Abstract
Multi-domain proteins have many advantages with respect to stability and folding inside cells. Here we attempt to understand the intricate relationship between the domain-domain interactions and the stability of domains in isolation. We provide quantitative treatment and proof for prevailing intuitive ideas on the strategies employed by nature to stabilize otherwise unstable domains. We find that domains incapable of independent stability are stabilized by favourable interactions with tethered domains in the multi-domain context. Stability of such folds to exist independently is optimized by evolution. Specific residue mutations in the sites equivalent to inter-domain interface enhance the overall solvation, thereby stabilizing these domain folds independently. A few naturally occurring variants at these sites alter communication between domains and affect stability leading to disease manifestation. Our analysis provides safe guidelines for mutagenesis which have attractive applications in obtaining stable fragments and domain constructs essential for structural studies by crystallography and NMR.
Genomes of many organisms encode large number of proteins with multiple domains1. Multi-domain proteins have an evolutionary advantage over large single domain proteins with respect to folding2. Further, multiple domains provide proteins with both structural3,4,5 (domain motion/interaction) and functional plasticity (involving new functional sites/binding/regulatory/allosteric sites). The evolution of various domain combinations in proteins is primarily governed by major recombination events (Duplication/ insertion/ deletion/ transposition)6,7,8,9. Further the evolutionary selection pressure for the newly generated domain combination is governed by functional advantage provided to the organism10. An ideal fitness function which could model the evolution of domains requires a biophysical term and a functional term10,11,12. The biophysical term serves to channel the sequence to remain in the stability zone (ΔGfolding ) for the proteins to function normally under physiological conditions12. The functional term is a combination of the overall efficiency of the reactions co-ordinated by the given sequence13,14. The domain stabilities and the strength of inter-domain interactions in a multi-domain protein have an enormous influence on the functioning of a multi-domain protein15,16,17. The inter-domain communications facilitate proper functioning of most multi-domain proteins18,19. The most common way to understand the emergent effects of domain-domain interactions on stability and folding employs the comparative analysis of energetics of the constituent domains in isolation along with the native full length form of the protein. A very common situation of obtaining single domains of multi-domain proteins with stable (native) structure for X-ray or NMR analysis is often non trivial. No clear guidelines are currently available for designing constructs coding for single domain of a multi-domain protein that is supported by a thorough understanding of origin of destability of domain structures of multi-domain proteins when considered in isolation. Till date the intricate relationship between the domain stability, inter-domain interactions and independent stable existence remains obscure. In our current study, using biophysical analysis of the domain stability and their interactions in multi-domain proteins, we aim to understand the basis of inability of certain domains to exist independently with native fold. The reasons for not observing certain sequences to exist independently in the extant proteins are manifold. 1. Innate nature of the sequence which cannot fold into a stable unit/conformation. 2. Such sequences have not been explored by evolution and hence are not observed in extant proteins. 3. Such sequences can exist stably where these proteins have evolved to interact with other proteins in an obligate fashion (Homo/hetero oligomers). 4. Incompleteness of the data: Current sequenced genomes lack the domain of interest as a single domain protein. We explore the possibility of lack of foldability and/or stability of domains of multi-domain proteins to exist independently. We further quantify the contributions of domain-domain interactions and solvent-protein interactions in order to understand how far these factors affect the stability in isolation. We also address how lack of inter-domain interactions in stable single domain homologues has contributed to differential selection pressure of non-interacting surface residues and subsequent evolution of these domain folds. Information from stable single-domain homologues of multi-domain proteins has been tailored with energetics calculations in order to arrive at guidelines for safe residue substitutions by site-directed mutagenesis to artificially stabilize a single domain of a multi-domain protein with native fold, but, in isolation.
Results
Stability of domains in single and multi-domain proteins
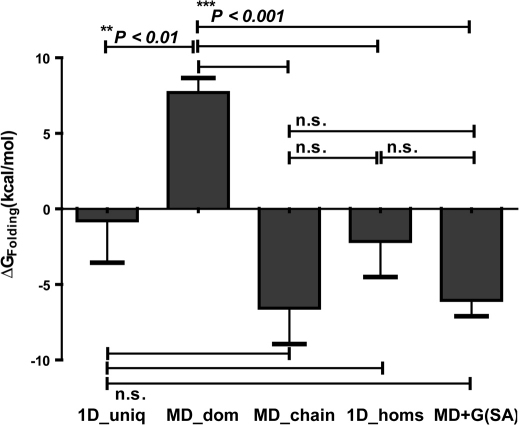
We grouped all the protein structures into four main datasets, viz; MD_doms, MD_chains, 1D_homs and 1D_uniq (See Table 1) corresponding to domains of multi-domain proteins, full-length gene products of multi-domain proteins, single domain homologues of domains in multi-domain proteins and a control, independent data set of single domain proteins respectively. We computed the stabilities (ΔGFolding) of each of the proteins/domains in these data sets using the empirical effective energy function, FoldX20 (See Methods). We found that domains of multi-domain proteins considered in isolation (MD_doms; 7.70 ± 0.96 Kcal/mol) are significantly less stable in comparison to (1) full length polypeptide chains (MD_chains; −6.57 ± 2.37 Kcal/mol) (Unpaired t-test: t = 6.603; df = 1282; P = 5.89E-11), (2) single domain homologues of domains in multi-domain proteins (1D_homs; −2.15 ± 2.34 Kcal/mol) (Unpaired t-test: two-tailed; t = 4.534; df = 1115; P = 6.41E-06) and (3) the control dataset of single-domain proteins (1D_uniq; −0. 78 ± 2.77 Kcal/mol) (Unpaired t-test: two-tailed; t = 3.237; df = 996; P = 1.25E-03) (Figure 1). On the contrary the stabilities of full length multi-domain proteins (MD_chains) is not very different from that of independently stable single domain homologues (1D_homs) (Unpaired t-test: t = 1.244; df = 687; P = 0.2139) and control set of single domain proteins (1D_uniq; −0. 78 ± 2.77 Kcal/mol) (Unpaired t-test: t = 1.309; df = 568; P = 0.1910) highlighting the probable importance of inter-domain interaction energy involved in stabilizing otherwise unstable domains (MD_doms).
Table 1. The Datasets used in the current study.
| Category | No of proteins/domains | Description |
|---|---|---|
| MD_doms | 428 | Domains of multi-domain proteins. |
| MD_chains | 856 | Full length proteins containg 2 domains each. |
| 1D_homs | 261 | Single domain homologous of MD_doms (Same SCOP Family). |
| 1D_uniq | 151 | Single domain proteins as per the present knowledge (No homologues in multi-domain proteins could be identified and belong to differnt homologous families) |
| Used as control in calculations. |
Figure 1. Free energies of folding for single and multi-domain proteins.
