Abstract
The sensitivity of the Roche COBAS Monitor v.2.0 test was slightly better than that of the Bayer bDNA 3.0 test, but the Monitor test underquantified specimens by 2.5- to 10.6-fold even at relatively low hepatitis C virus RNA concentrations. Dilution prior to assay minimally decreased the reproducibility of the Monitor assay, leading to the recommendation for all specimens to be diluted 1:100 prior to testing by this assay.
The role of quantitative HCV RNA assays in the diagnosis and management of chronic hepatitis C is expanding. Several large-scale clinical studies have indicated that a high baseline hepatitis C virus (HCV) RNA level was associated with a decreased likelihood of a sustained virologic response (SVR) to treatment with alpha interferon in combination with ribavirin (2, 15) or with pegylated alpha interferon plus ribavirin (5, 14). Subsequently, a lack of viral clearance within the first 12 weeks of therapy was found to identify individuals unlikely to undergo an SVR (7-10, 13, 17, 21). A more recent study found that a 2-log decrease in the HCV RNA titer after 12 weeks of therapy predicted an SVR (5). For these reasons, the National Institutes of Health Consensus Development Conference Statement on the Management of Hepatitis C: 2002 stated that testing for such an early virologic response “should be a routine part of monitoring patients with genotype 1.”
Since it is likely that future studies will reveal important new indications for measuring HCV RNA levels in HCV-infected patients, we sought to systematically compare the performance of the Bayer Quantiplex HCV RNA 2.0 assay (bDNA 2.0), the Bayer VERSANT HCV RNA 3.0 assay (bDNA 3.0; Bayer Diagnostics, Emeryville, Calif.), and the Roche COBAS Amplicor HCV Monitor v.2.0 assay (Roche Molecular Systems, Branchburg, N.J.).
The analytical sensitivities and linear ranges of the bDNA 3.0, bDNA 2.0, and Monitor assays were evaluated by using the World Health Organization (WHO) First International HCV RNA standard (18) and the Acrometrix HCV RNA nucleic acid panel (Acrometrix, Berkeley, Calif.,), respectively. A logistic regression on the results from the WHO standard indicated that the analytical sensitivity of the Monitor assay (undiluted) was slightly greater than that of the bDNA 3.0 assay (P = 0.012). The fitted regressions predicted that 95% of the results should be positive at 732 IU/ml in the Monitor assay and 817 IU/ml in the bDNA 3.0 assay (95% confidence limits for the difference, 24 to 166 IU/ml) (data not shown). The most striking finding obtained by using the Acrometrix panel was the departure from linearity of the Monitor assay at high viral titers (>5.5 to 6 log10 HCV RNA; data not shown). Additional Monitor testing of high-titer clinical archive specimens of genotypes 1, 2, and 3 confirmed this departure from linearity, but no evidence of genotype bias was noted (as previously described [1, 3, 12, 16]) (data not shown). Analysis of clinical specimens was approved by the University of Washington Institutional Review Board.
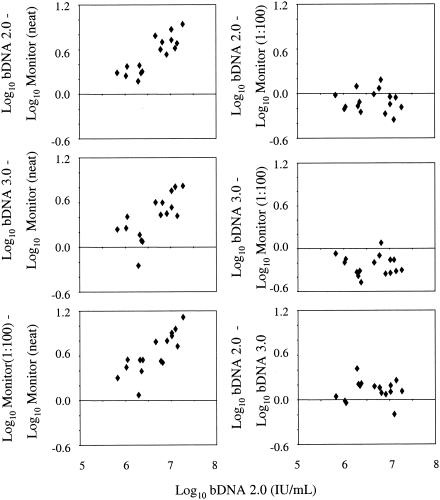
Further assessment of HCV RNA quantification was performed by using 16 high-titer clinical specimens. Monitor test performance was evaluated both undiluted and after 1:100 dilution using serial 1:10 dilutions into plasma that was HCV RNA negative by PCR. The same samples were assayed undiluted in the bDNA 2.0 and bDNA 3.0 assays. Differences between the log-transformed results from two assays are plotted against the log-transformed results from the bDNA 2.0 assay in Fig. 1. Results from the bDNA 2.0 assay were converted from equivalents per milliliter to international units per milliliter by using the manufacturer's recommended conversion factor of 5.8 Eq/IU; thus, the claimed linear range was converted from 200,000 to 120,000,000 Eq/ml to 34,500 to 20,700,000 IU/ml. For the bDNA 3.0 assay, the manufacturer's claimed linear range is 3,200 to 40,000,000 copies/ml or 615 to 7,690,000 IU/ml. (NB, a bDNA 3.0 copy is not the same as a bDNA 2.0 equivalent, according to the manufacturer.) With a single exception, estimates from the