mucins induce bacterial aggregation that prevents the bacteria from attaching to the tooth or oral epithelial cell surface and promotes their removal upon swallowing (Kolenbrander et al. 2010). The interaction of oral bacterial species with salivary mucins is specific, and the same host receptors that facilitate bacterial removal by aggregation and swallowing also initiate ecological succession on the tooth surface which is coated by saliva.
Perhaps the most unique and significant host protection mechanism in the periodontium is the constant transit of neutrophils from the underlying highly vascular periodontal tissue, through the connective and epithelial cell layers and into the gingival crevice. It has been calculated that approximately 30,000 polymorphonuclear neutrophils (PMNs) transit through periodontal tissue every minute (Schiott and Loe, 1970), which facilitates nearly constant contact between host neutrophils and the dental plaque biofilm. The high incidence of periodontitis in those individuals with low circulating neutrophils or congenital defects in neutrophil extravazation provides strong evidence that this neutrophil transit is a key component of periodontal innate defense (Darveau, 2010). Accordingly, the structure of the periodontal tissue surrounding the tooth surface is fashioned such that neutrophils can transit through the tooth surface and inhibit biofilm growth. In particular, a specialized epithelium, termed the junctional epithelium surrounds the tooth surface and forms the “junction” between the inanimate tooth and host tissue. The junctional epithelium is highly porous with large intracellular spaces and it contains no tight junctions and a lower number of desmosomes than the adjacent oral or succular epithelium (Bosshardt and Lang, 2005). It also expresses a higher concentration of the cell adhesion molecule CEACAM1, which through homophilic binding to itself may serve as the major cell adhesion molecule in this tissue (Heymann et al., 2010). Furthermore, clinically healthy junctional epithelial tissue expresses high levels of IL-8, a potent neutrophil chemoattractant, that draws neutrophils to the adjacent dental plaque biofilm. Other host defense mediators associated with neutrophil exit from the vasculature and transit through the connective tissue, such as ICAM-1 and E-selectin, are also expressed in the appropriate tissues in clinically healthy periodontal tissue (Tonetti et al. 1998).
We are just beginning to learn the relative contributions of oral commensal bacteria or host directed expression programs to the highly specialized tissue organization and orchestrated expression pattern of select inflammatory mediators in clinically healthy periodontal tissue. Early histological studies revealed that germ free (GF) and specific pathogen free (SPF) mice were very similar with respect to the unique junctional epithelium architecture and that both GF and SPF contained neutrophils in the junctional epithelium (Heymann et al., 2010). This has been confirmed subsequently in a study that examined the expression of CEACAM1 in GF and SPF rats and mice (Heymann et al., 2010). It was found that expression of this cell adhesion molecule, which is expressed developmentally during tooth eruption, localizes to the developing junctional epithelium where it is postulated to serve an important role in structural integrity. Therefore, commensal bacteria are not required for its expression. Likewise, another study examined the expression of secretory leukocyte protease inhibitor (Slpi) in GF and SPF mice (Hayashi et al., 2010). This defense protein, which protects the host from host-mediated protease-induced tissue damage, was found to be highly expressed in the junctional epithelium, an area where a strong inflammatory process is occurring, yet the oral commensal community was not required for its expression. However, commensal colonization has been shown to influence the periodontal innate host defense status. A pilot study revealed that SPF mice contain higher levels of IL-1β (Dixon et al., 2004), an inflammatory cytokine normally associated with inflammation. Perhaps more importantly, IL-11β was found in the GF mice underscoring the contribution of “inflammatory” cytokines in normal tissue homeostasis programs in the absence of a microbial stimulus.