Mean values of stabilities (ΔGFolding) of domains and proteins in the four given classes (1D_uniq, MD_doms, MD_chains and 1D_homs; See Table 1). Each bar identifies the mean ± s.e.m. within that class. The significance was accessed between all pairs by using unpaired students t-test. MD_doms are significantly less stable in comparison to (1) MD_chains (Unpaired t-test: t = 6.603; df = 1282; P = 5.89E-11), (2) 1D_homs (Unpaired t-test: two-tailed; t = 4.534; df = 1115; P = 6.41E-06) and (3) 1D_uniq (Unpaired t-test: two-tailed; t = 3.237; df = 996; P = 1.25E-03). ΔGint,(SA) for a domain in multi-domain system and ΔGfolding of the same domain is comparable to the ΔGfolding for 1D_homs (Unpaired t-test: two-tailed; t = 1.689; df = 1115; P = 9.15E-02) and MD_chains (Unpaired t-test: two-tailed; t = 2.344; df = 1282; P = 8.15E-01).
Inter-domain interactions and effects on stability
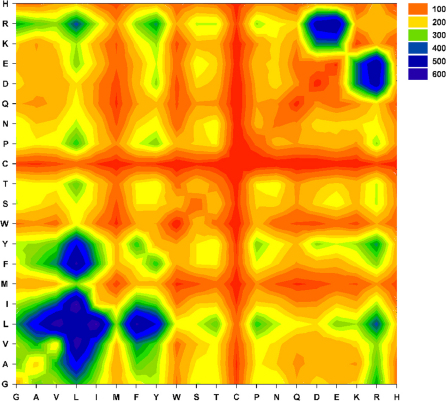
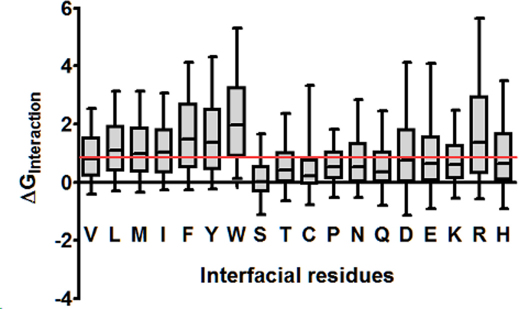
We observed that domain-domain interactions (See methods) across the interface primarily comprised of short range (van der Waal's) contacts between the hydrophobic residues (Figure 2). Most of the contacts are made by Leu, Ile, Phe, Ala and Tyr (dark blue contours in bottom left zone in Figure 2). Few interfaces are observed to be studded with salt bridges (especially Arg-Asp/Glu and Lys-Asp/Glu) (Figure 2; top right quadrant). The strengths of these interactions are proportional to the observed interaction interface surface area (Linear regression: r2 = 0.9273; P < 0.0001; ΔGInt = −0.0157*SAInterface + 5.092). in silico Alanine scanning revealed that the residues (Primarily Hydrophobic) involved in most frequent interactions across the interface are also the most energetically favoured interactions in the domain-domain interfaces (Figure 3), thereby acting as hotspots in the domain-domain interface. Figure 3 also shows that change in interaction energies due to Ala mutations is correlated with the volume/size of the interface residues in the wild type.
Figure 2. Interaction preferences in the domain-domain interface of multi-domain proteins.
Shown is the contour plot of frequencies of pair wise residue contacts between the two-interacting domains of a set of two-domain proteins (n = 428) . A distance based approach was used to determine the van der Waal's contacts between the two interacting domains (See methods). Note the preponderance of non-polar contacts and salt bridges (Bottom left quadrant and top right quadrant).
Figure 3. Effect of in silico Alanine Scanning mutagenesis of interfacial residues on the energy of association between the interacting domains.
Each of the interfacial residues of the 428 interfaces was mutated to Alanine in silico and the change in the interaction energies between the two domains was computed. All residues except Glycines and Alanines were mutated in this analysis. Most Ala-substitutions in the interface were found to be destabilizing. We find that the interface stability is majorly governed by large hydrophobic interactions (involving L, M, I, F, Y and W) and few polar contacts (involving R, D and E), as we observe the medians values of ΔGInteraction for these mutations are above the 1kcal/mol (Red line) threshold.
Solvation effects and stability
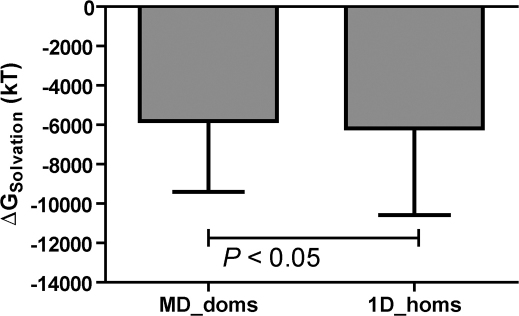
To understand precisely the contribution of solvent effects on the stabilization of domains, we computed the solvation energies explicitly (See methods) and found that the solvation energies are significantly less favourable (Paired t-test: two-tailed; t = 2.262; df = 704; P = 2.41E-02) for domains of multi-domain proteins considered in isolation (−5819 ± 135.4 kT Mean ± s.e.m.) in comparison to single domain proteins (−6177 ± 165.7 kT) (Figure 4). We find that the sum of expected interaction energy (given the precise surface area of interaction) (ΔGint,(SA)) for a domain in multi-domain system (obtained from the linear fit in Supplementary Figure S2) and ΔGfolding of the same domain (−6.047 ± 1.05 kcal/mol) is comparable to the ΔGfolding for the single domain homologues (−2.156 ± 2.34 kcal/mol) (Unpaired t-test: two-tailed; t = 1.689; df = 1115; P = 9.15E-02) and the complete multi-domain chain (Unpaired t-test: two-tailed; t = 2.344; df = 1282; P = 8.15E-01) (Figure 1).
Figure 4. Free energy of solvation of domains of multi-domain proteins and single domain homologues .
The solvation free energy computed as the difference in the reaction field energy (See methods) of the protein when the external dielectric is changed from ε = 1(air/vacuum) to ε = 80 (water) using the Delphi solvation model. The figure shows that the MD_doms have significantly higher solvation free energy in comparison to their single domain homologues (Paired t-test ; t = 2.262, df = 704, P = 0.0240).