Monitor (undiluted) assay were lower than those from the other three assays (Fig. 1, left three panels). The differences between the assay results depicted in the left three panels increased with the viral titer, which probably reflects an increasing departure from linearity in the Monitor (undiluted) assay results with increasing HCV RNA concentrations. In contrast, no evidence of variation was found in the difference between the bDNA 2.0 and Monitor (1:100) assay results with increasing viral titers (Pearson correlation, r = −0.07, P = 0.80; upper right panel) or between the bDNA 3.0 and Monitor (1:100) assay results (r = −0.06, P = 0.82; middle right panel). On average, results from the bDNA 2.0 assay were 0.10 log10 (20.5%) lower than those from the Monitor (1:100) assay, while those from the bDNA 3.0 assay were 0.23 log10 (41%) lower than those from the Monitor (1:100) assay (bDNA 2.0 assay versus Monitor [1:100] assay, P = 0.017; bDNA 3.0 assay versus Monitor [1:100] assay, P < 0.0001). Finally, the difference between the two versions of the bDNA assay did not vary over the range of viral titers tested (r = −0.007, P = 0.98). On average, the results from the bDNA 2.0 assay were 0.13 log10 (35%) higher than the results from the bDNA 3.0 assay. In sum, the bDNA assays consistently produced higher HCV RNA values than the Monitor assay with undiluted specimens; when Monitor test specimens were first diluted 1:100, this finding was reversed.
FIG. 1.
Evaluation of high-titer clinical specimens with Bayer bDNA 2.0, Bayer bDNA 3.0, Roche COBAS Monitor v. 2.0 undiluted (neat), and Roche COBAS Monitor v. 2.0 diluted 1:100. Log10-transformed bDNA 2.0 HCV RNA values are shown on the x axis. Differences between the HCV RNA values obtained by the bDNA 2.0, bDNA 3.0, and COBAS Monitor 1:100 assays compared to values obtained with the COBAS Monitor undiluted assay are shown in the left panels. Differences in HCV RNA levels obtained by the bDNA version 2.0 and 3.0 and COBAS Monitor 1:100 assays are shown in the right panels.
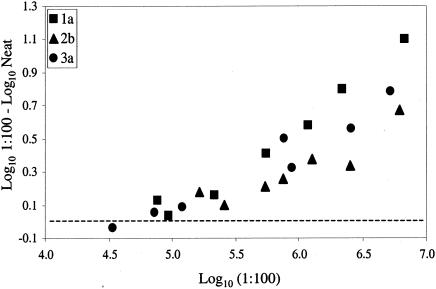
A more detailed investigation of the effect of specimen dilution prior to quantification with the Monitor test was performed by using three clinical specimens, one each of genotypes 1, 2, and 3, that were serially diluted in HCV-negative plasma. Each dilution was then treated as a separate specimen that was assayed undiluted and after 1:100 dilution. The difference between the log-transformed results obtained with and without 1:100 dilution is plotted against the log-transformed result from the 1:100 dilution in Fig. 2. The difference between the results from undiluted and diluted samples increased with increasing titers in the diluted sample, which likely reflects the increasing departure from linearity in the results from undiluted specimens with increasing HCV RNA concentrations. These results indicate that a substantial departure from linearity may occur in a result that falls well below the 500,000-IU/ml (5.7-log10) cutoff that has been recommended as the value above which dilution has been recommended in recent publications (6, 12, 16) and by the manufacturer's updated package insert. For example, estimates from three of the genotype 1a specimens increased from 212,000, 310,000, and 345,000 IU/ml when they were assayed undiluted to 552,000, 1,180,000, and 2,170,000 IU/ml, respectively, when they were assayed after 1:100 dilution.
FIG. 2.
Dilution of clinical specimens leads to improved accuracy of quantification by the Roche COBAS Monitor v. 2.0 assay. Three patient specimens were quantified with the COBAS Monitor assay, either diluted 1:100 with HCV RNA-negative plasma or undiluted (Neat). Diluted values were multiplied by the dilution factor, log10 transformed, and plotted on the x axis. The difference between the log10-transformed undiluted and log10-transformed diluted samples (multiplied by the dilution factor) was calculated and plotted on the y axis. A difference of up to 1 log was found between values obtained with undiluted or diluted samples. The zero point on the y axis is indicated by the dashed line.
