Abstract
The complexity and heterogeneity of tumours have hindered efforts to identify commonalities among different cancers. Furthermore, because we have limited information on the prevalence and nature of ubiquitous molecular events that occur in neoplasms, it is unfeasible to implement molecular-targeted cancer screening and prevention. Here, we found that the FEAT protein is overexpressed in most human cancers, but weakly expressed in normal tissues including the testis, brain, and liver. Transgenic mice that ectopically expressed FEAT in the thymus, spleen, liver, and lung spontaneously developed invasive malignant lymphoma (48%, 19/40) and lung-metastasizing liver cancer (hepatocellular carcinoma) (35%, 14/40) that models human hepatocarcinogenesis, indicating the FEAT protein potently drives tumorigenesis in vivo. Gene expression profiling suggested that FEAT drives receptor tyrosine kinase and hedgehog signalling pathways. These findings demonstrate that integrated efforts to identify FEAT-like ubiquitous oncoproteins are useful and may provide promising approaches for cost-effective cancer screening and prevention.
Although our understanding of the molecular mechanisms of carcinogenesis has greatly improved, this knowledge has not lead to the identification and development of effective tools for cancer screening and prevention 1. Approximately 30–40% of all cancer deaths are preventable, and this estimation is based on indirect measures that do not interfere with carcinogenesis, such as dietary modifications, lifestyle changes, minimizing carcinogen exposure, and vaccination against oncogenic viruses 1. More advanced chemoprevention measures such as tamoxifen, raloxifene, finasteride, and celecoxib 1 and the eradication of Helicobacter pylori are only available to high-risk groups for particular cancers.
In order to reach the long-term goal of establishing molecular-targeted cancer screening and prevention, it is important that we explore, characterize, and catalogue a distinct subclass of cancer genes 2 that are involved in diverse cancers. However, the systematic approaches that have been used to identify cancer genes, such as sequencing protein-coding exons 3,4,5,6,7,8,9, whole genome sequencing 10,11, and paired-end sequencing to comprehensively identify somatic rearrangements 12, have only further emphasized the marked heterogeneity and complexity of human neoplasms and have not successfully identified commonalities among cancers. Driver mutations that contribute to the development of human cancers 13 are highly variable among different types of cancer and among individual tumours of the same type. Thus, it is still unknown if there are oncogenic molecules that are commonly altered in diverse cancers.
There is accumulating evidence that cancers have heterogeneous combinations of deregulated cancer genes 13,14 and that signalling pathways rather than individual genes are the targets in tumorigenesis 15. Although some canonical signalling pathways are universally deregulated in cancers, different components of these pathways can be affected in different tumours 3,5,6,7,8,9,15. The proteins that are commonly overexpressed in cancers are predominantly thought to reflect “peripheral” changes 2,15 that result from neoplastic phenotypes (i.e., augmented metabolic and homeostatic processes such as glycolysis, macromolecular synthesis, and DNA replication) 16,17 and the resulting “stress phenotype” 18. Thus, these proteins have not been considered as targets for cancer therapy and prevention. However, this presumption has not been rigorously tested in vivo 19.
In the present study, we found that FEAT protein (faint expression in normal tissues, aberrant overexpression in tumours) is uniformly overexpressed in a variety of human cancers. Remarkably, FEAT transgenic mice indicated that FEAT potently drives tumorigenesis in vivo. Expression microarray analyses suggested that FEAT induces oncogenic pathways. The significance of overexpressed genes in cancer is increasingly recognized as potential leads for a variety of diagnostic and therapeutic approaches 6,7,20. Additional studies that identify and characterize FEAT-like oncogenic proteins will hopefully advance molecular-targeted cancer screening and prevention.
Results
Biochemical purification of FEAT protein
On analyses of molecules regulating nuclear apoptosis in a cell-free system 21,22, we noticed that the apoptosis-inducing activity was attenuated in the middle of active fractions, concomitant with the appearance of two polypeptides with apparent molecular masses of 74 and 66 kDa (Supplementary Fig. 1). This suggested that apoptosis inhibitors were copurified with apoptosis inducers. Microsequencing revealed that the 74 kDa protein is a rat homologue of human CGI-01 protein 23 that is encoded by METTL13 (methyltransferase like 13) gene (also known as KIAA0859). CGI-01, renamed FEAT, contains two S-adenosylmethionine-binding motifs (SAM-binding motifs) that are characteristic of methyltransferases and related enzymes (see Fig. 1f) 24, and the structure is well conserved across species (Supplementary Fig. 2). Capture compounds mass spectrometry using S-adenosylhomocysteine has detected the Arabidopsis thaliana orthologue of FEAT (At2g31740) 25, suggesting that FEAT can bind SAM. We did not detect protein methyltransferase activity, spermidine/spermine synthase activity, or ubiquinone synthase activity (Supplementary Fig. 3) in full-length or truncated FEAT proteins (Supplementary Note). Further studies are required to determine whether FEAT has enzymatic activities.
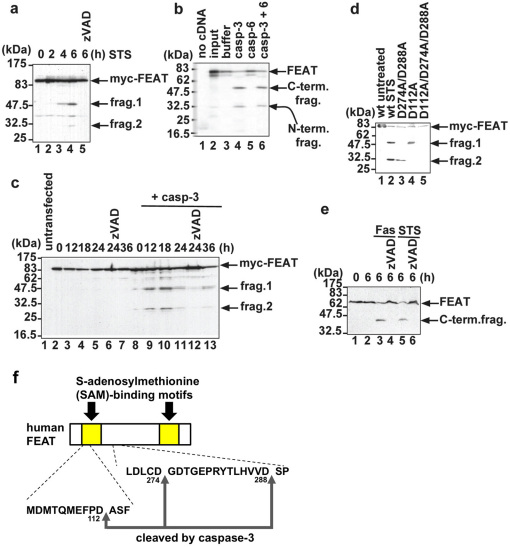
Figure 1. FEAT is a substrate for caspase-3.
(a) Cleavage of N-terminal Myc-tagged FEAT (myc-FEAT) in apoptotic cells. COS-7 cells expressing myc-FEAT were treated with 1 µM staurosporine (STS) for the indicated times. Lane 5: cells pretreated for 30 min with 100 µM zVAD-fmk, a broad spectrum caspase inhibitor. (b) In vitro transcribed/translated [35S]-labelled FEAT was cleaved by purified caspase-3, but not by caspase-6. (c) Caspase-3 is mainly responsible for FEAT cleavages. MCF-7 cells expressing myc-FEAT alone or together with procaspase-3 (+ casp-3) were treated with 1 µM STS for the indicated times. (d) Mutating caspase-3 cleavage sites abrogates FEAT cleavages. COS-7 expressing wild-type (wt) or mutant myc-FEAT were treated for 6 h with 1 µM STS (lane 2 to 5). Immunoblots (a, c, d) were probed with an anti-Myc antibody. (e) Cleavage of endogenous FEAT in apoptotic cells. Jurkat T cells preincubated for 1 h without or with 100 µM zVAD-fmk were treated for 6 h with 100 ng/ml CH-11 agonistic anti-Fas antibody or 1 µM STS. The immunoblot was stained with the anti-FEATΔN antibody. (f) Schematic diagram of the human FEAT structure and caspase-3 cleavage sites.