Residue changes in single domain proteins and their implications on domain stability
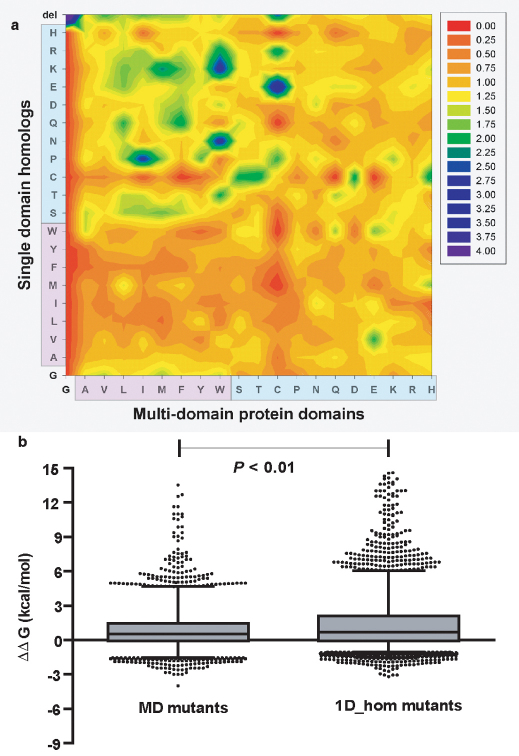
We quantified the sequence divergence among the corresponding protein pairs. We observed that there is considerable divergence (Mean sequence identity = 23.0% ±15.4) in the sequences of the domains of multi-domain proteins and their respective single domain homologues. To understand how sequence divergence of 1D_homs from the MD_doms, results in differential stability, we quantified residue changes specifically in the interface of domains of multi-domain proteins. We measured the propensities for each of the 400 possible mutation effects in these interfaces (See methods) (Figure 5a). By comparing domains of multi-domain proteins with their single domain homologues we note that most interfaces have been changed considerably with non-polar residues in the interface substituted by polar residues in the single domain homologues. We observe a clear electrostatic surface change in the domain pairs (Figure 5a: top left quadrant) for the regions corresponding to domain-domain interfaces. We further quantified the effect of these mutations by mutating in silico the non conserved interface sites of the multi-domain protein domain into the corresponding topologically equivalent residues of single domain homologues (Figure 5b). We performed swapping mutagenesis in silico by substituting residues in surface patches of the single domain homologues that are topologically equivalent to domain-domain interface of multi-domain proteins by the topologically equivalent residues in the domain-domain interface of multi-domain proteins and vice versa. We found that the swapped mutations showed no specific relationship among homologous domains (Pearson correlation: r2 = 0.02092; n = 5019; P < 0.0001); single domain homolog mutants are more destabilized (ΔΔG = 1.54 ± 3.49 Kcal/mol) than the multi-domain protein domain mutants (ΔΔG = 0.885 ± 1.76 Kcal/mol) taken out of multi-domain context (Paired t-test: two-tailed; t = 12.60; df = 5018; P = 7.34E-36) (Figure 5b).
Figure 5. Substitution preferences in single-domain homologues of multi-domain protein domains and the effect of swapping mutagenesis on the domain stabilities of multi and single domain proteins.
(a) Structure based sequence alignment between single domain-homologues and multi-domain protein domains were generated, and used to calculate the propensity of interface mutations (See methods). Observe the preferences for polar to apolar substitution frequencies in going from single-domain homologues to MD_doms. For e.g. the propensity of W to K substitutions in the interface. (b) The MD mutants represents the distribution of single point mutations (n = 5019), where each of the non-conserved interfacial residue was mutated to the corresponding residue found in the respective single domain homologue in the structure based sequence alignments. The 1D_hom mutants represents a set of single point mutations (n = 5019) on single domain proteins where the residues corresponding to the interface in the multi-domain protein homologue were mutated to those found in the MD_doms in the structure based sequence alignments. MD_mutants were significantly less destabilizing (Paired t-test; t = 12.60; df = 5018; P < 0.001) and show no relation (Spearman rank correlation test: r2 = −0.067; P = 1.50E-06) to stability effects in 1D_hom mutants.
Selection of interfacial residues and guidelines for domain stabilization
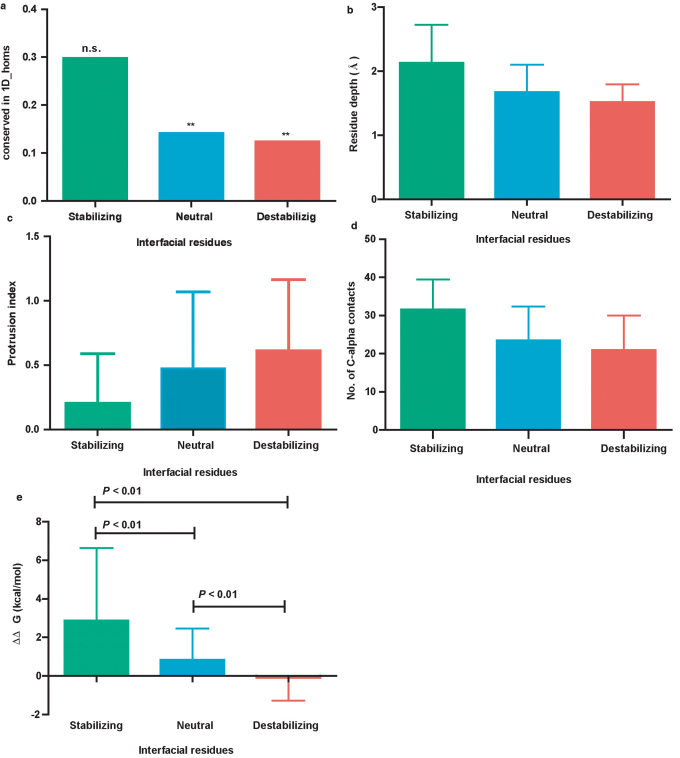
In order to identify potential key sites in the exposed interface patch of the multi-domain protein domains that are mutable to enhance the stability of the domain, we classified the interfacial residues into stabilizing; neutral and destabilizing (See methods). This classification is based on the free energy contribution of the individual residue type towards the overall domain free energy for folding in isolation (See Supplementary Fig. S3 online). Stabilizing and destabilizing residues are those with energy contribution towards the overall domain stability are respectively higher than and lower than a 95% confidence limit of the contribution of a typical surface residue to overall stability. Contribution of neutral residues is not different from that of a typical surface residue. We computed residue conservation in single domain homologues in positions which are topologically equivalent to the interface positions of domains in multi-domain proteins. We contend that the residue conservation in the single domain homologues is dependent on the class of interfacial residues (Stabilizing/Neutral/Destabilizing) which they belong and not on the frequency of observation of these three classes. We observed a bias in the proportion of residues conserved for these classes (χ2 = 80.42, df = 2, P = 3.44E-18, n = 1574; the a priori expected proportions of stabilizing, neutral and destabilizing residues are 0.25, 0.95 and 0.25 respectively) in the corresponding single domain homologues (Figure 6a). Stabilizing residues are conserved better (29.84%; n = 94) than the neutral (14.19%; n = 1452) and destabilizing residues (12.44%; n = 225) (Figure 