To further investigate the possibility that some samples are not accurately quantified at RNA concentrations below the 500,000-IU/ml cutoff, samples from 32 treatment nonresponders were tested on the Monitor assay both undiluted and after 1:100 dilution. Eleven of the 32 samples were quantified undiluted as <500,000 IU/ml (minimum viral titer, 217,000 IU/ml). HCV RNA levels in the same specimens diluted 1:100 were found to be 2.5- to 10.6-fold higher (median, 3.4) than the values obtained with undiluted samples. Taking into account the standard deviation of the diluted assay (Table 1), a 1:100 result >2.6-fold over the undiluted result would indicate a difference due to a departure from linearity; 10 of the 11 specimens were quantified 1:100 at >2.6-fold greater than the undiluted result. The remaining 21 specimens were quantified undiluted as >500,000 IU/ml (maximum, 1,620,000 IU/ml). Again, results from the 1:100 dilutions of these samples were 2.4- to 11.1-fold higher (median, 5.7) than the results obtained with the same samples tested undiluted (data not shown).
TABLE 1.
Reproducibility of the Bayer bDNA 3.0 and COBAS Roche Monitor v.2.0 assays
| Assay | Median HCV RNA levela | No. of samples × no. of replicatesc | Intraassay SDa | Assay SDa,b |
|---|---|---|---|---|
| bDNA 3.0 | 5.59 | 5 × 7−8 | 0.055 | 0.075 |
| Monitor undiluted | 5.57 | 5 × 8−11 | 0.067 | 0.119 |
| Monitor 1:100 | 5.57 | 5 × 8−11 | 0.137 | 0.170 |
Log10 number of HCV RNA international units per milliliter.
Combination of intraassay and interassay variations.
Values for numbers of replicates are ranges.
The underestimation of HCV RNA levels in this range by the Monitor quantitative assay is particularly important, given the finding by several large clinical trials that subjects with viral titers greater than 800,000 IU/ml are less likely to experience an SVR to antiviral therapy (2, 5, 14, 15). No such departure from linearity was detected with the Bayer bDNA 3.0 assay. Previous studies have demonstrated limited linearity in the Monitor assay (1, 6, 11), which was presumably corrected by retesting diluted samples when the result is greater than the 500,000-IU/ml high-end cutoff recommended by the manufacturer. Our data clearly demonstrate that this procedure does not completely correct the problem. Additional detailed analysis would be required to determine a lower threshold above which all samples should be diluted when the Monitor assay kit is used. Thus, we conclude that this saturation effect could best be avoided by diluting all specimens 1:100 with HCV RNA-negative plasma or serum prior to testing with the Monitor assay. Doing so would also reduce the likelihood of falsely concluding that a patient lacked a 2-log drop in the HCV RNA level after 12 weeks of therapy and was therefore a treatment nonresponder.
Intra- and interassay variations for the bDNA 3.0 and Monitor assays were assessed by using clinical specimens of genotypes 1a, 1b, 2a, 2b, and 3a with titers of 144,000 to 1,570,000 IU/ml by the bDNA 2.0 assay (Table 1). One sample of each genotype was tested two or three times in each of three runs on the bDNA 3.0 assay and three to five times in each of three runs on the Monitor assay. In an effort to minimize the impact of dilution errors on assay variation, the diluted specimens were obtained from single 1:100 dilutions rather than serial 1:10 dilutions. The overall assay standard deviation was calculated as the square root of the sum of the intraassay and interassay variances. Intraassay standard deviations from the bDNA 3.0 and Monitor (undiluted) assays were similar, but the overall assay standard deviation from the Monitor (undiluted) assay was slightly greater than the overall standard deviation from the bDNA 3.0 assay. Diluting the samples 1:100 increased both the intraassay and overall assay standard deviations. However, the increase in the overall assay standard deviation was caused almost entirely by the increase in the intraassay component; this was most likely due to a combination of dilution “error” and the 100-fold lower level of HCV RNA being quantified (compared to the undiluted samples) (4 D. Brambilla, S. Granger, and J. Bremer, 7th Conf. Retrovir. Opportunistic Infect., abstr. 774, 2000). Although the dilution procedure incurs a slight increase in within-run variation, it would avoid any possibility that samples were being underquantified in an era when viral titer criteria are used to guide the implementation of antiviral therapies and to judge the likelihood of an SVR.