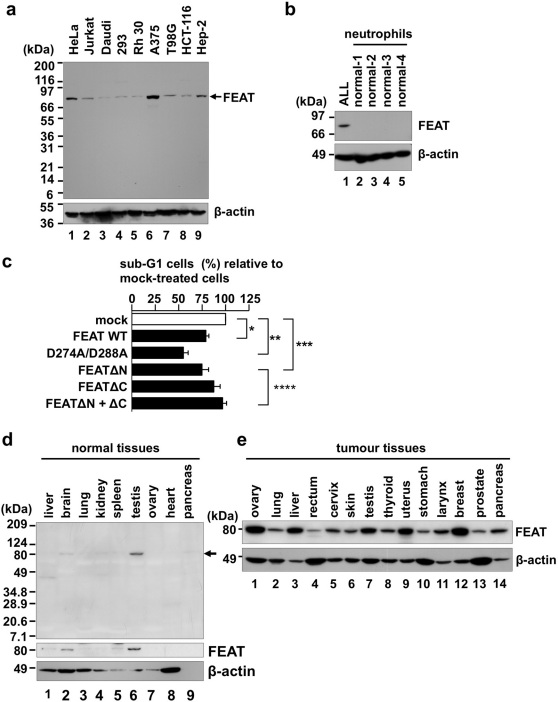
Figure 2. FEAT is overexpressed in human cancers.
(a) The immunoblot (IMB-105) contains lysates (10 µg protein/lane) from the following human cancer-derived cell lines: HeLa, uterine cervical carcinoma; Jurkat, T-cell leukaemia; Daudi, Burkitt lymphoma; 293, embryonal kidney transformed by adenovirus type 5; Rh 30, rhabdomyosarcoma; A375, malignant melanoma; T98G, glioblastoma; HCT-116, colon carcinoma; and Hep-2, larynx carcinoma. Immunofluorescence microscopy using the same antibody revealed that FEAT is diffusely localized in the cytoplasm and nucleus of HeLa cells (Supplementary Fig. 5; Supplementary Note). (b) Absence of FEAT expression in normal neutrophils. Peripheral blood mononuclear cells from a patient with acute lymphoblastic leukaemia (ALL) and neutrophils from four normal volunteers were analyzed by immunoblotting. (c) FEAT attenuates spontaneous neutrophil apoptosis. His-tagged wild-type (WT) or mutant FEAT proteins were introduced into neutrophils by protein transduction. Apoptotic neutrophils with a hypodiploid DNA content were analyzed by flow cytometry and normalized to that of cells incubated with irrelevant proteins (mock) (*, P = 0.0012, n = 9; **, P = 0.0001, n = 9; ***, P = 0.0192, n = 4; ****, P = 0.0135, n = 4; paired t-test). Error bars, s.e.m. (d) Multiple Tissue Blot (Human, WB46, 75 µg protein/lane) was probed with anti-FEATΔN (uppermost panel; FEAT is indicated by an arrow), anti-FEATΔC (middle panel), and anti-actin (lowermost panel) antibodies. (e) Human Tumor Tissue Blot (WB51). The proteins loaded (50 µg/lane) were derived from the following: ovary stromal sarcoma, lung adenocarcinoma, hepatocellular carcinoma, rectal adenocarcinoma, cervical squamous cell carcinoma, skin malignant melanoma, testis embryonal carcinoma, thyroid follicular carcinoma, uterine adenocarcinoma, stomach adenocarcinoma, laryngopharynx squamous cell carcinoma, breast ductal carcinoma, prostate hyperplasia, and pancreatic adenocarcinoma. (a, b, and e) Blots were probed with anti-FEATΔN (upper panels) and anti-actin (lower panels) antibodies.
FEAT is cleaved by caspase-3
Exogenously-expressed FEAT was cleaved during staurosporine-induced (STS-induced) apoptosis of COS-7 cells (Fig. 1a). In vitro transcribed/translated FEAT was cleaved by caspase-3, but not by caspase-6 (Fig. 1b). Purified His-tagged FEAT was cleaved by purified caspase-3. FEAT was minimally cleaved in apoptotic MCF-7 cells, which are deficient in caspase-3 26, and coexpression of procaspase-3 led to efficient cleavages of FEAT (Fig. 1c). Site-directed mutagenesis studies (Fig. 1d) revealed caspase-3 cleavage sites in human FEAT. Endogenous FEAT was cleaved in Jurkat T cells undergoing Fas- and STS-induced apoptosis (Fig. 1e). These results indicated that caspase-3 cleaves FEAT in apoptotic cells (Fig. 1f). D112 and D288 are well conserved across species (Supplementary Fig. 2; Supplementary Note), suggesting that caspase cleavages of FEAT play critical role(s) in organisms.
FEAT attenuates apoptotic cell death and the antiapoptotic activity is abrogated upon caspase-3-mediated cleavages
Cleavages of antiapoptotic kinases and phosphatases by caspases fine-tune apoptosis through terminating prosurvival signalling and generating proapoptotic peptide fragments 27. We therefore assessed whether FEAT or its caspase-3-cleaved fragments affect apoptosis. Ex vivo experiments using plasmid transfection and RNA interference suggested that FEAT can impede apoptosis (Supplementary Fig. 4; Supplementary Note). We searched for cell types that do not express FEAT, found that almost all cancer-derived cell lines express FEAT (Fig. 2a), and decided to use neutrophils (Fig. 2b).
Protein transduction 28 of wild-type FEAT (FEAT WT) and FEATΔN (amino acid 289–699), a fragment generated by caspase-3 cleavage, significantly attenuated spontaneous apoptosis in neutrophils (Fig. 2c). The D274A/D288A mutant was more potent than FEAT WT or FEATΔN. In contrast, FEATΔC (amino acid 1–274), N-terminal fragment generated by the cleavage between the SAM-binding motifs, did not affect apoptosis, and the addition of FEATΔC interfered with the antiapoptotic function of FEATΔN (FEATΔN + ΔC). Taken together, the results are consistent with the notion that FEAT has the ability to attenuate apoptosis, which is abrogated by caspase-3 cleavages between the SAM-binding motifs.
FEAT is aberrantly overexpressed in human cancer tissues
We noticed that FEAT corresponds to the TGACCTCCAG tag that is used in the serial analysis of gene expression (SAGE) studies of human transcriptomes, which has been linked to a transcript that is uniformly elevated in human colon, brain, breast, and lung cancers and melanoma compared with the corresponding normal tissues 29. Consistent with these classifications, immunoblotting analyses of normal human tissues showed weak FEAT expression only in the testis, brain, and liver (Fig. 2d), which correlated with mRNA expression 23. In marked contrast, FEAT protein was moderately to highly expressed in a wide range of human cancer tissues (Fig. 2e), suggesting that FEAT is a ubiquitous protein involved in tumour biology.
FEAT upregulation is oncogenic in vivo
To assess whether FEAT upregulation in human cancers contributes to tumorigenesis, we generated transgenic mice that express human FEAT under a promoter that is active in a wide range of tissues (Supplementary Note). Immunoblotting showed expression of FEAT in the thymus, spleen, liver, and lung of transgenic mice (Fig. 3a). Thymocytes from transgenic mice showed significant decreases in Fas ligand- and glucocorticoid-induced cell death (Fig. 3b), indicating that transgenic expression of FEAT attenuates apoptosis.