6a). Furthermore, binomial tests between proportion of residues conserved and their expected proportion are significant only for neutral and destabilizing residues (P < 0.0001, the expected conservation frequency for the binomial tests were computed as the 23% of total residues for each class, as the mean sequence identity between the MD_doms and 1D_homs is 23%. Figure 6a). To understand how this differential conservation has resulted we probed into the mechanisms by which a surface residue contributes to the overall stability. We measured three parameters, 1. Residue depth (Å) 2. Residue protrusion index and 3. Number of Cα—Cα contacts. We find that the stabilizing residues are located more deeply (Figure 6b) (on the average the residue depth is 2.13 Å ± 0.59) than the destabilizing residue positions (average residue depth: 1.51 Å ± 0.27). This resulted in having substantial intra-protein contacts (Figure 6d). Further the protrusion of these residues into the solvent is also considerably less, rendering locally compact structures (Figure 6c). We further tested the effect of point mutations on every interfacial residue site (See methods). We substituted each residue into 19 other residue types in silico using FoldX and computed the ΔΔG of the mutations. We found that the stabilizing residues in the locations of isolated domains equivalent to domain interface when mutated, lead to more destabilization of the domain than the neutral (Mann-Whitney U-test: U = 318000; rank sum : Stabilizing = 968900, Neutral = 22860000, P < 0.0001 ) and destabilizing residue sites (Mann-Whitney U-test: U = 3217; rank sum : Stabilizing = 40250, Destabilizing = 12400; P < 0.0001) (Figure 6e). Further the destabilizing residues when mutated have a larger chance of stabilization (% of stabilizing mutations i.e. with negative ΔΔG are 9.5, 27.28 and 55.55 when stabilizing, neutral and destabilizing residues were respectively mutated).
Figure 6. Variation in conservation of interfacial residues driven by differential local environments resulting in differential stability effects upon in silico mutagenesis.
(a) Proportion of identical residues observed at topologically equivalent positions of the single domain homologous proteins and corresponding multi-domain protein interfaces. There is a bias in the proportion of residues conserved for these classes (χ2 = 80.42, df = 2, P < 0.0001, n = 1574; the expected proportions of stabilizing, neutral and destabilizing residues were 0.25, 0.95 and 0.25 respectively) in the corresponding single domain homologues. Bionomial tests between proportion of residues conserved and their expected proportion are significant only for Neutral and Destabilizing residues (P < 0.0001, the expected conservation frequency for the binomial tests were computed as the 23.00% of total residues for each class, as the mean sequence identity between the MD_doms and 1D_homs is 23%). (b) The extent of burial as measured by the depth of the interfacial residue from the nearest water accessible atom. (c) Protrusion index is a function describing the local compactness at a residue position and also an indicator of local shape (Flatness of interface). We find that the unstable residues are mostly protruding away from the centre of mass of the protein. (d) Stability contribution of residue as accounted by the number of Cα-Cα contacts within a 12Å sphere centered around the Cα of each of the interfacial residues. (e) Energetic effects of single site in silico mutagenesis of the interfacial residues of domains of multi-domain proteins. Each of the interface residue sites (Classified as Stabilizing /Neutral /Destabilizing) were mutated to 19 other residues. Stabilizing site mutants; n = 3591 (189 × 19) are significantly destabilizing than the neutral site (Mann-Whitney U-test: U = 318000; rank sum : Stabilizing = 968900, Neutral = 22860000 P < 0.0001 ) n = 127566 (6714× 19) and destabilizing sites (Mann-Whitney U-test: U = 3217; rank sum : Stabilizing = 40250, Destabilizing = 12400; P < 0.0001) mutants; n = 2565 (135× 19). This shows that domains in isolation are most often stabilized when destabilizing interface sites are mutated.
nsSNPs in the domain-domain interfaces
To understand the effect of single amino-acid substitutions in the interface, we analyzed the non-synonymous Single Nucleotide Polymorphism's (nsSNPs) in the interfaces of multi-domain proteins. We specifically studied human multi-domain proteins and identified nsSNP in the domain-domain interfaces (See Supplementary Table S1 online). We identified 33 nsSNPs in the domain-domain interface of the 14 human proteins from our dataset (See Supplementary Table S1 online) and studied their effect on the structure and stability of the interface (See methods). Most of the destabilizing changes (14/33; ΔΔG >1.0 Kcal) are observed where a wild type non-polar residue is mutated to a polar residue due to the base substitution (See Supplementary Table S1 online). This pattern has been observed in one third of the nsSNPs studied (See Supplementary Table S1 online). Also larger destabilizing effects are noted when those residue sites involved in ligand-binding (8/33 cases studied) are mutated.
