Abstract
Heliothis zea nudivirus-1 (HzNV-1) is an insect virus previously known as Hz-1 baculovirus. One of its major early genes, hhi1, is responsible for the establishment of productive viral infection; another gene, pag1, which expresses a non-coding RNA, is the only viral transcript detectable during viral latency. Here we showed that this non-coding RNA was further processed into at least two distinct miRNAs, which targeted and degraded hhi1 transcript. This is a result strikingly similar to a recent report that herpes simplex virus produces tightly-regulated latent specific miRNAs to silence its own key early transcripts. Nevertheless, proof for the establishment of viral latency by miRNA is still lacking. We further showed that HzNV-1 latency could be directly induced by pag1-derived miRNAs in cells infected with a pag1-deleted, latency-deficient virus. This result suggests the existence of a novel mechanism, where miRNAs can be functional for the establishment of viral latency.
Heliothis zea nudivirus 1 (HzNV-1) is a baculiform insect virus with a circular double-stranded DNA genome. This viral genome has been sequenced and was found to encode approximately 154 open reading frames1,2,3,4,5. This virus was originally regarded as a member of the Baculoviridae family, but due to its lack of an occlusion body and low sequence homology to baculoviruses6, it is now temporarily re-classified with other non-occluded viruses as a new Nudivirus genus7,8. The HzNV-1 virus has a relatively broad host range and has been reported to infect many insect cell lines1,9,10,11.
HzNV-1 virus can establish distinct latent and productive infection cycles in insect cell cultures3,12,13,14. It thus provides a convenient model system for the study of molecular switches between these two viral infection cycles. During productive infection, the virus generates more than 100 transcripts, and high titers of virus progeny are produced. In this infection stage, the majority of Tn-368 and Sf-21 insect host cells are killed leaving a small percentage of the cells become latently infected3,13. During viral latency, the persistency-associated gene 1 (pag1), is the only viral gene transcript detected4. In this infection stage, virus genomes exist either as episomes or are inserted into the host genome, and persist through many cell passages without the releasing of viral particles14. Occasionally, viruses can be released from very small proportions of latently-infected cells (usually less than 0.2%), resulting in the death of these cells. This small quantity of continuously-released virions results in the presence of low viral titers (around 103 pfu/ml) in the culture medium of the latently-infected cells4,14.
Although pag1 is the only transcript detectable during latent viral infection, it is also expressed during productive viral infection. Uniquely, the transcript of pag1 is a non-coding RNA, previously also referred to as the persistency-associated transcript (PAT1)3,4, and was found to be involved in the establishment of latent HzNV-1 virus infection3,4. The pag1 has several interesting features. Sequence analysis of pag1 revealed abundant direct and inverted repeats. The transcript of pag1 is not translated, as it does not associate with polysome4,5, and was later shown to be a nuclear RNA4,5. Although we can not rule out the possibility that pag1 transcript is an intron of a larger transcript, the fact that the direct upstream sequence of pag1 coding region is a promoter suggesting that this is more likely to be an independent transcript3,5.
During productive viral infection, an abundant 6.2 kb transcript is expressed from a gene located in the fragment of HzNV-1 Hind III-I (hhi1) of the viral genome3,4,15,16. hhi1 is a viral gene expressed very early during viral productive infection (0.5 h post viral infection, hpi), and it was shown to involve in viral re-activation from latency3,16. It was also shown that the suppression of hhi1 expression can switch viral infection from productive to latent infection3,16. In contrast with the expression of most immediate-early genes of baculovirus, the proper expression of hhi1 requires viral factors15.
Recently the herpes simplex virus (HSV-1) was shown to use the miRNAs derived from a latent transcript to block the function of early genes, without the proper function of early transcripts, it is speculated that virus may enter latency17, however, a proof is still lacking. In this study, we found that HzNV-1 can serve as a convenient system to investigate such possibility. We provided evidences showing that the pag1 is capable of down-regulating hhi1 transcript in this virus. Subsequent experiments showed that pag1 functions through the expression of two microRNAs (miRNAs), which target to and degrade hhi1 transcript. These miRNAs could be detected in viral infected cells and our data indicated that the miRNA derived from pag1 can establish latent viral infection in the cells.