Therefore, much more needs to be learned concerning if and how oral commensal bacteria contribute to the highly orchestrated inflammatory response seen in this tissue. Not only do we need to better understand how alterations in cytokine levels by commensal bacteria in the absence of disease may positively or negatively affect periodontal tissue homeostasis programs, but also there have been no studies that have directly examined the molecular mediators associated with neutrophil transit in germ free mice. This represents a significant gap in our knowledge. If these mediators are solely developmentally expressed, as in the case of the epithelial cell adhesion molecule CEACAM1 and secretory leukocyte protease inhibitor (Slpi), then what are the host signals that regulate their expression in such a highly organized fashion? Likewise, if additional germ free studies reveal that commensal bacteria contribute to the neutrophil transit process, it may represent novel therapeutic avenues to “augment” innate defense with the use of probiotic approaches. The contribution of oral commensal bacteria to expression of defensins, CD14, lipopolysaccharide binding protein, all of which have been shown to be present in clinically healthy tissue, also needs to be determined.
Periodontitis/Bacterial Interactions
Periodontitis, similar to inflammatory bowel disease, is a clinical syndrome that can have multiple etiologies. The generally accepted view is that periodontitis results from the interaction between a microbial challenge derived from the subgingival biofilms on the tooth surface and a deregulated host response in the periodontal tissues (Page and Kornman, 1997). Disruption of this interaction through debridement of the tooth surfaces, supplemented occasionally by antibiotic delivery, is the standard and in most instances broadly effective treatment strategy.
The complexity of the sub-gingival microbiota has hindered the identification of the precise microbial etiology of periodontitis although very strong correlations between the amount and composition of the dental plaque biofilm and disease have been described (Socransky et al., 1998). Furthermore, extensive microbial compositional analysis, based originally on culture techniques and subsequently extended by large scale DNA:DNA hybridization methodologies, has identified potential periopathogens, designated the red complex. Examination of potential virulence characteristics shared by red-complex bacteria, Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola, has not yielded clear associations with disease. However, one shared attribute is their ability to either inhibit or evade innate host responses. This has led to the speculation that the strong association of these bacteria with diseased sites may be related to their ability to disrupt periodontal innate defense functions facilitating untoward host interactions with the entire dental plaque community (Darveau, 2010). Dental plaque communities obtained from either healthy or diseased sites are both potent activators of TLR2 and TLR4 (Yoshioka et al. 2008) and are therefore capable of disrupting established host homeostasis programs (Bosshardt and Lang, 2005). Nevertheless, “red” complex bacteria can be found in clinically healthy sites, albeit at lower numbers, indicating that their presence alone is not responsible for disease.
Adult type chronic periodontitis appears to be truly a microbial community - associated disease. Consistent with this, it has been reported that the stability of the dental-plaque microbial composition maybe a good predictor of periodontal health and that changes in this community are associated with changes in the clinical status of the adjacent tissue (Kumar et al., 2006). However, the factors that lead to changes in the plaque microbiota that are associated with periodontitis are not known. A greater understanding of potential “triggers” that initiate these changes by either altering innate defense function or selecting for a different microbial community may be obtained from examination of environmental and endogenous factors that are associated with an increased incidence of periodontitis. These include oral hygiene, smoking, obesity, stress and potential genetic associations (Stabholz et al., 2010). Almost certainly there are multiple potential mechanisms by which normal host homeostasis may be disrupted, eliciting alterations in either the host protective status or the microbial composition or both. Understanding the effects of these risk factors on the microbial/host relationship should uncover additional mechanisms by which host homeostasis can be detrimentally disrupted.