Discussion
To be able to fold independently, the free energy for folding (ΔGfolding) for domains should be favourable. The FoldX20 computed free energies (See Methods and Supplementary fig. S4 online) showed that domains of multi-domain proteins in isolation (MD_doms) are significantly less stable in comparison to (1) full length polypeptide chains (MD_chains), (2) single domain homologues of domains in multi-domain proteins (1D_homs) and (3) the control dataset of single-domain proteins (1D_uniq) (Figure 1). This could be due lack of stabilization forces within the isolated domain, as we observe that the extent of stability of full length multi-domain proteins (MD_chains) is not very different from that of independently stable single domain homologues (1D_homs). This finding highlights the importance of inter-domain interaction energy involved in stabilizing otherwise unstable domains (MD_doms). The hydrophobic contacts predominate in the domain-domain interface (Figure 2). A few observed salt bridges are likely to provide specificity and shape complementarities required for stable interactions across the interface (Figure 2). As the predominant force driving the interface formation is hydrophobic, we expect a linear relationship between the interface surface area and the interaction energy (See Results). In silico Ala scanning can show the relative importance of a residue in forming an interface. The energy change greater than 1 kcal/mol upon Alanine mutation, indicates that the WT residue at that site contribute significantly to the overall interaction energy across the interface. The most interacting residues in (mostly hydrophobic and few polar) the domain-domain interface also show drastic destabilization (> 1kcal/mol; Figure 3) in terms of interaction free energies, when mutated to Alanine, thereby acting as hotspots. These hotspot residues stabilize the interaction and the full length proteins. In isolation the unstable nature of domains of multi-domain proteins (MD_doms), to exist independently (Figure 1), is believed to be due to lack of sufficient solvation energy21,22,23,24,25. The domains of multi-domain proteins as shown before (Figure 2) have a large number of hydrophobic residues at the domain-domain interface which are exposed in the absence of domain-domain interaction, resulting in poor solvation of these domains and high unfavourable ΔGfolding. Indeed it has been implicated in the hydrophobic collapse26 during the folding of multi-domain proteins. Supplementary Figure S1 shows an example of a multi-domain protein from our dataset highlighting the importance of hydrophobic character in the domain-domain interface. These extended flat hydrophobic surfaces between the domains, forces many multi-domain proteins to collapse and fold cooperatively. This process is driven by a quick desolvation of the interface during folding25. This is shown in our empirical free energy computations using FoldX (See Supplementary Fig. S1 online). We further corroborate our initial energy computations by computing explicit solvation energy (Figure 4). This clearly showed us that reduced solvation makes the isolated domains (MD_doms) unstable (Figure 1 and Figure 4). We hypothesize that multi-domain proteins compensate this loss of solvation energy by interacting favourably with their tethered domains, thus increasing the overall ΔGfolding. The interaction energy between two interfaces scales linearly with the interaction surface area (See Supplementary Fig. S2 online). In theory the interaction energy required to compensate the loss of solvation effects can be computed from this linear fit. This when added to the free energy of the isolated domain makes it favourable (Figure 1) and comparable to the free energy of folding of single domain proteins (Figure 1). This shows that the stabilization of an isolated unstable domain can be enhanced by incorporation of suitable residues to result in improved interaction energy. This also indicates that the interaction energy computed from the linear relationship with exposed surface area is approximately equal to the additional solvation energy of 1D_homs and the inter-domain interaction energy of the full-length multi-domain proteins (MD_chains). Moreover, this shows that the unstable nature of domains can be modulated by engineering interactions in the domain-domain interface patch.
The single domain homologues adopt the same fold and topology (i.e. belong to the same family in SCOP database) as the domains in multi-domain proteins, but have enhanced stability (Figure 1), in spite of high structural similarity (Average Dali Z-scores: 15.73±9.44; n = 705). We showed that there is considerable sequence divergence between the corresponding protein pairs and the single domain homologues have specific mutations at topologically equivalent sites corresponding to the domain-domain interface (Figure 5). These mutations make the surface more polar and amenable for solvation, thereby enhancing the overall free energy (Figure 4). Previous studies showed that polar-apolar residue substitutions in homologous proteins are less frequent than other patterns and are implicated in amyloidogenesis27. We have already showed that the single domain homologues have more favourable solvation free energy (∼ 350 kT difference) than the domains of multi-domain proteins (Figure 4). This result is a direct outcome of the observed residue changes in the interface (Figure 5a). The energetic effects of swapped mutations were more drastic on single domain homologues in comparison to the multi-domain protein mutants (Figure 5b). This shows that choice of residues (alterations and conservation) at interface sites is governed by stability contributions. Stabilizing mutations (non-polar to polar) are preferred at interface sites of domains of multi-domain proteins to aid in their independent existence (Figure 5b and Figure 6a). The single domain homologues are stable and belong to the same SCOP family as the isolated unstable multi-domain protein domain. These residue changes primarily modulate the surface electrostatics of the domains (Figure 5a) resulting in favourable solvation free energy (Figure 4). This pattern of specific residue changes observed in this analysis can aid in successful design of fragments and modulating domain stability in isolation. We propose a classification scheme to support this idea (See Supplementary Fig. S2 online). It is known from previous studies28,29 that surface residues are less conserved in proteins and conservation of non functional residues is generally attributed to constraints in the structural fold30. Stability contributes significantly to the overall fitness function, and hence we would expect those sites with residue contribution to the overall stability low to be less conserved in homologous proteins9,31. The observed substitution frequencies and hence the nature of conservation at a given site would be proportional to the energetic effects of the residues' contribution towards the overall stability32,33. For a given domain-domain interfacial residue, we expect higher conservation of stabilizing residues in comparison to neutral and destabilizing residues. Here we showed that the energy based classification of interfacial residues clearly segregates the residues into those under differential selection pressure (Figure 6). The differential selection pressure is the direct outcome of the differential contribution towards stability. This resulted in different extent of conservation of the three classes. Further, effects on stability of the domains show a gradation when these three classes are mutated independently. The loss of free energy upon mutation thus is site dependent (specific class) and is more drastic for the stabilizing residues. This clearly shows that destabilization of a structure caused by a residue is not tolerated as it is mutated almost always. We also observed a trend in the residue depth, number of Cα—Cα contacts and the protrusion of a residue to be dependent on the three classes. Stabilisation is achieved by deeper residues having more contacts and less protrusion. This classification is thus shown to have a mechanistic basis. Using in silico mutagenesis we probed the plasticity of these sites to accommodate different residues and modulate stability. Most point mutations on proteins in such studies are destabilizing34. We propose that the site-specific nature of the classified residues determines the extent of stabilization/destabilization upon mutation. We found striking difference in the proportion of mutants gaining stability when destabilizing residues positions were mutated to the other 19 residues. Thus there are more chances of enhancing domain stability by selectively mutating destabilizing interfacial residues to naturally occurring substitutions found in their equivalent sites of single domain homologues.
Natural variations/mutations at the domain-domain interfaces can be studied by analysing the nsSNPs. Small variations in the amino acids at the domain-domain interface can have large functional consequences35,36. Most monogenic and several polygenic diseases have been known to be caused by nsSNPs37,38,39. The effect of nsSNP can be deleterious or neutral depending on the extent to which the variant amino acid affects the protein structure39,40 and function41,42. The severity of the nsSNP had been shown to be directly correlated with its effect on protein structure and stability43,44. Few of the observed nsSNPs (9/33) in the domain-domain interface have been studied by computational and experimental methods (See Supplementary Table S1 online)45,46,47,48. We believe that deleterious nsSNPs in the domain-domain interfaces act by breaking/altering the communication between the two interacting domains, required for proper folding/stability, thus leading to non-functional proteins followed by disease manifestation.