The establishment of latent viral infection through miRNA-producing non-coding viral RNA represents a simple yet highly effective way for HzNV-1 to achieve latent viral infection in the host cells. Although different from HzNV-1, a further prove for the miRNA to establish viral latency in HSV-1 may be still needed, the similarity between HSV and HzNV-1 in using miRNA derived from latent-specific transcripts to control early transcript, and subsequently result viral latency represents an interesting viral convergent evolution. Furthermore, miRNA as a switch for the transition of productive to latent viral infections may be a widely-spreaded phenomenon of virus/host interactions in nature.
Results
hhi1 expression is negatively regulated by pag1
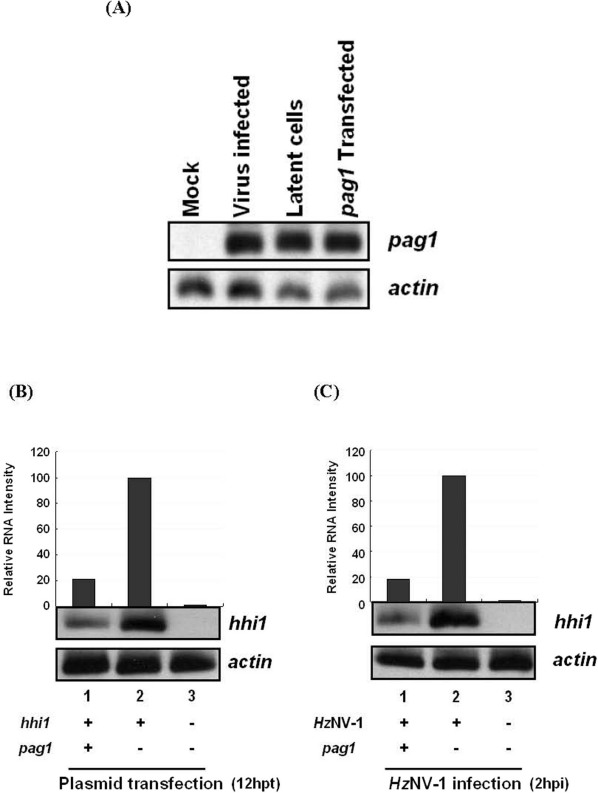
In order to study the interactions of pag1 and hhi1, two expression plasmids, pKSpP1 and pKShHI, were constructed. The former used pag1 and the latter used hsp 70 promoters to express pag1 and hhi1 transcripts, respectively. Northern analysis showed that pag1 was not only expressed in the productive and latent viral infected cells, it was also properly expressed in pKSpP1 transfected cells (Fig. 1A). Both plasmids were then co-transfected into SF-21 cells to identify any possible interactions. The hhi1-expressing vector pKShH1 was also transfected into SF-21 cells with an empty vector (pBluescript II KS−, Stratagene) as a control. Northern blot analysis showed that the level of hhi1 expression decreased dramatically in the presence of pag1 (Fig. 1B). To confirm that pag1 can negatively down-regulate hhi1 expression during HzNV-1 infection, cells were first transfected with pag1-expression vector pKSpP1 and the transfected cells were subsequently infected with HzNV-1. Similarly, hhi1 expression from viral infection was suppressed by the transfection of pKSpP1 (Fig. 1C). In this experiment, the level of hhi1 transcript was detected at 2 hours post HzNV-1 infection (hpi), because, according to our previous study, this is the timing at which hhi1 expression reaches to the highest level16.
Figure 1. Interactions of hhi1 and pag1 by northern analysis.
(A) The expression of pag1 transcripts in the productive infected, latent infected, and pKSpP1 transfected cells were probed. (B, C) pKShHI was either co-transfected with pKSpP1 (B), or co-infected with HzNV-1 (C), then subjected to northern analysis using hhi1 probe. Actin signal were used as a loading control.
Prediction of possible pag1 miRNA precursors
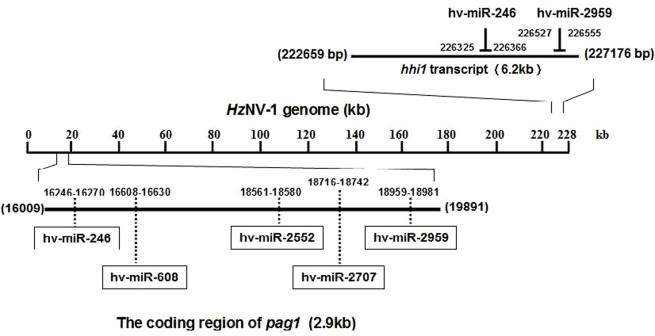
Previously, sequence analysis of pag1 revealed the presence of abundant direct and inverted repeats with no significant open reading frames (ORFs) in its coding region4,5. In this experiment, our analysis showed that pag1 transcript contains abundant stem-loop structures. Therefore, it is possible that pag1 transcript may produce miRNAs to suppress the expression of hhi1. In order to explore this possibility, 23 possible miRNAs were first predicted from pag1 coding region and five of them were predicted to target to hhi1 coding region (Fig. 2).