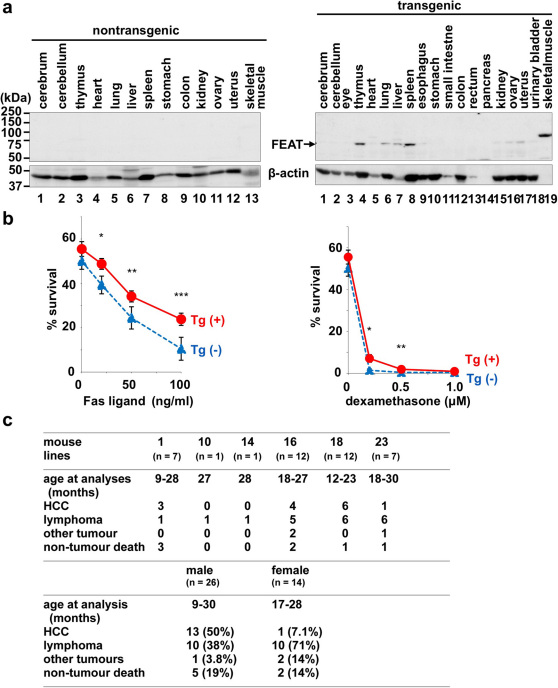
Figure 3. Analyses of FEAT transgenic mice.
(a) FEAT protein expression in nontransgenic (left panel) or transgenic (right panel) mouse organs. Similar results were obtained with first generation offspring of three distinct founders. The blot (30 µg protein/lane) was probed with anti-FEATΔN (upper panels) and anti-actin (lower panels) antibodies. (b) Cell death of thymocytes is attenuated in FEAT transgenic mice. Survival of thymocytes isolated from transgenic mice (Tg (+)) (red, solid lines) or nontransgenic littermates (Tg (-)) (blue, dashed lines) were analyzed by flow cytometry (means ± s.e.m.) after treatment for 24 h with Fas ligand (left panel) (*, P = 0.02492; **, P = 0.0497; ***, P = 0.0226, n = 5; paired t-test) or dexamethasone (right panel) (*, P = 0.0331; **, P = 0.0411, n = 6; paired t-test). (c) Tumours in founder, first and second generation FEAT transgenic mice. HCC, hepatocellular carcinoma.
After 12 month of age, the transgenic mice began developing hepatocellular carcinoma (HCC), as supported by immunohistochemical analyses of α-fetoprotein and albumin, and malignant lymphoma (Fig. 3c and Supplementary Fig. 6). HCC was also observed in a 9-month-old male transgenic mouse (Supplementary Fig. 6), suggesting that the hepatocarcinogenesis can be initiated earlier. In contrast, none of the nontransgenic littermates developed HCC, and lymphoma was observed in nontransgenic littermates with a lower incidence (18%, 3/17). Consequently, the transgenic mice developed tumours or died earlier than the nontransgenic littermates (Fig. 4a). HCC and lymphoma were observed in the offspring of four distinct founders (Fig. 3c), arguing against the possibility that the tumorigenesis resulted from integration of the transgene in endogenous cancer genes in the mouse genome. The murine HCC recapitulated the strong male predilection (Fig. 3c) that is observed in human patients 30.
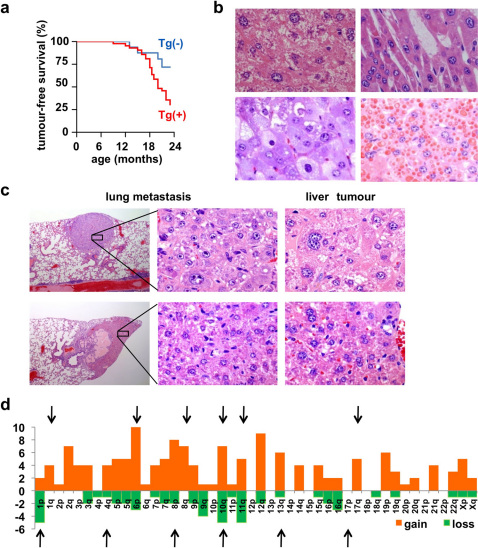
Figure 4. Hepatocellular carcinoma (HCC) in FEAT transgenic mice.
(a) Kaplan-Meier curve for tumour-free survival of FEAT transgenic mice (Tg (+)) (red) (n = 40) and nontransgenic littermates (Tg (-)) (blue) (n = 17) (P = 0.038, log-rank test). (b) Various histological subtypes of HCC. Well-differentiated tumours composed of dysplastic hepatocytes with atypical large polyploid nuclei and anisokaryosis with compact (upper left panel) and trabecular (upper right panel) growth patterns, and variants such as a clear-cell type (left lower panel) and HCCs with cytoplasmic inclusion bodies (right lower panel). (c) Morphology of neoplastic cells in lung metastases, which were similar to primary liver tumours. (b and c) hematoxylin and eosin (H&E) staining. Original magnifications are: ×115 (c, left panels) and ×600 (b; c, middle and right panels). (d) Syntenic human chromosomal regions corresponding to CNAs detected by array comparative genomic hybridization (array-CGH). Each column shows the number of genes present in each human chromosomal arm that was commonly gained (red) or lost (green) in HCCs of FEAT transgenic mice (n = 6). The arrows indicate the chromosomal arms that are gained or lost in human HCCs 41.
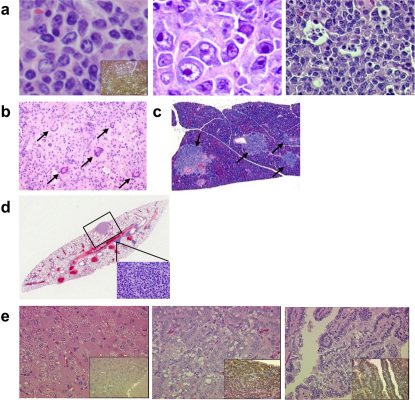
Murine HCCs displayed various histological subtypes similar to human HCCs (Fig. 4b). Lung metastases were detected in two mice with HCC (Fig. 4c). Most lymphomas belonged to diffuse large or Burkitt-like B-cell lymphoma (Fig. 5a), both of which expressed the B-cell marker CD45R (B220) (100%, 15/15) (inset) and had undergone clonal rearrangement of the immunoglobulin genes (83%, 5/6). These are the types of lymphoma that are most common in human patients. None of the examined lymphomas were positive for the T-cell marker CD3 (0%, 0/7). CD45R- and CD3-negative polymorphic variants with giant cells were also observed (Fig. 5b). The lymphomas were highly invasive and often infiltrated the pancreas (Fig. 5c).
Figure 5. Lymphoma and co-occurrence of other tumours in FEAT transgenic mice.
(a) Microscopic appearance of B-cell lymphomas from FEAT transgenic mice. Centroblastic (left panel) or immunoblastic (middle panel) variants of diffuse large B-cell lymphoma and Burkitt-like B-cell lymphoma with a starry sky appearance (right panel). (b) Giant cells (arrows) in polymorphic lymphoma. (c) Infiltration of lymphoma in the pancreas (arrows). (d) A section of the lung with metastases of hepatocellular carcinoma (HCC) (rectangle, corresponding to left upper panel in Fig. 4c) and lymphoma (inset). (e) HCC (left panel), and a lung adenocarcinoma (middle and right panels) that developed in a transgenic mouse, which also harboured lymphoma. (a–e) H&E staining. Original magnifications are: ×600 (a), ×115 (b), ×60 (c), ×15 (d), and ×400 (e). The insets show immunohistochemical staining for a B-cell marker CD45R (B220) (a), and prosurfactant protein C, a marker of type II pneumocytes, which was absent in the HCC and present in the lung adenocarcinoma (e).