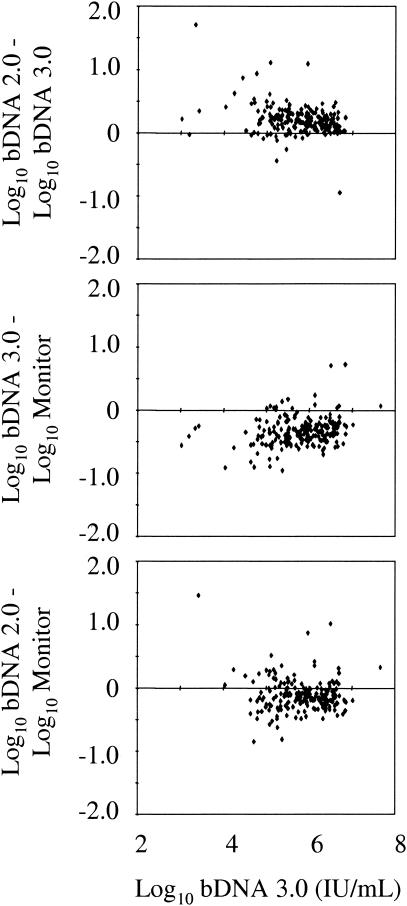
To assess whether the three quantitative assays yielded similar HCV RNA results, 280 of 284 consecutive clinical specimens assayed by the bDNA 2.0 and 3.0 assays were also tested with the Monitor assay. All 280 specimens were tested first at a 1:100 dilution by the Monitor assay and retested undiluted if the first assay yielded negative results. Two of the 284 consecutive specimens were also retested with the bDNA 3.0 assay after a 1:100 dilution of the sample was made with plasma that was HCV RNA negative by PCR, since the undiluted result was above the upper limit of linearity of the bDNA test (7,690,000 IU/ml). Seventy-four of 284 specimens were excluded because they were HCV RNA negative by the qualitative Roche COBAS Amplicor HCV test, v. 2.0 (Roche Molecular Systems), performed in accordance with the manufacturer's instructions (12). Another 16, 14, and 18 specimens were excluded from the respective pairwise comparisons between the bDNA 2.0 and bDNA 3.0 assays, the bDNA 3.0 and Monitor assays, and the bDNA 2.0 and Monitor assays because at least one of two assay results was below the limit of detection or the Monitor assay was not performed (Fig. 3). The log10-transformed HCV RNA titers of the specimens tested ranged from 4.50 to 7.92 (median, 6.04) IU/ml in the bDNA 2.0 assay. One hundred ninety-one (68%) assays of diluted specimens were positive in the Monitor assay; the other 89 samples were retested undiluted. The great majority of differences between assay results were within 0.5 log10 (81.1 to 95.3%) or 1 log (98.5 to 100%), as recently reported by Shiffman et al. (20). The median difference between the bDNA 2.0 and bDNA 3.0 results was 0.20 log10, while the median difference between the bDNA 3.0 and bDNA 2.0 assays and the Monitor assay were −0.34 and −0.13 log10, respectively. Similar to the high-titer specimen comparison, the bDNA 2.0 and combined Monitor assays (primarily from 1:100 dilution) demonstrated consistently higher HCV RNA results than the bDNA 3.0 assay.
FIG. 3.
Differences between assay results from consecutive clinical specimens. Log10-transformed bDNA 3.0 HCV RNA values are shown on the x axis. Differences between the log10-transformed HCV RNA values obtained from the same clinical specimens by the bDNA 2.0, bDNA 3.0, and COBAS Monitor assays are indicated on the y axis.
The recent calibration of quantitative assays against the WHO HCV RNA standard represents a significant advance for HCV RNA quantification. However, our data are consistent with those of other groups, suggesting that HCV RNA quantitative results obtained by different assays may not necessarily be interchangeable, although the results are expressed in the same units (international units per milliliter) (1, 6, 19). We therefore recommend that the same quantitative assay be used longitudinally (preferably in the same laboratory) if patients are to be monitored by using such data, to minimize any confusion over changes in the viral level over time.