Figure 2. Mapping of the predicted miRNAs in pag1 transcript.
Schematic drawing of the pag1 transcript to show its 5 predicted miRNAs against hhi1, and 2 miRNAs, hv-miR-246 and hv-miR-2959, proved to be expressed by virus in the infected Sf21 cells.
Proper expression of miRNAs from pag1 is confirmed in productively and latently infected cells, and also in cells transfected with pag1 plasmid
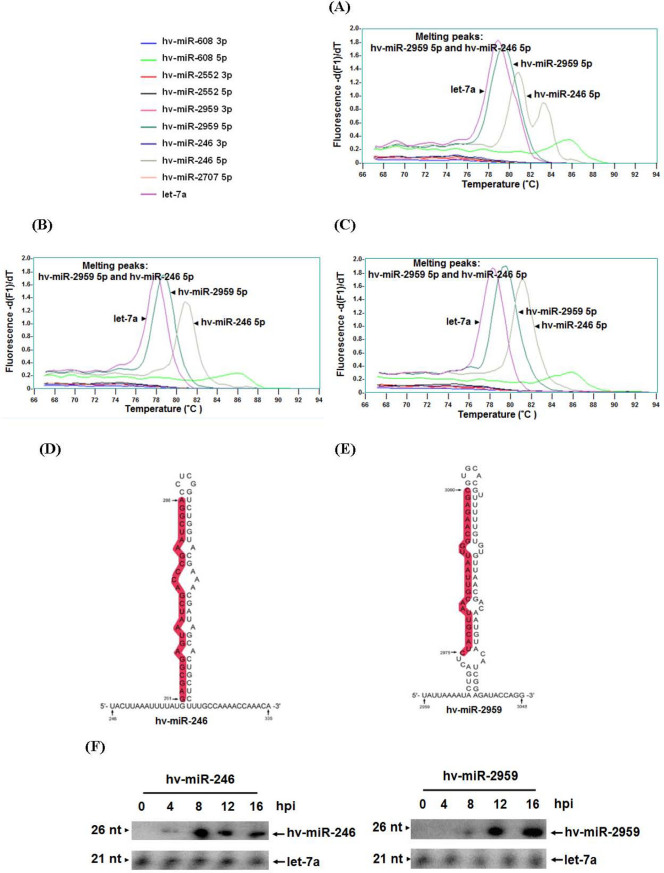
To confirm whether the above predicted miRNAs truly come from pag1 and are expressed in infected Sf-21 cells, a stem-loop PCR18,19 was performed to analyze the miRNA expression in pag1-transfected cells. The pag1-transfected cells were harvested at 12 hpi and total RNA was extracted. The cDNAs of miRNAs were obtained using miRNA stem-loop RT primers (Table 1). Stem-loop PCR analysis was performed using these cDNAs as the templates with specific miRNA primers designed based on our predictions. In order to investigate whether if any of the predicted miRNAs are expressed in viral infected cells, total RNAs were extracted from productive and latent viral infected cells and subsequently analyzed by stem-loop PCR. Real-time PCR products were also separated by gel electrophoresis and visualized by EtBr staining as a further control to show proper expression of different PCR products (Fig. 3 and Supplemental Fig.1). The result showed that two miRNAs, hv-miR-246 5P and hv-miR-2959 5p, were expressed in the HzNV-1 productively infected cells (Fig. 3A), HzNV-1 latently infected cells (Fig. 3B), and pag1-transfected cells (Fig. 3C). These two miRNAs were cloned and the predicted secondary structures of hv-miR-246 5P and hv-miR-2959 5p precursors are shown in Fig. 3D, and Fig. 3E, respectively. Mature miRNA are shadowed in red and the numbers on nucleotides indicating their corresponding positions to pag1 transcript.