One transgenic mouse developed both HCC and lymphoma (Fig. 5d), and another transgenic mouse harboured three tumours, i.e., lymphoma, HCC, and lung adenocarcinoma (Fig. 5e), as confirmed by the expression prosurfactant protein C (insets). In addition, one mouse had lymphoma and rhabdomyosarcoma (Supplementary Fig. 6). Immunohistochemistry showed that FEAT expression is higher in HCCs than adjacent liver tissues. Sequencing analyses showed that there were no mutations in the FEAT transgene in HCCs (0%, 0/5), suggesting that structural changes in FEAT are not required for tumorigenesis. Unlike the CF-1 and C3H inbred strains, the occurrence of HCC is unusual in C57BL/6 mice 31. Thus, FEAT is a unique oncoprotein that potently induces both HCC and malignant lymphoma on the C57BL/6 inbred strain background.
FEAT transgenic mice as a relevant model for human HCCs
Mouse models of cancers with extensive physiological and molecular similarities to human patients can be exploited to determine the causes of human cancers, to devise the strategy for cancer prevention, and to develop and test new treatments 32. Inflammation plays critical roles in the initiation and promotion of human and murine HCCs 33. To evaluate the possibility that FEAT promotes hepatocarcinogenesis by inducing prolonged inflammatory responses such as autoimmune hepatitis, we examined whether FEAT transgenic mice with HCC have background inflammatory lesions in the liver. Only sparse low-grade perivasculitis with minimal tissue destruction was observed in 50% (7/14) of the liver adjacent to HCC in FEAT transgenic mice, while a similar degree of perivasculitis was detected in 35% (6/17) of the liver from nontransgenic littermates (Supplementary Fig. 7). The observations did not support the idea that FEAT causes HCC through inflammatory mechanisms.
We next assessed whether the HCCs in FEAT transgenic mice closely model genetic events in human HCCs. The loss of p53 tumour suppressor function is a key event in certain subsets of human HCCs 34. In particular, HCCs caused by aflatoxin characteristically have the S376A mutation in p53. Mutations in the open reading frame (ORF) of the p53 mRNA were not detected in mouse HCCs (0%, 0/7). MDM2 amplification, which is observed in some human HCCs 35, was unlikely because immunohistochemistry did not reveal increased MDM2 in the mouse HCCs compared with adjacent normal liver tissues (0%, 0/5). Activation of Wnt signalling pathway through mutations in ß-catenin or Axin that is found in a small population of human HCCs 36 was unlikely because of the absence of the nuclear localization of ß-catenin (0%, 0/6) and of any mutations in the hotspots in the ß -catenin gene (0%, 0/9). Overall, HCCs in the transgenic mice do not correspond to certain minor subsets of human HCCs 36.
To evaluate whether the mouse HCCs harbour chromosomal changes similar to human patients, the amplification (gain) or deletion (loss) of genomic regions (copy number alterations, CNAs) were analyzed by microarray-based comparative genomic hybridization (array-CGH). A genome-wide view of large-scale CNAs showed that the murine HCCs had marked genomic instability, with more gains than losses (Supplementary Fig. 8) as observed in human HCC 37, while the liver from a transgenic mouse without HCC had minimal CNAs. This suggested that genomic fragility developed during the process of hepatocarcinogenesis rather than because of direct effects of the FEAT transgene.
Tandem duplications that range in size from 3 kb to greater than 1 Mb is the most commonly observed architectures of rearrangement in a recent study of human breast cancer genomes 12. High-resolution array-CGH revealed gain or loss of such small chromosomal segments in the murine HCCs. The small-scale CNAs involved 18 known cancer genes (http://www.sanger.ac.uk/genetics/CGP/Census/) (MDS1, PDGFRA, PIK3R1, JAZF1, WHSC1L1, HOOK3, PCM1, MLLT3, PTEN, CBL, ERCC5, ERCC4, CYLD, CBFB, BRIP1, MLLT1, TMPRSS2, NF2) (Supplementary Table 1). TTN 4, SKP2 38, EED1 39, and PVT1 40 have also been implicated in oncogenesis. To directly compare the small-scale CNAs in mouse and human HCCs, we listed focal CNAs that were shared among the murine HCCs and identified human chromosomal loci syntenic to the CNAs (Supplementary Table 1). The syntenic regions covered most of the chromosomal changes that were previously implicated in human HCCs 41 (Fig. 4d) and exhibited marked similarities with array-CGH data of human HCCs 37. This cross-species synteny implies that the carcinogenesis in these mice closely mimics that in human patients and indicates that the FEAT transgenic mouse may be a highly relevant model of human HCCs 42.
Molecular bases for oncogenic functions of FEAT
The oncogenic potential of FEAT in transgenic mice seemed to be disproportionate to the moderate ability to attenuate apoptosis (Fig. 2c, Fig. 3b, and Supplementary Fig. 4). NIH3T3 cells that overexpressed FEAT, FEATΔN, or FEATΔC did not consistently form colonies in soft agar, suggesting that the assay for anchorage-independent growth can fail to detect potently tumorigenic genes. This supported the limitation of ex vivo screening for cancer genes and the much broader relevance of genetic engineering and in vivo analyses of mice 19.
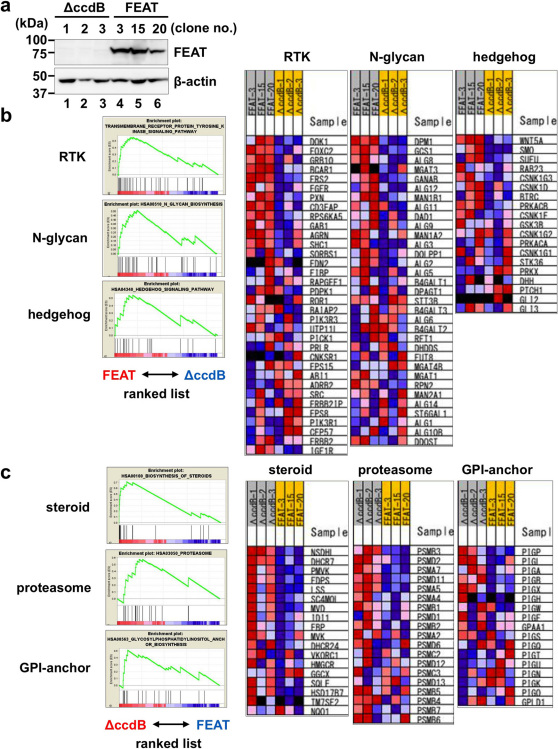
To evaluate whether FEAT has cellular functions relevant in oncogenesis, FEAT was overexpressed in NIH3T3 cells, which only weakly express FEAT protein (Fig. 6a), and the alterations in genome-wide transcriptional profiles were analyzed by microarrays. Gene set enrichment analysis (GSEA) 43 revealed that FEAT overexpression increases the signatures of receptor tyrosine kinase (RTK) and hedgehog signalling pathways (Fig. 6b and Supplementary Table 2), which are known to play major roles in the development and maintenance of cancers 44,45,46. The results suggested that the ability to drive multiple oncogenic pathways underlies the robust tumorigenic potential of FEAT in vivo.
Figure 6. Genome-wide expression profiling of NIH3T3 cells overexpressing human FEAT.
(a) Immunoblot analyses of NIH3T3 cell clones stably transfected with a control plasmid (ΔccdB-1, ΔccdB-2, and ΔccdB-3) and human FEAT cDNA (FEAT-3, FEAT-15, and FEAT-20). The blot was probed with anti-FEATΔN (upper panel) and anti-actin (lower panel) antibodies. Signatures enriched in FEAT-overexpressing (FEAT-3, -15, and -20) (b) or control (ΔccdB-1, -2, and -3) (c) NIH3T3 cells as assessed by gene set enrichment analysis. Enrichment plots (left panels) and heat maps (right panels) display gene sets with enrichment score (ES) > 0.5 and false discovery rate (FDR) q-value < 0.25. RTK, receptor tyrosine kinase. GPI, glycosylphosphatidylinositol.