The sensitivities and specificities of the three quantitative assays were also evaluated by using data from the 284 consecutive clinical specimens described above (Table 2). Specimens were considered true positives if they were positive in both bDNA assays and either Monitor assay (diluted or undiluted). Ninety-one samples with negative results from either bDNA assay or both Monitor assays were classified as HCV positive or negative after retesting with the qualitative Roche COBAS Amplicor HCV test. These data demonstrate the improved clinical specificity of the newer Bayer bDNA 3.0 assay (95.9%) over version 2.0 (70.3%), with similar sensitivities (95.2 versus 94.3%), from consecutive specimens received in a high-volume clinical laboratory. The Monitor assay performed slightly better than the bDNA 3.0 assay in both clinical sensitivity (97.6%) and specificity (100%), although these differences were not significant. Compared to those of other groups who have studied the performance characteristics of these two assay versions, our data demonstrate a lower specificity for the bDNA 2.0 assay than the previously reported 96% (3). The surprising number of false-positive results (n = 22) was obtained in five separate runs, although 9 of the 22 were from a single run. Three of 21 samples were positive when they were retested with the bDNA 2.0 assay. The 22nd sample was not retested owing to insufficient volume. The 22 false positives had values ranging from 33,500 to 963,000 (median, 100,000) IU/ml. Two factors could have affected these results. One is that multiple operators in a high-throughput clinical laboratory generated the bDNA 2.0 assay results over multiple runs. In addition, a single lot of reagents was used during the testing period. Finally, the specificity of the bDNA 3.0 assay (95.9%) was similar to those found by Beld et al. (96.8%) (1) and Trimoulet et al. (98.2%) (22).
TABLE 2.
Clinical sensitivities and specificities of three HCV RNA quantitative assays tested on 284 consecutive clinical samples
| Assay | No. of samples positive by assay/total (% sensitivity) | No. of samples positive by assay/total (% specificity) |
|---|---|---|
| bDNA 2.0 | 198/210 (94.3) | 52/74 (70.3) |
| bDNA 3.0 | 200/210 (95.2) | 71/74 (95.9) |
| COBAS Monitor | 201/206 (97.6) | 74/74 (100) |
In summary, our study demonstrates the improved clinical specificity of the updated, Food and Drug Administration-approved bDNA 3.0 assay over version 2.0 to a level that approaches but does not surpass that of the Monitor assay. In addition, the Monitor assay had slightly greater analytical sensitivity than the bDNA 3.0 assay. The bDNA 3.0 assay showed excellent reproducibility and dynamic range. The Monitor (undiluted) and bDNA 3.0 assays were similar in terms of within- and between-assay variations and lack of bias for quantification of HCV genotypes 1, 2, and 3. In a subset of HCV RNA specimens, a saturation effect was noted with the Monitor (undiluted) assay, so that specimens were initially misquantified below 500,000 IU/ml. This effect was not seen with the bDNA 3.0 assay in this range. Dilution of specimens 1:100 prior to analysis with the Monitor assay corrected this test error. Although dilution was found to increase assay variance, the overall increase in accuracy obtained with dilution justifies the recommended dilutional algorithm for the Monitor assay. When this procedure was used, the Monitor assay gave HCV RNA values consistently higher than those obtained with the bDNA 3.0 assay. Finally, the kits tested here performed adequately but future quantitative assays will aim to improve both the sensitivity and the linear range of HCV RNA detection.
Acknowledgments
This study was supported in part by NIH contracts N01-DK-9-2318 and N01-DK-9-2328, HALT-C Clinical Trial-Virology Laboratory and HALT-C Clinical Trial-Data Coordinating Center, respectively. We acknowledge the generous contribution of COBAS Amplicor HCV Monitor Test v.2.0 and COBAS Amplicor HCV Test v. 2.0 kits from Roche Molecular Systems.
Finally, we thank all of the members of the University of Washington Viral Hepatitis Laboratory for ongoing contributions, with special thanks to Christina Yoshihara for assistance in preparation of the manuscript.