Table 1. Oligonucleotides used in this study.
| microRNA name | primer name | sequence |
|---|---|---|
| hv-miR-246-3P | 2463pForward primer | AACCGTTTCTCGTCACGATAGCA |
| 2463pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACCATG | |
| hv-miR-246-5p | 2465pForward primer | GGCTCCAGGCTAAGCCCAGCT |
| 2465pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCTCGCC | |
| hv-miR-608-3P | 6083pForward primer | GGTGGTTCTCAAATTCAGAGCT |
| 6083pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACATTACG | |
| hv-miR-608-5P | 6085pForward primer | CAACGGCTATGATGTCACGGTTTTG |
| 6085pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCCATGG | |
| hv-miR-2552-3P | 25523pForward primer | TCCACACGTTCTGAAAACGACG |
| 25523pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTCTAAT | |
| hv-miR-2552-5P | 25525pForward primer | TAACACCTTCTTAACTTATGTT |
| 25525pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTTTCTC | |
| hv-miR-2959-3P | 29593pForward primer | AGAGGCTACATGTAACAGCAATTGT |
| 29593pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAAAACA | |
| hv-miR-2959-5P | 29595pForward primer | CACGTGCGAGAACGGTTAATTGCA |
| 29595pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGATGCA | |
| hv-miR-2707-5P | 27075pForward primer | GAGACACATGCTCATAGCGTAACG |
| 27075pUPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCAACAA | |
| let-7a | 7-Forward primer | GCCGCTGAGGTAGTAGGTTGTA |
| 7-UPL RT primer | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAACTAT |
Figure 3. Cloning and analysis of the predicted miRNA by stem-loop PCR and northern blot.
Stem-loop PCR was performed to clone and analyze the proper expression of the predicted miRNAs in (A) HzNV-1 productively infected cells, (B) pag1-transfected cells, and (C) HzNV-1 latently infected cells. (D, E) Predicted secondary structures of hv-miR-246 5P (D), and hv-miR-2959 5p (E), precursors. (F) Small RNAs harvested from HzNV-1 productively infected cells at various time points were analysis by northern blots with probes against predicted HzNV-1 miRNAs (top panels) or let-7a miRNA as a positive control (bottom panels).
The melting curve for hv-miR-246 5p had two peaks in the HzNV-1 productively infected cells (Fig. 3A). We cloned these two fragments into cloning vector and analyzed their sequences. The sequence of the upper band did not match to the predicted miRNA and therefore is a non-specific product. The molecule size of the upper band is also lager than the predicted miRNA. In these experiments, proper expression of let-7a miRNA gene was performed as a positive control20. To further confirm the results from stem-loop PCR, northern blot analysis showed that hv-miR-246 and hv-miR-2959 became detectable in productively infected cells at 4 hpi and 8 hpi, respectively (Fig. 3F). However, they were not detected in mock infected cells (data not shown). These results indicated that the pag1 transcript was indeed processed into miRNAs during the infection of HzNV-1 (Fig. 3F).
hhi1 expression can be down-regulated by pag1 miRNAs
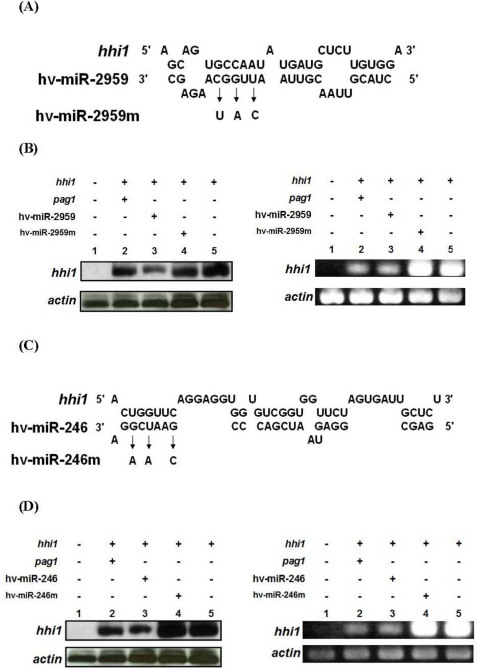
To further test whether these two miRNAs target hhi1 and suppress its transcripts, these miRNAs were separately transfected into Sf-21 cells along with a hhi1-expressing vector and the levels of hhi1 transcript were analyzed by northern blot analysis at 12 hpi. Both miRNAs were able to reduce the amount of hhi1 transcript within the cells (Fig. 4). The sequences of hv-miR-246 and hv-miR-2959 are mapped to nucleotides 226325 to 226366 and 226527 to 226555 of the hhi1 coding region, respectively (Fig. 2). To confirm if the matching of miRNA sequences to hhi1 was important, i.e., acting as targeting sites, we constructed mutant miRNAs with three base-pair mutations in the matching regions for these two miRNAs (Fig. 4A, 4C). The mutational substituted bases are indicated by arrows. Northern analysis (Fig. 4B, 4D; left panels) and RT-PCR (Fig. 4B, 4D; right panels) showed that these two mutant miRNAs did not affect the levels of hhi1 transcript (Fig 4B, 4D), suggesting that our predicted target positions are crucial for functioning. In these experiments, the levels of actin transcript were used as a loading control.