FEAT is upregulated in most human cancer cells
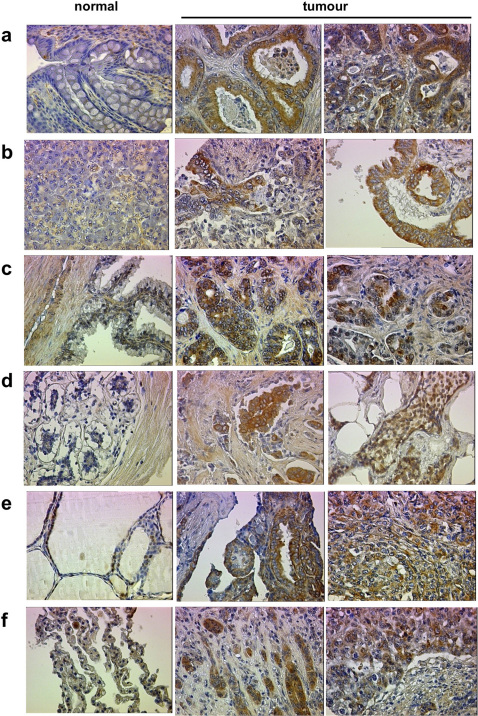
Previous studies using expression microarrays reported that FEAT (KIAA0859, CGI-01) mRNA is upregulated in uterine and ovarian cancers (http://www.freepatentsonline.com/20050244843.html), superficial bladder cancer (https://www.oncomine.org), adenocarcinomas of the stomach 47, and retinoblastoma 48. To further assess the range of human cancers in which FEAT may be involved in oncogenesis, we examined FEAT levels in various human cancers and their subtypes using Tissue Arrays. Immunohistochemistry detected significant upregulation of FEAT in colon (46%, 17/37), pancreatic (61%, 11/18), prostate (39%, 7/18), breast (51%, 18/35), ovary (24%, 6/25), thyroid (47%, 9/19), and non-small-cell lung cancers (64%, 14/22) (Fig. 7 and Supplementary Fig. 9; Supplementary Table 3). FEAT was upregulated in cancer cells but not nonneoplastic stromal cells or normal cells adjacent to cancer tissues. Thus, FEAT upregulation is common and widespread in human cancers, suggesting that this phenomenon is a fundamental process in the development of most cancers.
Figure 7. Overexpression of FEAT in diverse human cancer cells.
Immunohistochemical staining of Tissue Arrays using anti-FEAT ΔN antibody. (a) Normal colon tissue, grade I and III adenocarcinoma of the colon. (b) Normal pancreas tissue, grade I and II adenocarcinoma of the pancreas. (c) Normal prostate tissue, Gleason score 1 and 3 adenocarcinoma of the prostate. (d) Normal breast tissue, infiltrating ductal and lobular carcinomas of the breast. (e) Normal thyroid tissue, grade I papillary and grade III follicular carcinomas of the thyroid. (f) Normal lung tissue, grade III adenocarcinoma and grade III squamous cell carcinoma of the lung. Original magnification: × 400. The following Tissue Arrays were used: High density multiple organ cancer and normal tissue microarray, MC5001; Breast cancer tissue microarray with self-matching adjacent normal tissue and normal tissue controls, BR721; Colon adenocarcinoma (combination of marginal and normal), BC05021; and Liver carcinoma (combination of cancer, cancer adjacent and normal tissue), LV803.
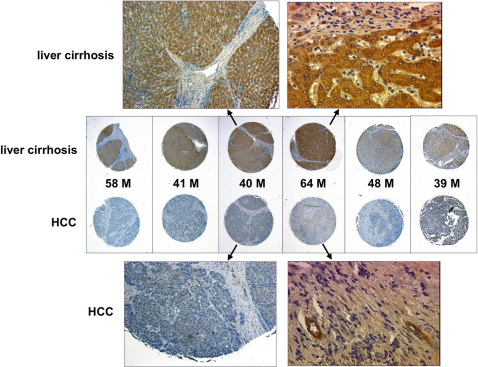
Notably, FEAT overexpression can also precede neoplastic transformation in human HCCs; hepatocytes that were adjacent to HCC in liver cirrhosis expressed high levels of FEAT (Fig. 8), most likely reflecting ongoing carcinogenesis in these cirrhotic livers. FEAT overexpression was also observed in intraductal carcinoma in situ of breast (Supplementary Fig. 9) 49, an early phase preceding invasion. These observations suggest that FEAT functions from preneoplastic or early neoplastic processes of diverse human cancers.
Figure 8. FEAT is overexpressed in liver cirrhosis adjacent to human hepatocellular carcinoma (HCC).
FEAT expression in HCC and the adjacent cirrhotic liver was compared for several patients using a Tissue Array (LV803). The age and gender (M, male) of the patients are indicated. Original magnifications are: ×115 (left uppermost panel, left lowermost panel), ×400 (right uppermost panel, right lowermost panel), and ×40 (middle panels).
Discussion
One of the most feasible and promising approaches for cancer prevention and screening is to target a common event that occurs in most tumours. Despite these potential therapeutic advantages, it is still poorly understood whether a crucial molecular event commonly occurs in the early oncogenesis of most cancers. The present study demonstrates that an unrecognized protein, FEAT, is highly oncogenic in vivo. Remarkably, this prominent promoter of tumorigenesis is aberrantly overexpressed in most human cancers starting in the early phases of tumorigenesis. Chromatin immunoprecipitation studies with mouse embryonic stem cells (ESCs) indicate that oncogene products such as c-Myc 50,51,52, N-Myc 51, and E2F1 51 are bound to the promoter of the mouse Mettl13 gene (also known as 5630401D24Rik). Amplicons in various cancers including HCC 37, malignant lymphoma, and high-risk multiple myeloma (http://www.freepatentsonline.com/y2008/0274911.html) involve the human METTL13 gene at 1q24.3. These observations might explain how FEAT protein is upregulated in tumours. Further studies are needed to fully elucidate the regulation of FEAT expression in normal and neoplastic tissues.
FEAT was originally purified from rat livers as a protein that inhibits nuclear apoptosis in vitro. Interestingly, FEAT homologues were not detectable in unicellular eukaryotes such as yeasts and green algae (Chlamydomonas reinhardtii), suggesting that the functions of FEAT are unique to multicellular organisms. Interestingly, mouse FEAT belongs to the Myc module in mouse ESCs that is responsible for the similarity between ESCs and cancer cells 53, implicating FEAT as a link between cancer and stem cell biology. Ex vivo experiments confirmed that FEAT attenuates apoptotic cell death. However, gene expression profiling revealed that FEAT also affects various cell signalling and metabolic pathways (Fig. 6b and 6c; Supplementary Table 2). Furthermore, in a recent genome-wide linkage analysis, genetic variations in the human METTL13 gene have been associated with increased susceptibility to postpartum mood syndrome 54. CpG-island microarray analyses of frontal cortex tissues have revealed higher DNA methylation close to the METTL13 gene among bipolar disorder females and psychosis females 55. Therefore, it will be important to perform integrated studies to fully elucidate how the multifunctional properties of FEAT contribute to tumorigenesis.