In conclusion, often, a domain of a multi-domain protein interacts with other domains in the same protein primarily by hydrophobic residues in the inter-domain interface, thereby stabilizing an otherwise unstable domain. Independently existing stable single domain homologues of these domains undergo selective non-polar to polar mutations which modulate the solvation and hence stabilize the fold. Here we also showed that the sub-set of residues in the inter-domain interface (self domain destabilizing residues) of multi-domain proteins can be selectively mutated to aid in the formation of stable single domain constructs with the ability to fold, which otherwise might form inclusion bodies inside cells. This is of potential importance to crystallography and NMR where routinely protein domains of large multi-domain proteins are generated to solve the domain structure in isolation. The present study suggests strategies for domain stabilization in such cases. Naturally occurring variants (nsSNPs) in the inter-domain interface often contribute to disease states by altering the extent and nature of interactions between the tethered domains.
Methods
Dataset generation: wild-type and mutant protein/domains
We obtained a set of single and two-domain monomeric proteins from PDB. Monomeric proteins have been identified by consulting PiQSi49 and PISA50. We used domain boundaries as proposed by SCOP database51. These structures were filtered using resolution cut-off of 2.5 Å and sequence identity cut-off of 40%. The SCOP families corresponding to multi-domain proteins have been used to identify single domain homologues. A control dataset of non-redundant single domain proteins from completely different families has also been formed. The summary of the datasets used in the current analysis is provided in Table 1. The lists of PDB and SCOP accession codes are provided in the Supplementary information (Datasets). All the structures have been optimized at 298K by using the optimize tool available in FoldX (version 3.0 beta)20 before free energy computations. The in silico modelling of mutations have been performed using mutate module of FoldX. Every interfacial residue has been mutated to 19 other residue types and the change in the free energy for folding between the mutant and wild-type (ΔΔGfolding) has been computed. This was then used to study the stabilizing and destabilizing mutations in the interface. In silico Alanine scanning mutagenesis has been performed for interfacial residues and the changes in interaction energy (ΔGInteraction = GInt,Ala − GInt,WT) between the two interacting domains have been computed.
Energy computation and stabilities of wild type & mutant domains/proteins
We used an empirical effective energy function FoldX52 to compute the ΔGFolding for all the proteins. The FoldX energy function's stability module computes ΔGFolding and also provides a residue level contribution to the overall ΔGFolding . The FoldX energy function was originally fitted for the ΔΔG computations for mutations52 to be used as a modelling tool. In order to be able to compare ΔGFolding values of large datasets of proteins, we benchmarked FoldX calculations using experimental information. The free energies for folding computed using FoldX have been compared to experimentally determined free energies obtained from Protherm database53 for a set of test proteins (n = 35) (See Supplementary Information: Datasets). We filtered the Protherm database to obtain this test set of proteins, with known 3D-structure, which unfolded reversibly in a 2-state manner in the presence of chaotropic agents at 298K. Further, the FoldX calculations have been done at the same temperature and pH as the unfolding experimental conditions. Significant correlation (Spearman rank correlation: r = 0.5679; P = 4.00E-04) has been noted between the FoldX computed and experimentally determined free energy changes (See Supplementary Fig. S4 online). This encouraged us to use FoldX to compute free energy for folding for all the four datasets at 298K and pH 7.0 and also compare the distributions of ΔGFolding. Electrostatic free energy and consequently the solvation free energy for both multi-domain protein domains as well as their single-domain homologues have been computed independently (Figure 4). Solvation energy was computed by using the solution of non-linear Poisson–Boltzmann equation implemented in program Delphi54,55, as the difference between total grid energies in water and vacuum. The internal dielectric (εint) of 4 has been used for the protein's interior and external dielectric (εext) of 1 and 80 has been used in vacuum and solvent computations respectively.
Domain-domain interaction preferences & interaction hotspots at interfaces
Inter-domain interfaces have been recognized using the FoldX complex analysis module. To quantify interactions across the domain-domain interface, a distance based approach has been used. Residues across the interface with distances below the cut-off have been considered as interacting. The cut-off distance is computed as the sum of van der Waal's radii of interacting atoms plus 0.5Å56. The van der Waal's radii for the protein atoms were taken from Chothia (1975)57. Using this information a pair-wise interaction matrix has been computed to identify most frequent interacting pairs in the interface (Figure 2). In order to quantify the importance of these pair-wise interactions, we also computed the interaction energy between the two interacting domains for all the proteins. In silico Alanine scanning mutagenesis at all non-Ala and non-Gly interface sites has been performed to compute the strength of interaction hot spots. The interaction hot spots have been identified by an increased interaction energy ( >1 Kcal/mol destabilization) between two interacting domains on mutation into an Alanine (Figure 3).
Classification of interfacial residues
Inter-domain interfacial residues have been classified into stabilizing neutral and destabilizing. This classification is based on energy contribution towards the overall stability (ΔGfolding) of the domain taken in isolation for the free energy computations. All the interfacial residues energy contributions have been assessed using FoldX stability module. We used a residue-dependent cut-offs for classifying the interface residues into the above three classes. These cut-offs were obtained for all the twenty residue types using the control dataset (1D_uniq). The 1D_uniq (Table 1) was used to compute the free energy contribution of all the surface residues. These were then segregated into the 20 residue types. The energy values obtained have been tested for normality and fitted to normal distributions to compute mean (µ) and standard deviation (σ). This has been performed for all the 20 amino-acid types. The lower (µ − 1.98σ) and upper (µ + 1.98σ) limits of the 95% confidence interval of these distributions have been used as lower and upper cut-offs for the classification. This enabled us to overcome the bias in the FoldX computed energy-values for certain amino acids. If the interface residue energy is less than µ − 1.98σ, or more than µ + 1.98σ, then it is classified as a stabilizing residue and destabilizing residue respectively. All the residues with intermediate energy contribution are classified as neutral (See Supplementary Fig. S3 online).
Residue conservation and changes at interface
We quantified the preferred mutations occurring in the interface region by measuring the propensities of all 400 possible mutations. The structures of domains of multi-domain proteins and their respective single-domain homologues have been aligned using DALI58. The structure based sequence alignments thus generated have been used to compute the observed changes in the interface. The propensity of a particular mutation to occur in the interface of a single domain protein (Pi→j, where, i and j are topologically equivalent sites in multi-domain protein domain and single domain homologues respectively) has been measured as the ratio between the frequency of observed mutation in the interface (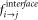 ) and the frequency of same mutation in the data set over the entire sequence (
) and the frequency of same mutation in the data set over the entire sequence (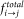 ).
).
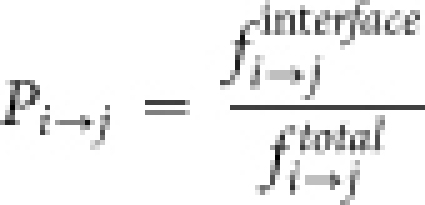 |
Pi→j > 1 indicates most preferred substitutions (Figure 5a) in the single domain homologues.