Figure 4. Down-regulation of hhi1 expression by pag1, hv-miR-246, and hv-miR-2959.
(A, C) sequences of hv-miR-2959 and hv-miR-246 were shown, and miRNAs with mutations were denoted as hv-miR-246m and hv-miR-2959m, separately. (B, D) levels of hhi1 transcript in various treatments were analyzed by northern hybridization (left panel) and RT-PCR (right panel).
pag1 miRNA functions in establishing latent viral infection
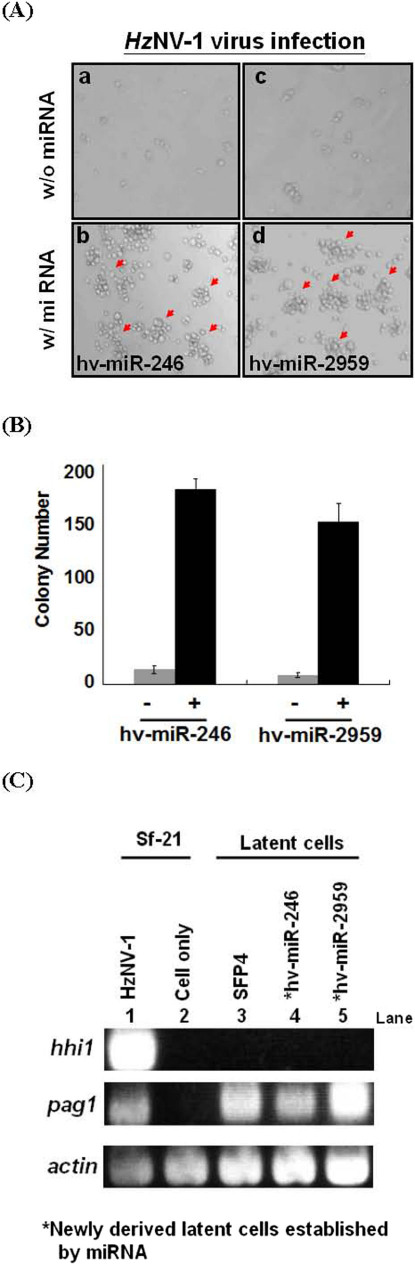
Previously, we showed that pag1 gene can promote the establishment of latent viral infection4. To test whether these miRNAs alone are enough to promote latent viral infection, Sf-21 cells were first transfected with miRNA followed by HzNV-1 infection. The number of colonies was then recorded at 12 dpi. Most of the Sf-21 cells died when they were infected with HzNV-1 virus alone and only a small percentage of cells became latently infected (Fig. 5A). However, the number of latently-infected cell colonies increased dramatically upon transfection of individual hv-miR-246 or hv-miR-2959 (Fig. 5B). We also analyzed the gene expression of pag1 and hhi1 in HzNV-1 infected cells with or without miRNA treatments at 12 dpi (Fig. 5C). hhi1 expression could only be detected in HzNV-1 productively infected cells (Fig. 5C, lane 1) whereas pag1 expression could be detected in productive and latent cells SFP4 (Fig. 5C, lanes 1 and 3). Cells transfected with these two miRNAs, hv-miR-246 and hv-miR-2959, produced only pag1 transcript but not hhi1 transcript, this is an evidence of viral latency (Fig. 5C, lanes 4 and 5). These results indicate that miRNAs alone can function to establish or at least to strongly promote latent viral infection. Furthermore, pag1 transcript, but not that of hhi1, was expressed in the latent cells induced by the transfection of miRNA. Besides, these latent cells can be cultured for long-term passages. All these evidences indicate strongly that the establishment of latent viral infection by miRNA is not a transient effect; it is a long term latent viral infection in the cells.