REFERENCES
- 1.Beld, M., R. Sentjens, S. Rebers, C. Weegink, J. Weel, C. Sol, and R. Boom. 2002. Performance of the New Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS Amplicor HCV Monitor, version 2.0, assay. J. Clin. Microbiol. 40:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, and J. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 3.Detmer, J., R. Lagier, J. Flynn, C. Zayati, J. Kolberg, M. Collins, M. Urdea, and R. Sanchez-Pescador. 1996. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J. Clin. Microbiol. 34:901-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erice, A., D. Brambilla, J. Bremer, J. B. Jackson, R. Kokka, B. Yen-Lieberman, and R. W. Coombs. 2000. Performance characteristics of the QUANTIPLEX HIV-1 RNA 3.0 assay for detection and quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffmann, and J. Yu. 2002. Peg-interferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 6.Germer, J. J., P. J. Heimgartner, D. M. Ilstrup, W. S. Harmsen, G. D. Jenkins, and R. Patel. 2002. Comparative evaluation of the VERSANT HCV RNA 3.0, QUANTIPLEX HCV RNA 2.0, and COBAS AMPLICOR HCV MONITOR version 2.0 assays for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:495-500. (Erratum, 40:1885.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanley, J. P., L. M. Jarvis, J. Andrew, R. Dennis, P. C. Hayes, J. Piris, R. Lee, P. Simmonds, and C. A. Ludlam. 1996. Interferon treatment for chronic hepatitis C infection in hemophiliacs—influence of virus load, genotype, and liver pathology on response. Blood 87:1704-1709. [PubMed] [Google Scholar]
- 8.Hino, K., M. Okuda, T. Konishi, H. Ishiko, and K. Okita. 1995. Serial assay of hepatitis C virus RNA in serum for predicting response to interferon-alpha therapy. Dig. Dis. Sci. 40:14-20. [DOI] [PubMed] [Google Scholar]
- 9.Karino, Y., J. Toyota, M. Sugawara, K. Higashino, T. Sato, T. Ohmura, T. Suga, Y. Okuuchi, and T. Matsushima. 1997. Early loss of serum hepatitis C virus RNA can predict a sustained response to interferon therapy in patients with chronic hepatitis C. Am. J. Gastroenterol. 92:61-65. [PubMed] [Google Scholar]
- 10.Kleter, G. E., J. T. Brouwer, R. A. Heijtink, S. W. Schalm, and W. G. Quint. 1993. Detection of hepatitis C virus RNA in patients with chronic hepatitis C virus infections during and after therapy with alpha interferon. Antimicrob. Agents Chemother. 37:595-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konnick, E. Q., M. Erali, E. R. Ashwood, and D. R. Hillyard. 2002. Performance characteristics of the COBAS Amplicor hepatitis C virus (HCV) monitor, version 2.0, international unit assay and the National Genetics Institute HCV Superquant assay. J. Clin. Microbiol. 40:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, W. M., K. R. Reddy, M. J. Tong, M. Black, D. J. vanLeeuwen, F. B. Hollinger, K. D. Mullen, N. Pimstone, D. Albert, and S. Gardner. 1998. Early hepatitis C virus RNA responses predict interferon treatment outcomes in chronic hepatitis C. Hepatology 28:1411-1415. [DOI] [PubMed] [Google Scholar]
- 14.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 15.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 16.Mellor, J., A. Hawkins, and P. Simmonds. 1999. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J. Clin. Microbiol. 37:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orito, E., M. Mizokami, K. Suzuki, K. Ohba, T. Ohno, M. Mori, K. Hayashi, K. Kato, S. Iino, and J. Y. Lau. 1995. Loss of serum HCV RNA at week 4 of interferon-alpha therapy is associated with more favorable long-term response in patients with chronic hepatitis C. J. Med. Virol. 46:109-115. [DOI] [PubMed] [Google Scholar]
- 18.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 19.Sherman, K. E., S. D. Rouster, and P. S. Horn. 2002. Comparison of methodologies for quantification of hepatitis C virus (HCV) RNA in patients coinfected with HCV and human immunodeficiency virus. Clin. Infect. Dis. 35:482-487. [DOI] [PubMed] [Google Scholar]
- 20.Shiffman, M. L., A. Ferreira-Gonzalez, K. R. Reddy, R. K. Sterling, V. A. Luketic, R. T. Stravitz, A. J. Sanyal, C. T. Garrett, M. De Medina, and E. R. Schiff. 2003. Comparison of three commercially available assays for HCV RNA using the international unit standard: implications for management of patients with chronic hepatitis C virus infection in clinical practice. Am. J. Gastroenterol. 98:1159-1166. [DOI] [PubMed] [Google Scholar]
- 21.Tong, M. J., L. M. Blatt, J. G. McHutchison, R. L. Co, and A. Conrad. 1997. Prediction of response during interferon alfa 2b therapy in chronic hepatitis C patients using viral and biochemical characteristics: a comparison. Hepatology 26:1640-1645. [DOI] [PubMed] [Google Scholar]
- 22.Trimoulet, P., P. Halfon, E. Pohier, H. Khiri, G. Chene, and H. Fleury. 2002. Evaluation of the VERSANT HCV RNA 3.0 assay for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]