Residue micro-environment and stability
The residue depth; DPX index (defined as the distance of a non-hydrogen buried atom from its closest solvent accessible protein neighbor), residue protrusion; CX value (defined as the ratio between the external volume and the volume occupied by the protein within a sphere of 10 Å centered at every interfacial atom. This has been averaged over all the atoms for a given interfacial residue) and number of Cα—Cα contacts within a 12 Å sphere for every interfacial residue has been computed and used to quantify the local environment of any given residue. The DPX and CX indices59 have been computed using web-servers developed at ICGEBnet Protein tools.
Analysis of natural variations in the domain-domain interfaces
In-order to understand the stability effects of natural variations in domain-domain interfaces; we have analyzed non-synonymous single nucleotide polymorphisms (nsSNPs) in the human proteins (Supplementary information Table S1). We identified the nsSNPs in these proteins from the mapping database (SNPdb to PDB) LS-SNP/PDB60. We identified 33 interfacial nsSNPs in 14 human proteins. We studied the effect of nsSNP by modeling the structure of variant proteins and computing the ΔΔGfolding using the FoldX build module. We also obtained other parameters such as ligand binding, solvent accessibility and conservation of the nsSNP sites from LSSNP/PDB60.
Author Contributions
RMB and NS wrote the main manuscript text and RMB prepared figures 1–6. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr. Madan Babu of LMB, Cambridge, UK for suggestions and encouragement. We thank Dr. O Krishnadev and Ms Smita Mohanty for discussions and suggestions. R.M.B is supported by a fellowship from Council of Scientific and Industrial Research, New Delhi. This research is supported by the Department of Biotechnology, New Delhi and also by the Mathematical Biology program sponsored by Department of Science and Technology, New Delhi.
References
- Ekman D., Bjorklund A. K., Frey-Skott J. & Elofsson A. Multi-domain proteins in the three kingdoms of life: orphan domains and other unassigned regions. J Mol Biol 348, 231–43 (2005). [DOI] [PubMed] [Google Scholar]
- Han J. H., Batey S., Nickson A. A., Teichmann S. A. & Clarke J. The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol 8, 319–30 (2007). [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kuroda Y. & Yokoyama S. Characteristics and prediction of domain linker sequences in multi-domain proteins. J Struct Funct Genomics 4, 79–85 (2003). [DOI] [PubMed] [Google Scholar]
- Munro A. W., Lindsay J. G., Coggins J. R., Kelly S. M. & Price N. C. Analysis of the structural stability of the multidomain enzyme flavocytochrome P-450 BM3. Biochim Biophys Acta 1296, 127–37 (1996). [DOI] [PubMed] [Google Scholar]
- Pang A., Arinaminpathy Y., Sansom M. S. & Biggin P. C. Interdomain dynamics and ligand binding: molecular dynamics simulations of glutamine binding protein. FEBS Lett 550, 168–74 (2003). [DOI] [PubMed] [Google Scholar]
- Apic G., Huber W. & Teichmann S. A. Multi-domain protein families and domain pairs: comparison with known structures and a random model of domain recombination. J Struct Funct Genomics 4, 67–78 (2003). [DOI] [PubMed] [Google Scholar]
- Fong J. H., Geer L. Y., Panchenko A. R. & Bryant S. H. Modeling the evolution of protein domain architectures using maximum parsimony. J Mol Biol 366, 307–15 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. 3rd, Beaussart F. & Bornberg-Bauer E. Domain deletions and substitutions in the modular protein evolution. Febs J 273, 2037–47 (2006). [DOI] [PubMed] [Google Scholar]
- Xia Y. & Levitt M. Roles of mutation and recombination in the evolution of protein thermodynamics. Proc Natl Acad Sci U S A 99, 10382–7 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. & Gough J. Genomic and structural aspects of protein evolution. Biochem J 419, 15–28 (2009). [DOI] [PubMed] [Google Scholar]
- Kondrashov A. S., Sunyaev S. & Kondrashov F. A. Dobzhansky-Muller incompatibilities in protein evolution. Proc Natl Acad Sci U S A 99, 14878–83 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Weinreich D. M. & Hartl D. L. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet 6, 678–87 (2005). [DOI] [PubMed] [Google Scholar]
- Riechmann L. & Winter G. Early protein evolution: building domains from ligand-binding polypeptide segments. J Mol Biol 363, 460–8 (2006). [DOI] [PubMed] [Google Scholar]
- Gong S. et al. Structural and functional restraints in the evolution of protein families and superfamilies. Biochem Soc Trans 37, 727–33 (2009). [DOI] [PubMed] [Google Scholar]
- He H. W., Feng S., Pang M., Zhou H. M. & Yan Y. B. Role of the linker between the N- and C-terminal domains in the stability and folding of rabbit muscle creatine kinase. Int J Biochem Cell Biol 39, 1816–27 (2007). [DOI] [PubMed] [Google Scholar]
- He H. W., Zhang J., Zhou H. M. & Yan Y. B. Conformational change in the C-terminal domain is responsible for the initiation of creatine kinase thermal aggregation. Biophys J 89, 2650–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker A., Huthmacher C., Ramirez F., Lengauer T. & Albrecht M. Functional evaluation of domain-domain interactions and human protein interaction networks. Bioinformatics 23, 859–65 (2007). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. The extra C-terminal tail is involved in the conformation, stability changes and the N/C-domain interactions of the calmodulin-like protein from pearl oyster Pinctada fucata. Biochim Biophys Acta 1784, 1514–23 (2008). [DOI] [PubMed] [Google Scholar]
- Yesylevskyy S. O., Kharkyanen V. N. & Demchenko A. P. Dynamic protein domains: identification, interdependence, and stability. Biophys J 91, 670–85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J. et al. The FoldX web server: an online force field. Nucleic Acids Res 33, W382–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P. & Goodarzi G. Folding of the multidomain human immunodeficiency virus type-I integrase. Protein Sci 3, 888–97 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanicolas J. & Kuhlman B. Computational design of affinity and specificity at protein-protein interfaces. Curr Opin Struct Biol 19, 458–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. Desolvation shell of hydrogen bonds in folded proteins, protein complexes and folding pathways. FEBS Lett 527, 166–70 (2002). [DOI] [PubMed] [Google Scholar]
- Liu Z. & Chan H. S. Solvation and desolvation effects in protein folding: native flexibility, kinetic cooperativity and enthalpic barriers under isostability conditions. Phys Biol 2, S75–85 (2005). [DOI] [PubMed] [Google Scholar]