Figure 5. Establishment of latent viral infection by miRNAs.
(A) Sf21 cells were transfected with or without miRNA followed by HzNV-1 infection. (B) Column representation of the results of panel (A). (C) Confirmation of hhi1 and pag1 expressions in various cells by RT-PCR.
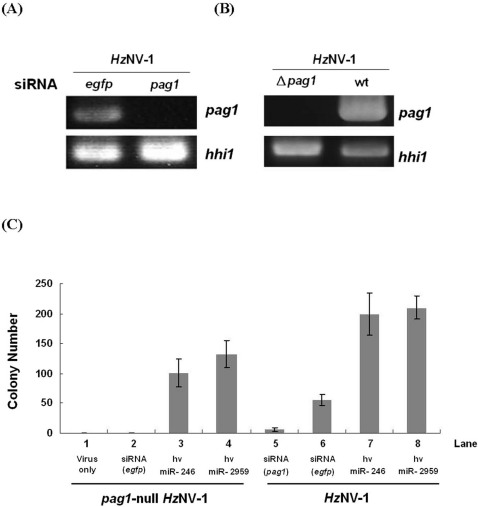
In these and previous experiments, we have observed that the number of latently-infected cell colonies increased upon miRNA or pag1 transfection. However, whether these miRNA or pag1 can only promote viral latency or are essential for the establishment of latent viral infection remain unknown. In order to confirm the function of the miRNA and pag1, we further constructed a pag1-null HzNV-1 virus for infection experiment. Sf-21 cells were infected with pag1-null virus or transfected with a siRNA targeting to pag1 transcript followed by HzNV-1 infection. RT-PCR confirmed that the suppression of pag1 gene expression by siRNA was successful (Fig. 6A) and also, no pag1 expression was detected from pag1-null HzNV-1 virus (Fig. 6B). Further experiments showed that upon knocking down (Fig. 6C, lane 5) or deletion of pag1 transcripts (Fig. 6C, lane 1), the number of latent colony dropped significantly, and none of the cell colony can survive two weeks after viral infection. However, the number of latently-infected cell colonies increased dramatically upon transfection of individual hv-miR-246 or hv-miR-2959 (Fig. 6C, line 3, 4, 7 and 8). These cells keep on viable for long time (at least 2 months up to now). These experiments demonstrated that pag1 and its derived miRNAs play a crucial role in the establishment of HzNV-1 latent viral infection.
Figure 6. Suppression of HzNV-1 viral latency by knocking down pag1 expression.
(A) RT-PCR showed that artificial siRNA can efficiently suppress pag1 expression in HzNV-1 infected cells. (B) pag1 expression is not detectable by RT-PCR in the pag1-null HzNV-1-infected cells. (C) Formation of latent colony is not observed by the infection of pag1-null HzNV-1.
Discussion
Previously, we found that hhi1 can activate the expression of many early HzNV-1 genes and reactivate HzNV-1 virus from latency16. We had also discovered that the transcript of pag1, a non-coding RNA, plays a critical role to block hhi1-induced apoptosis21 and to the establishment of latent viral infection4. In this study we found that pag1 transcript could suppress hhi1 expression remarkably through the targeting of hhi1 transcript by miRNAs. Interestingly, these two miRNAs, hv-miR-246 or hv-miR-2959, were individually sufficient for strong promotion and/or establishment of latent viral infection. The observation that pag1-null HzNV-1 virus was unable to establish latent infection further suggested that pag1 plays a key role in the establishment of latent infection.
Most miRNA target sites have perfect pairing to the seed region located near the miRNA 5′ end. Nevertheless, recent reports have also demonstrated that perfect pairing between target sequence and the miRNA can also take place near the 3′ end of miRNA22,23. Our results also showed that the 3′ ends of miRNAs derived from pag1 have perfect base pairings to the target sequences, and mutagenesis experiments showed that these pairing are critical for functioning. To further confirm that inhibition of hhi1 expression was mediated through miRNA pathway, the effects of silencing dicer1 and ago1 expressions (Supplemental Fig. 2A) on the production of pag1-derived miRNA and control let-7a were assessed in Drosophila S2 cells (Supplemental Fig. 2B). It is known that dicer1 is an important component of the miRNA progenesis; whereas, ago1 is not involved in the generation of mature RNAs, rather, is involved in the interaction between miRNA and target transcript. Thus, it is reasonable that levels of miRNAs hv-miR-246, hv-miR-2959 and let-7a were not affected by ago1 knockdown using stem-loop RT-PCR analysis (Supplemental Fig. 2B). We also found that hhi1 expression was not down-regulated by pag1 in either dicer1 or ago1 knocked down cells (Supplemental Fig. 2C), suggesting that miRNA pathway is essential for the production of pag1-derived miRNAs.