A potential problem with transgenic mouse models of cancer is that the transgene is expressed throughout the entire organ, while human cancers are thought to develop from a single mutated cell in the context of a relatively normal organ 32. FEAT transgenic mice did not have the premalignant lesions (ex. steatohepatitis, cholestatic hepatitis, and liver fibrosis) that typically precede HCCs in other mouse models and human patients with underlying hepatitis B or C virus infections, alcoholic liver injury, or nonalcoholic steatohepatitis 41. Thus, a possible limitation of the FEAT transgenic mouse model is that it cannot recapitulate the inflammatory mechanisms that underlie most human HCCs. Inflammatory responses leading to STAT3 signalling downstream of interleukin-6 have been implicated in the development of murine and human HCCs 33. Whereas FEAT induces malignant HCCs in mice that can metastasize to the lung, constitutive interleukin-6-STAT3 signalling can only induce benign hepatic adenomas 56, suggesting that the inflammatory mechanisms play central roles mainly at earlier phases in the evolution of HCCs. Immunohistochemical analyses of Tissue Arrays from human patients revealed that FEAT is diffusely overexpressed in hepatocytes in liver cirrhosis adjacent to HCC. FEAT upregulation in transgenic mice may bypass the preneoplastic processes that precede advanced liver cirrhosis. In compensation for the inability to recapitulate the earlier inflammatory phases of hepatocarcinogenesis, FEAT transgenic mice may help us investigate a ‘pure' later phase of carcinogenic processes that are not complicated by genetic alterations that are secondary to the preceding processes 57.
The relatively delayed development of HCC and lymphoma in transgenic mice (Supplementary Fig. 6) implies that FEAT is involved in promoting, rather than initiating oncogenesis. The genomic instability that was indicated by the array-CGH also suggests that multiple additional genetic alterations are required for tumorigenesis. The incidence of tumours in transgenic mouse lines was not correlated with differences in FEAT transgene expression among these lines (1 > 16 > 23 ≫ 18), suggesting that the level of FEAT protein is not a rate-limiting factor in tumorigenesis. In addition, Tissue Arrays showed no correlation between FEAT levels and tumour grades. Downregulation of FEAT in human HCC compared to the adjacent cirrhotic liver and the moderate ability of FEAT to attenuate apoptotic cell death in HeLa cells suggested that FEAT is not necessary to maintain cancers. Taken together, these results indicate that FEAT upregulation potently promotes the development of multiple tumours in vivo, mainly at the prodromal and early phases of carcinogenesis, and sets the stage for additional oncogenic processes. Spontaneous lymphoma, HCC, lung cancer, and rhabdomyosarcoma can occur in C57BL/6 mice, albeit at low rates. The occurrence of these specific types of tumours suggests that the FEAT transgene promoted the intrinsic tendency of C57BL/6 mice to develop tumours such as lymphoma. Therefore, it is likely that FEAT accelerates and enhances the intrinsic predispositions of certain humans to develop cancers.
Biochemical events that are common among cancers have been used to screen or monitor tumour development, spread, and viability, as exemplified by positron emission tomography (PET) for glucose uptake. Compared to alterations in other oncogenes and tumour suppressor genes 15, FEAT is upregulated in an unusually wide range of tumours. Moreover, Tissue Array analyses demonstrate that FEAT upregulation can be easily examined using formalin-fixed paraffin-embedded sections that are available in most community hospitals. MUC1 58 and survivin 59, which are overexpressed in diverse cancers, are currently being examined as targets for immunotherapy in clinical trials. Immunization with FEAT may trigger immune responses that eradicate precancerous lesions and early-stage tumours, preventing further development of invasive and metastatic cancers. Thus, FEAT might become the prototype of a subfamily of cancer genes that could lead to new methods for cost-effective cancer screening and prevention. The results of our study suggest that we should further explore FEAT-like ubiquitous oncoproteins and exploit the molecular features of these proteins in various clinical applications.
Methods
Reagents
Benzyloxycarbonyl-Val-Ala-Asp(OMe)-fluoromethylketone (zVAD-fmk) was obtained from Enzyme Systems. The pcDL-SRα-procaspase-3 plasmid was kindly provided by Dr. Fumiko Toyoshima-Morimoto (Institute for Virus Research, Kyoto University, Kyoto, Japan). The agonistic anti-Fas IgM monoclonal antibody (CH-11) was purchased from MBL. The INSTA-Blot membrane (IMB-105) was obtained from IMGENEX. Multiple Tissue Blot (Human) (WB46) and Human Tumor Tissue Blot (WB51) were obtained from Calbiochem (Merck). Tissue Arrays that were spotted with an array of formalin-fixed and paraffin-embedded specimens derived from various normal and tumour tissues were purchased from US Biomax.
Antibodies
The following primary antibodies were used: rabbit polyclonal antibodies against actin (A5060; Sigma), human c-Myc (Anti-Myc Tag, 06-549; Upstate, Millipore), albumin (A0001; DakoCytomation Denmark A/S), and prosurfactant protein C (AB3786; Chemicon International, Millipore); mouse monoclonal antibodies against human c-Myc (9E10; sc-40), HA (Y-11; sc-805), MDM2 (sc-965) (Santa Cruz Biotechnology), α-fetoprotein (AB3786; Vector Laboratories), and ß-catenin (610153; BD Transduction Laboratories, BD Biosciences).
Rabbit polyclonal antibodies against human FEATΔN and human FEATΔC were produced and affinity-purified by Operon Biotechnologies (Tokyo, Japan). Detailed characterization of the antibodies is described in Supplementary Methods.
Determination of caspase cleavage sites
The candidate aspartic acid residues encoded by FEAT cDNA in the pBluescript SK(-) plasmid (Stratagene) were mutated to alanine using the GeneEditor in vitro Site-Directed Mutagenesis System (Promega). In vitro transcription/translation of the cDNAs was performed using the TNT T7/T3 Coupled Reticulocyte Lysate System (Promega) and Tran35S-label (MP Biomedicals). The [35S]-labelled recombinant wild-type or mutant FEAT proteins were incubated with purified recombinant caspase-3 for 2 h at 37°C, resolved by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by autoradiography.
Isolation of neutrophils, protein transduction, and analysis of spontaneous apoptosis
The PTD-4 sequence (YARAAARQARA) optimized for protein transduction 60 was introduced between the His-tag and N-terminus of the wild-type or mutant FEAT proteins. The proteins encoded by the pET-16b plasmids (Novagen, Merck) were expressed in BL21(DE3)pLys Escherichia coli (Novagen, Merck) and purified with Ni2+-NTA agarose (Qiagen) under denaturing conditions.
Neutrophils were isolated from the peripheral blood of healthy adult volunteers using two-step Percoll (GE Healthcare Bio-Sciences) gradients. Neutrophils were resuspended in RPMI-1640 medium (Sigma) supplemented with 10% FBS (GIBCO, Invitrogen, Life Technologies), 50 U/ml penicillin, and 50 µg/ml streptomycin (GIBCO, Invitrogen, Life Technologies) (FBS-RPMI) and then treated with 1 µM of the purified proteins for 15 min at 37°C. After the treatment, the cells were diluted with FBS-RPMI and then incubated for 24 h at 37°C. Neutrophil apoptosis was assessed by ethanol fixation, propidium iodide staining, and analyses of the cells with hypodiploid DNA content using the FACScan flow cytometer and CellQuest software (BD Biosciences). Cell death was also confirmed by flow cytometric analyses of cells stained with fluorescein isothiocyanate-conjugated (FITC-conjugated) annexin V and propidium iodide using the ApoAlert annexin V Apoptosis Detection kit (Clontech Laboratories, Takara Bio).