- Camacho C. J., Kimura S. R., DeLisi C. & Vajda S. Kinetics of desolvation-mediated protein-protein binding. Biophys J 78, 1094–105 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Huang X., Margulis C. J. & Berne B. J. Hydrophobic collapse in multidomain protein folding. Science 305, 1605–9 (2004). [DOI] [PubMed] [Google Scholar]
- Broome B. M. & Hecht M. H. Nature disfavors sequences of alternating polar and non-polar amino acids: implications for amyloidogenesis. J Mol Biol 296, 961–8 (2000). [DOI] [PubMed] [Google Scholar]
- Shanahan H. P. & Thornton J. M. An examination of the conservation of surface patch polarity for proteins. Bioinformatics 20, 2197–204 (2004). [DOI] [PubMed] [Google Scholar]
- Caffrey D. R., Somaroo S., Hughes J. D., Mintseris J. & Huang E. S. Are protein-protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci 13, 190–202 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G. C. & Stadler P. F. Solvent exposure imparts similar selective pressures across a range of yeast proteins. Mol Biol Evol 26, 1155–61 (2009). [DOI] [PubMed] [Google Scholar]
- Tokuriki N. & Tawfik D. S. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19, 596–604 (2009). [DOI] [PubMed] [Google Scholar]
- Kragelund B. B. et al. Conserved residues and their role in the structure, function, and stability of acyl-coenzyme A binding protein. Biochemistry 38, 2386–94 (1999). [DOI] [PubMed] [Google Scholar]
- Hamill S. J., Cota E., Chothia C. & Clarke J. Conservation of folding and stability within a protein family: the tyrosine corner as an evolutionary cul-de-sac. J Mol Biol 295, 641–9 (2000). [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Stricher F., Schymkowitz J., Serrano L. & Tawfik D. S. The stability effects of protein mutations appear to be universally distributed. J Mol Biol 369, 1318–32 (2007). [DOI] [PubMed] [Google Scholar]
- Bauer-Mehren A., Furlong L. I., Rautschka M. & Sanz F. From SNPs to pathways: integration of functional effect of sequence variations on models of cell signalling pathways. BMC Bioinformatics 10 Suppl 8, S6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Michonova-Alexova E. & Alexov E. Approaches and resources for prediction of the effects of non-synonymous single nucleotide polymorphism on protein function and interactions. Curr Pharm Biotechnol 9, 123–33 (2008). [DOI] [PubMed] [Google Scholar]
- Stitziel N. O., Binkowski T. A., Tseng Y. Y., Kasif S. & Liang J. topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res 32, D520–2 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R. & Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22, 2729–34 (2006). [DOI] [PubMed] [Google Scholar]
- Dobson R. J., Munroe P. B., Caulfield M. J. & Saqi M. A. Predicting deleterious nsSNPs: an analysis of sequence and structural attributes. BMC Bioinformatics 7, 217 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. F. et al. Genome bioinformatic analysis of nonsynonymous SNPs. BMC Bioinformatics 8, 301 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lopez J. et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis Cartilage 17, 321–7 (2009). [DOI] [PubMed] [Google Scholar]
- Di Giusto D. A. et al. Plasminogen activator inhibitor-2 is highly tolerant to P8 residue substitution–implications for serpin mechanistic model and prediction of nsSNP activities. J Mol Biol 353, 1069–80 (2005). [DOI] [PubMed] [Google Scholar]
- Rajasekaran R. et al. Effect of deleterious nsSNP on the HER2 receptor based on stability and binding affinity with herceptin: a computational approach. C R Biol 331, 409–17 (2008). [DOI] [PubMed] [Google Scholar]
- Wohlrab H. The human mitochondrial transport/carrier protein family. Nonsynonymous single nucleotide polymorphisms (nsSNPs) and mutations that lead to human diseases. Biochim Biophys Acta 1757, 1263–70 (2006). [DOI] [PubMed] [Google Scholar]
- Hentati A. et al. Human alpha-tocopherol transfer protein: gene structure and mutations in familial vitamin E deficiency. Ann Neurol 39, 295–300 (1996). [DOI] [PubMed] [Google Scholar]
- Gough C. A., Gojobori T. & Imanishi T. Cancer-related mutations in BRCA1-BRCT cause long-range structural changes in protein-protein binding sites: a molecular dynamics study. Proteins 66, 69–86 (2007). [DOI] [PubMed] [Google Scholar]
- Ory P. A., Clark M. R., Kwoh E. E., Clarkson S. B. & Goldstein I. M. Sequences of complementary DNAs that encode the NA1 and NA2 forms of Fc receptor III on human neutrophils. J Clin Invest 84, 1688–91 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali-Osman F., Akande O., Antoun G., Mao J. X. & Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem 272, 10004–12 (1997). [DOI] [PubMed] [Google Scholar]
- Levy E. D. PiQSi: protein quaternary structure investigation. Structure 15, 1364–7 (2007). [DOI] [PubMed] [Google Scholar]
- Krissinel E. & Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–97 (2007). [DOI] [PubMed] [Google Scholar]
- Murzin A. G., Brenner S. E., Hubbard T. & Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247, 536–40 (1995). [DOI] [PubMed] [Google Scholar]
- Guerois R., Nielsen J. E. & Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320, 369–87 (2002). [DOI] [PubMed] [Google Scholar]
- Gromiha M. M. et al. ProTherm, Thermodynamic Database for Proteins and Mutants: developments in version 3.0. Nucleic Acids Res 30, 301–2 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F. & Zhou H. X. Electrostatic contribution to the binding stability of protein-protein complexes. Proteins 65, 87–102 (2006). [DOI] [PubMed] [Google Scholar]
- Oron A., Wolfson H., Gunasekaran K. & Nussinov R. Using DelPhi to compute electrostatic potentials and assess their contribution to interactions. Curr Protoc Bioinformatics Chapter 8, Unit 8 4 (2003). [DOI] [PubMed]
- Keskin O., Tsai C. J., Wolfson H. & Nussinov R. A new, structurally nonredundant, diverse data set of protein-protein interfaces and its implications. Protein Sci 13, 1043–55 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Structural invariants in protein folding. Nature 254, 304–8 (1975). [DOI] [PubMed] [Google Scholar]
- Holm L., Kaariainen S., Wilton C. & Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics Chapter 5, Unit 5 5 (2006). [DOI] [PubMed]
- Vlahovicek K., Pintar A., Parthasarathi L., Carugo O. & Pongor S. CX, DPX and PRIDE: WWW servers for the analysis and comparison of protein 3D structures. Nucleic Acids Res 33, W252–4 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M., Diekhans M., Lien S., Liu Y. & Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics 25, 1431–2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information