HzNV1 virus is an insect-specific virus. In our studies, we found it shares striking similarity in the gene regulation networks during viral latency with the vertebrate virus, HSV-124. Despite the high numbers of viral transcripts produced during productive viral infections, both viruses silence the expression of all genes except the expression of latency-associated transcript (LAT) in HSV-125, and pag1 transcript in HzNV-13,4, during latent viral infection. It is interesting to note that HzNV-1 is a baculiform insect virus and HSV, on the other hand, is an icosahedra virus. They are quite distinct in shape and genomic sequences and are obviously not evolved from the same origin. Nevertheless, LAT is a latency-associated transcript, which contains a non-coding intron25 and pag1 transcript is similarly a non-coding transcript predominately expressed during viral latency4. Such striking similarity between these transcripts strongly suggests that the strategy for producing a non-coding RNA during latency is not an isolated viral infection phenomenon restricted to the HSV of vertebrates.
LAT was previously identified as an 8.3 kb transcript expressed from the genome of HSV22. This RNA is spliced and yields a stable intronic non-coding region of 2 kb and an unstable exonic RNA. This unstable exonic RNA was further processed to produce several miRNAs, which are found to suppress protein expression of early HSV genes, ICP0 and ICP417,26. More recently, two sRNAs of 62 nt and 32 nt, but not miRNA, were predicted to be located on the 2 kb stable intronic region. Shen et al. (2009) found that these sRNAs play a role through an unidentified mechanism to inhibit apoptosis and productive infection27. Although these authors showed that these miRNAs or sRNAs were likely responsible for latent HSV infection due to the suppression of two important early genes, direct proof of latent establishment due to these miRNAs or sRNAs is still lacking.
In our results, due to HzNV-1 can establish latent viral infection in insect cells, we were able to show that HzNV-1 virus establishes latent viral infection through the suppression of the hhi1 transcript using non-coding transcript pag1. We also showed that even miRNAs from pag1 transcript alone are sufficient to establish latent viral infection. Since the transiently transfected miRNA is sufficient for the triggering of latent viral infection using pag1-null HzNV-1, suggesting that once entering viral latency, viral gene expression is some how restricted without the need of a continuous supply of pag1 transcript for hhi1 suppression.
Our experiments showed that, although HzNV-1 virus is not a herpes virus and pag1 transcript has no sequence homology with LAT, shutting-off of all genes except the expression of one non-coding transcript during latency of these two viruses is a striking convergent evolution of viral-host interactions. In addition, the use of miRNAs derived from the non-coding RNA for similar functions by two unrelated viruses suggests that miRNA can be an important and effective mechanism for switching between productive and latent infections in a wide-range of host cells. Furthermore, pag1 transcript is a stable non-coding RNA consistently observed during productive and latent viral infections3,4, which resembles the intronic stable non-coding RNA of LAT. Although in the study of herpes virus, miRNA was not found from the stable non-coding RNA of LAT, we showed that pag1 transcript can generate functional miRNAs.