Generation and breeding of FEAT transgenic mice
All animal experiments were approved by the Animal Experiment Committee at the Chiba Cancer Center. Injection of the DNA fragment containing the H-2Kd (MHC Class I) promoter, FEAT cDNA, β-globin intron, and SV40 poly (A) signal into fertilized eggs collected from superovulated C57BL/6CrSlc females, implantation into pseudo-pregnant mice, breeding, and weaning were performed by Japan SLC (Shizuoka, Japan).
The founder mice were screened for integration of human FEAT cDNA by Southern blotting using genomic DNA purified from tails. Briefly, 0.5 cm of mouse tails were incubated overnight at 55°C in 0.5 ml lysis buffer (10 mM TrisHCl, pH 7.5, 50 mM ethylenediamine tetraacetic acid (EDTA), 0.5% SDS, 100 mM NaCl, and 100 µg/ml proteinase K). After adding 0.5 ml UltraPure phenol:chloroform:isoamyl alcohol (Invitrogen, Life Technologies), the samples were rotated for 30 min at room temperature, and then centrifuged for 2 min at 7,000 rpm. The aqueous phase was recovered and treated similarly with 0.5 ml chloroform. The genomic DNA was precipitated by adding 0.4 ml isopropanol, and then washed with 70% ethanol and dissolved in 0.2 ml TE buffer (10 mM TrisHCl, pH 8.0, and 1 mM EDTA). After digesting with BamHI and capillary transfer to Nytran SuPerCharge nylon membranes using the TurboBlotter Rapid Downward Transfer Systems (Schleicher & Schuell Bioscience, Whatman, GE Healthcare Bio-Sciences), the integrated cDNA was detected using the AlkPhos Direct Labeling and Detection System (GE Healthcare Bio-Sciences).
Thereafter, expression of the transgene in each mouse was determined by polymerase chain reaction (PCR) analysis using DNA isolated from the tail by a simplified protocol. Briefly, 0.5 cm of tail was incubated overnight at 55°C in 100 µl lysis buffer (20 mM TrisHCl, pH 7.5, 100 mM EDTA, 0.5% Tween-20 (Bio-Rad), and 100 µg/ml proteinase K), followed by the addition of 900 µl distilled water. For a 20 µl PCR reaction using KOD Dash DNA polymerase (TOYOBO), 0.5 µl of the lysate was used as a template with the following primers: 5′-TGGCTCTTTGGCATGGATGA-3′ (forward) and 5′-TATGACATCGTAGCAAGGCC-3′ (reverse). Sperm were obtained from the epididymides of transgenic mice and cryopreserved by Kyudo Co., Ltd. (Tosu, Japan).
Transgenic mice and their nontransgenic littermates were bred and maintained in a specific pathogen-free (SPF) animal facility at the Chiba Cancer Center Research Institute. Transgenic lines were maintained by crossing male transgenic mice to normal female C57BL/6 mice (Charles River Laboratories). Mice that had tumours or were in a moribund state were euthanized and necropsied.
Apoptotic cell death of mouse thymocytes
Thymocytes were isolated by grinding the dissected thymus with the plunger of a 1-ml sterile syringe in FBS-RPMI at room temperature. The cells were left untreated or treated with Fas ligand (rhsSuperFasLigand) (Alexis) or dexamethasone (Nacalai Tesque) for 24 h at 37°C in a humidified atmosphere containing 5% CO2. Cell death was quantified by staining the cells with annexin V-CFS (R&D Systems) and propidium iodide (Calbiochem, Merck), and then analyzing the cells by flow cytometry.
Microscopic analyses of mouse tissues
After dissection, the mouse organs were fixed with 3.7% formaldehyde in phosphate-buffered saline, sliced into 2-mm thick sections, placed into tissue cassettes (Tissue-Tek Uni-Cassette; Sakura Finetek Japan), and immersed in fresh fixative. Further processing and interpretation of the microscopic pathology were performed by Narabyouri Research Co., Ltd. (Nara, Japan). Briefly, fixed tissues were dehydrated through a series of ethanol (20% to 100%) and xylene solutions and embedded in paraffin using an automatic tissue processor, sectioned with a microtome, deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E). Immunohistochemical staining for CD45R (B220) and CD3 was performed with an Autostainer (DAKO).
Array-CGH
The quality of genomic DNA was assessed by electrophoresis of 200 ng DNA on a 0.8% agarose gel containing 0.5 µg/ml ethidium bromide. Only DNA preparations without smearing were used for subsequent steps. DNA labelling and hybridization to 244k slide format 60-mer oligonucleotide microarrays for array-CGH (Agilent Mouse Genome CGH Microarray Kit 244A) were performed using genomic DNA from mouse HCCs as experimental samples and genomic DNA from the liver of a normal C57BL/6 mouse as a reference sample. The microarrays were scanned using a Micro Array Scanner, and data were extracted using Feature Extraction software and analyzed using CGH analytics 3.4 software according to the manufacturer's instructions (Agilent Technologies).
Gene expression profiling with microarrays
The ORF of human FEAT cDNA was subcloned into the pEF-DEST51 vector (Invitrogen, Life Technologies). NIH3T3 cells were transfected with pEF-DEST51-FEAT using HilyMax (Dojindo Laboratories). After 7 days of selection with 10 µg/ml blasticidin S (Kaken Pharmaceutical), colonies were picked and screened for clones in which FEAT protein was overexpressed. The ccdB gene in the pEF-DEST51 vector was deleted to construct the pEF-DEST51-ΔccdB plasmid, which was stably transfected into NIH3T3 cells to obtain control cell lines.
Total RNA was extracted from control (ΔccdB-1, -2, and -3) and FEAT-overexpressing NIH3T3 cell lines (FEAT-3, -15, and -20) using the RNeasy Mini kit (Qiagen). Gene expression profiling was performed with Sentrix Mouse WG-6 v2 BeadChip Array (Illumina) at the Research Support Center, Graduate School of Medical Sciences, Kyushu University. Raw gene expression data were first subjected to average normalization using BeadStudio 3.0 software. Average signals with detection P-values (based on Illumina replicate gene probes) > 0.01 were removed from analyses. GSEA was performed using the software available via the world wide web site (http://www.broadinstitute.org/gsea/).
Immunohistochemistry
Paraffin-embedded sections were deparaffinized with Clear-Advantage (Polysciences), rehydrated, treated with Citrate-based Antigen Unmasking Solution (Vector Laboratories), and stained using the ImmPRESS kit (Vector Laboratories) and ImmPACT Chromogen (Vector Laboratories) according to the manufacturer's instructions. The slides were counterstained with Mayer's Hematoxylin (Merck), and the coverslips were mounted with Gel/Mount aqueous mounting medium (Biomeda). The stained tissues were observed by the Zeiss Axioskop 2 plus microscope and images were acquired using the Zeiss AxioCam camera controlled by AxioVision software (Carl Zeiss MicroImaging). Images were also captured using the Leica MZ16 FA stereomicroscope, Leica DFC300 FX digital camera, and Leica IM500 Image Manager software (Leica Microsystems).