In contrast to adult type chronic periodontitis, in one particular instance of aggressive periodontitis affecting adolescents of African descent, there is evidence to suggest that a single specific microbial etiology may be responsible for the development of disease. Aggregatibacter actinomycetemcomitans is a gram-negative rod which produces a leucotoxin that specifically lyses human neutrophils. The organism displays significant genetic diversity but one particular clone, referred to as JP2, has a number of genetic variations that distinguish it from other clonal types, including a 530 base-pair deletion in the promoter region of the leucotoxin gene operon. As a result, the JP2 clone produces significantly enhanced levels of leucotoxin compared to the other lineages of this bacterium which could theoretically lead to an enhanced potential to disrupt the immune defences of the periodontium. Population genetic analysis by multi-locus sequencing of A. actinomycetemcomitans strains from geographically dispersed individuals suggest that the JP2 clone originally emerged as a distinct genotype in the Mediterranean part of Africa over 2000 years ago and subsequently spread to West Africa from where it was transferred to North and South America by the trans-Atlantic slave trade in the 16th –18th centuries. Remarkably, despite its now global dissemination, the JP2 clone still remains exclusively associated with individuals of West African descent indicating a strong host tropism effect (Haubek et al., 2008). Whilst the prevalence of aggressive periodontitis in adolescents is normally less than 1%, it is far higher in individuals of North and West African descent. In a recent longitudinal study of the disease in Moroccan adolescents, 61 of 428 (14.3%) individuals who were periodontally healthy at baseline had developed disease after 2 years. Moreover, in this population, individuals who carried the JP2 clone at baseline were far more at risk of developing disease than those who carried non-JP2 clones of this bacterium (Relative Risk 18.0 vs 3.0) (Haubek et al., 2008). Hence the JP2 clone of A. actinomycetemcomitans has the characteristics of a traditional bacterial pathogen albeit in a host restricted background.
Summary and Future Directions
While a correlation between a specific clonal type of A. actinomycetemcomitans and aggressive periodontitis in a select human population has been observed, the relationship between the highly characterized periopathogenic microbial community and chronic adult type periodontitis, the most common form of disease, remains to be determined. Additional characterization of the composition of the microbiota in periodontal health versus disease may lead to the description of additional species which one could presumptively associate with disease causation. For example in one investigation (Kumar et al., 2005), 16S rRNA gene cloning and sequencing identified several novel disease associated organisms including Peptostreptococcus stomatis, Filifactor alocis and species drawn from the Desulfolobulus, Dialister and Synergistetes genera. The potential contribution of clonal types and the influence of environmental triggers in altering the oral microbial composition need to be further explored to determine how a healthy microbiotia is altered into one associated with a destructive host response. Incorporation of polymicrobial approaches into in vitro and in vivo systems to examine the potential of bacterial–bacterial communication events on the host response are also required. Finally, an analysis of microbial metagenomic libraries derived from different clinical states may provide an alternative method to determine the functional characteristics of the entire microbial consortium which are required to either maintain the homeostasis of health or develop disease in the periodontal tissues.
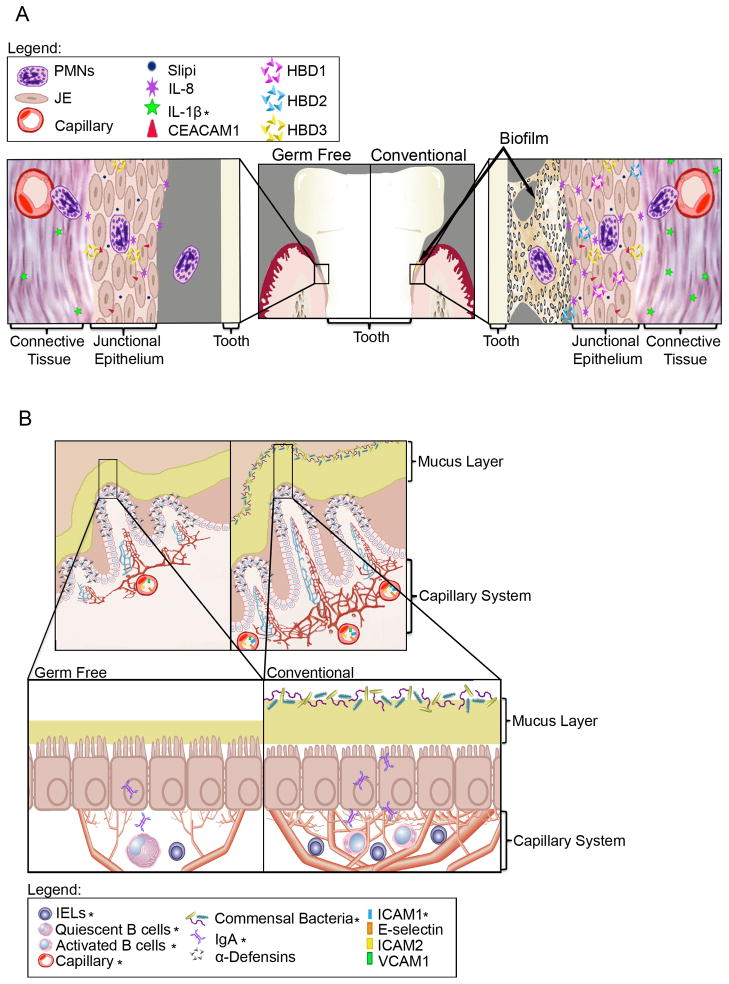
Figure 1.
A. Current knowledge of microbial influence on the junctional epithelium (JE) based on cumulative data from human and mouse studies. The architecture of JE tissue and presence of PMNs are similar between germ-free and conventional mice. Several molecules appear to change dramatically with the addition of bacteria but many are unchanged (Darveau, 2010). B. Overview of current knowledge of microbial influence on the intestinal epithelium. The architecture of the intestinal tissue is changed markedly with the addition of bacteria; the crypts are deeper, the capillary network is more extensive, the mucus layer is reduced, cilia are shorter and many differences are seen with immune cells and molecules as indicated (Hooper, 2004). The figure indicates the relative location and abundance of innate immune cells/molecules. (* indicates changes due to microbial interactions confirmed in germ free studies).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Selected Reading
- Bosshardt DD, Lang NP. J Dent Res. 2005;84:9–20. doi: 10.1177/154405910508400102. [DOI] [PubMed] [Google Scholar]
- Darveau RP. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DR, Reife RA, Cebra JJ, Darveau RP. J Periodontol. 2004;75:1486–1492. doi: 10.1902/jop.2004.75.11.1486. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Firestone ND, Gross EL, Difranco JM, Hardman JH, Vriesendorp B, Faust RA, Janies DA, Leys EJ. PLoS One. 2011;6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubek D, Ennibi OK, Poulsen K, Vaeth M, Poulsen S, Kilian M. Lancet. 2008;371:237–242. doi: 10.1016/S0140-6736(08)60135-X. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Matsunaga T, Yamamoto G, Nishii K, Usui M, Yamamoto M, Tachikawa T. J Periodontal Res. 2010;45:618–625. doi: 10.1111/j.1600-0765.2010.01276.x. [DOI] [PubMed] [Google Scholar]
- Heymann R, Wroblewski J, Terling C, Midtvedt T, Obrink B. J Periodontol. 2001;72:454–460. doi: 10.1902/jop.2001.72.4.454. [DOI] [PubMed] [Google Scholar]
- Hooper LV. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Nat Rev Microbiol. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Kumar PS, et al. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Microbiol Mol Biol Rev. 2009 Sep;73(3):407–50. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RC, Kornman KS. Periodontol 2000. 1997;14:9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x. [DOI] [PubMed] [Google Scholar]
- Schiott CR, Loe H. J Periodont Res. 1970;5:36–41. doi: 10.1111/j.1600-0765.1970.tb01835.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Stabholz A, Soskolne WA, Shapira L. Periodontol 2000. 2010;53:138–153. doi: 10.1111/j.1600-0757.2010.00340.x. [DOI] [PubMed] [Google Scholar]
- Tonetti MS, Imboden MA, Lang NP. J Periodontol. 1998;69:1139–1147. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- Wade WG. J Clin Periodontol. 2011;38(Suppl 11):7–16. doi: 10.1111/j.1600-051X.2010.01679.x. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Yoshimura A, Kaneko T, Golenbock DT, Hara Y. J Periodontol. 2008;79:920–928. doi: 10.1902/jop.2008.070516. [DOI] [PubMed] [Google Scholar]
- Zaura E, Keijser BJ, Huse SM, Crielaard W. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, et al. Oral Biofilm Architecture on Natural Teeth. PLoS ONE. 2010;5(2):e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]