Author Contributions
AT designed the project. AT, DT, AN, and KTani designed experiments. AT, HT, KTakahashi, TT, KM, AI, OK, and KY performed a significant amount of the experimental work. AT, MO, and TK performed most of the data collection and data analysis. AT wrote the main manuscript text and prepared figures and tables. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank T. Honjo, I. Chung-Okazaki, A. Shirahata, T. Hashimoto, C. F. Clarke, P. Gin, W. Frommer, X. Wang, F. Toyoshima-Morimoto, and A. K. Munirajan for reagents; H. Ohno for cell lines; M. Sugimoto, K. Igarashi, and the Research Support Center, Graduate School of Medical Sciences, Kyushu University for technical support; S. H. Kaufmann, T. Marumoto, and A. Suzuki for critically reading; K. Sakurai, Y. Nakamura, N. Shibano-Kondo, A. Kitabayashi-Akao, S. Ito, and M. Okada for technical assistance; T. Moriguchi, M. Taketo, T. Hori, T. Ichinohe, K. Yamamoto, M. Sasada, T. Uchiyama, A. Uemura, A. Kotani-Yoshida, T. Ozaki, H. Kageyama, S. Haraguchi, M. Mizoe-Amako, T. Nakamura, and E. Nishida for discussions and suggestions; J. Hirai, Y. Asano-Ashida, K. Miyata, M. Ushijima, and K. Maekawa for secretarial assistance. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Academic Research Grant from Kyoto University, and a grant from the Tokyo Biochemical Research Foundation.
References
- Bode A. M. & Dong Z. Cancer prevention research - then and now. Nat. Rev. Cancer 9, 508–516 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal P. A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006). [DOI] [PubMed] [Google Scholar]
- Greenman C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007). [DOI] [PubMed] [Google Scholar]
- Jones S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Brug M. P. & Wahlestedt C. Navigating genomic maps of cancer cells. Nat. Biotechnol. 28, 241–242 (2010). [DOI] [PubMed] [Google Scholar]
- Meyerson M., Gabriel S. & Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696 (2010). [DOI] [PubMed] [Google Scholar]
- Stephens P. J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton M. R., Campbell P. J. & Futreal P. A. The cancer genome. Nature 458, 719–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- Vogelstein B. & Kinzler K. W. Cancer genes and the pathways they control. Nat. Med. 10, 789–799 (2004). [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C. & Thompson C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji A. F., Nghiem P. & Schreiber S. L. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol. Cell 12, 271–280 (2003). [DOI] [PubMed] [Google Scholar]
- Luo J., Solimini N. L. & Elledge S. J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Wagner E. F. & Palmiter R. D. The origins of oncomice: a history of the first transgenic mice genetically engineered to develop cancer. Genes Dev. 21, 2258–2270 (2007). [DOI] [PubMed] [Google Scholar]
- Santarius T., Shipley J., Brewer D., Stratton M. R. & Cooper C. S. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 10, 59–64 (2010). [DOI] [PubMed] [Google Scholar]
- Lazebnik Y. A., Cole S., Cooke C. A., Nelson W. G. & Earnshaw W. C. Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J. Cell Biol. 123, 7–22 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A. et al. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc. Natl. Acad. Sci. USA 93, 8395–8400 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. H., Chou C. Y., Ch'ang L. Y., Liu C. S. & Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 10, 703–713 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan R. M. & Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 310, 417–427 (1994). [DOI] [PubMed] [Google Scholar]
- Wirsing L., Naumann K. & Vogt T. Arabidopsis methyltransferase fingerprints by affinity-based protein profiling. Anal. Biochem. 408, 220–225 (2011). [DOI] [PubMed] [Google Scholar]
- Janicke R. U., Sprengart M. L., Wati M. R. & Porter A. G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 273, 9357–9360 (1998). [DOI] [PubMed] [Google Scholar]
- Kurokawa M. & Kornbluth S. Caspases and kinases in a death grip. Cell 138, 838–854 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze S. R., Hruska K. A. & Dowdy S. F. Protein transduction: unrestricted delivery into all cells? Trends Cell Biol. 10, 290–295 (2000). [DOI] [PubMed] [Google Scholar]
- Velculescu V. E. et al. Analysis of human transcriptomes. Nat. Genet. 23, 387–388 (1999). [DOI] [PubMed] [Google Scholar]
- Naugler W. E. et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 317, 121–124 (2007). [DOI] [PubMed] [Google Scholar]
- Anisimov V. N., Ukraintseva S. V. & Yashin A. I. Cancer in rodents: does it tell us about cancer in humans? Nat. Rev. Cancer 5, 807–819 (2005). [DOI] [PubMed] [Google Scholar]
- Frese K. K. & Tuveson D. A. Maximizing mouse cancer models. Nat. Rev. Cancer 7, 645–658 (2007). [DOI] [PubMed] [Google Scholar]
- Grivennikov S. I., Greten F. R. & Karin M. Immunity, inflammation, and cancer. Cell 140, 883–899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S. P., Schwank J., Staib F., Wang X. W. & Harris C. C. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene 26, 2166–2176 (2007). [DOI] [PubMed] [Google Scholar]
- Toledo F. & Wahl G. M. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6, 909–923 (2006). [DOI] [PubMed] [Google Scholar]
- Laurent-Puig P. & Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene 25, 3778–3786 (2006). [DOI] [PubMed] [Google Scholar]
- Zender L. et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125, 1253–1267 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E. et al. Role of the F-box protein Skp2 in lymphomagenesis. Proc. Natl. Acad. Sci. USA 98, 2515–2520 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy J. L. et al. EDD, the human orthologue of the hyperplastic discs tumour suppressor gene, is amplified and overexpressed in cancer. Oncogene 22, 5070–5081 (2003). [DOI] [PubMed] [Google Scholar]
- Graham M. & Adams J. M. Chromosome 8 breakpoint far 3′ of the c-myc oncogene in a Burkitt's lymphoma 2;8 variant translocation is equivalent to the murine pvt-1 locus. EMBO J. 5, 2845–2851 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi P. A. & DePinho R. A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6, 674–687 (2006). [DOI] [PubMed] [Google Scholar]
- Chin L. & Gray J. W. Translating insights from the cancer genome into clinical practice. Nature 452, 553–563 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwind A., Fischer O. M. & Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer 4, 361–370 (2004). [DOI] [PubMed] [Google Scholar]
- Rubin L. L. & de Sauvage F. J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 5, 1026–1033 (2006). [DOI] [PubMed] [Google Scholar]
- Varjosalo M. & Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 22, 2454–2472 (2008). [DOI] [PubMed] [Google Scholar]
- Kim B. et al. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 63, 8248–8255 (2003). [PubMed] [Google Scholar]
- Gratias S. et al. Genomic gains on chromosome 1q in retinoblastoma: consequences on gene expression and association with clinical manifestation. Int. J. Cancer 116, 555–563 (2005). [DOI] [PubMed] [Google Scholar]
- Vincent-Salomon A. et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin. Cancer Res. 14, 1956–1965 (2008). [DOI] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J. & Orkin S. H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res. 18, 1177–1189 (2008). [DOI] [PubMed] [Google Scholar]
- Kim J. et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon P. B. et al. Genome-wide linkage and follow-up association study of postpartum mood symptoms. Am. J. Psychiatry 166, 1229–1237 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J. et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 82, 696–711 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouissou S. et al. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature 457, 200–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson S. S. & Grisham J. W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 31, 339–346 (2002). [DOI] [PubMed] [Google Scholar]
- Kufe D. W. Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer 9, 874–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri D. C. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 8, 61–70 (2008). [DOI] [PubMed] [Google Scholar]
- Ho A., Schwarze S. R., Mermelstein S. J., Waksman G. & Dowdy S. F. Synthetic protein transduction domains: enhanced transduction potential in vitro and in vivo. Cancer Res. 61, 474–477 (2001). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information