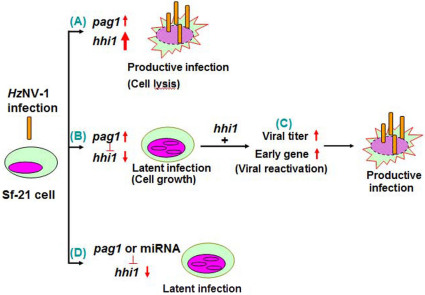
In Fig. 7, we summarize the interactions between hhi1 and pag1 to better illustrate how they function as molecular switches to determine should the viruses enter productive or latent infections in the host cells. The initial infection of HzNV-1 results in high levels of hhi1 expression (Fig. 5C, lane 1)16, and a moderate level of pag1 expressions (Fig. 5C, lane 1)3,4. The newly produced abundant hhi1 transcript can tolerate some RNA degradation caused by the miRNAs derived from pag1. Then, the hhi1 transcript, before degradation or survived from initial degradation, can quickly transactivate many viral early genes for productive viral infection to occur in most of the infected Sf21 cells (Fig. 7A)16. A low percentage of cells, by which hhi1 transcript is successfully suppressed by pag1 miRNA during initial viral infection, enter latency14, where the level of hhi1 transcript becomes undetectable while still maintain a moderate level of pag1 transcript (Fig. 7B; Fig. 5C, lanes 3–5)3. Viral re-activation of latent cells can be induced by introducing hhi1 transcript, which then stimulates HzNV-1 early genes expression, resulting in virus production (Fig. 7C)16. If pag1 gene4 or its derived miRNAs (Fig. 6C lanes 3,4) were transfected into the cells prior to the initial viral infection, the chance of cells entering latent viral infection becomes strongly boosted (Fig. 7D). Different from the function of miRNAs produced from LAT of the herpes virus, which blocks protein expression, miRNAs derived from pag1 transcript degrade the transcript of hhi1. More significantly, our evidences proved that pag1 transcript and its derived miRNAs can establish latent viral infection in the cells.
Figure 7. A comprehensive model for the establishment of productive and latent HzNV-1 viral infections through hhi1 and pag1 interactions.
Methods
Cells and virus
Spodoptera frugiperda IPLB-Sf-21 was incubated in TC-100 insect cell culture medium, which contained 10% of FBS at 26°C (Gibco BRL)28,29. Standard HzNV-l virus was derived by a serial dilution of the stock viral solution and isolated by plaque purification. Latently infected colonies were calculated as described in Wu et al.16. The titers of the virus clones were estimated by both Q-PCR30 and TCID5028,29.
Plasmid construction
We obtained pag1 and hhi1 coding regions by PCR and inserted them down-stream to the hsp 70 promoter (p-hsp)31 and pag1 promoter16 of plasmids pKSh and pKSp, respectively to result plasmids pKShHI and pKSpP1. The nucleotide sequence of the hhi1 gene was submitted to GenBANK, with an accession number of AF264019.
Computational prediction of viral miRNAs
The pag1 miRNA prediction was carried out using the complete gene sequence of pag1 (NC_004156) obtained from the National Center for Biotechnology Information. The pag1 miRNA prediction was performed using miRNA prediction database (Vir-Mir) (http://alk.ibms.sinica.edu.tw)32.
RNA isolation and northern blot analysis
2 × 105 cells/well were seeded in a 24-well culture plate (Corning) and then transfected with two plasmids, pKShHI and pKSpP1, 0.5 µg each, separately or collectively, using Cellfectin according to the manufacturer's protocol (Invitrogen). Sf-21 cells were inoculated at a multiplicity of infection (MOI) of 1 with HzNV-l virus and incubated at 26°C with gentle rocking. After absorption, total cellular RNA samples were extracted from productively-infected cells at the indicated time and from latently-infected SFP4 cells3,16 using RNeasy Mini Kit (Qiagen). Briefly, total RNA were extracted from virus-infected Sf-21 or SFP4 cells (Qiagen). Then, equal amounts of RNA samples (15 µg each) were loaded, blotted onto Hybond-N+ nylon transfer membrane (GE Healthcare), and hybridized with hhi1 or pag1 specific probes, which were labeled with DIG by PCR amplification of template HzNV-1 viral DNA using the PCR DIG Probe Synthesis Kit (Roche Applied Sciences) according to the manufacturer's instructions.
miRNA assay by stem-loop real-time PCR and northern blot
The total RNA samples from pag1 transfected cells were isolated using RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The miRNA cDNAs were synthesized from total RNAs using miRNA specific stem-loop primers (Table 1) according to criteria described previously18. Analysis of mature miRNA expression by RT–PCR was also carried out using a method described previously18. The cDNAs were diluted 10 times to perform PCR for expression confirmation and expression pattern analysis. Real-time PCRs were performed, respectively, in 20 µl mixture containing 1 µl cDNA, 0.5 µM forward and reverse primers, and 2 × SYBR Green qPCR master mix (Fermentas) with the following parameters: 94°C for 15 s, 60°C for 30 s and 72°C for 30 s. The real-time PCR products were detected by electrophoresis with 3% agarose gel containing ethidium and photographed under UV light. Expression of let-7a, a miRNA normally expressed in most of the cells20, was used as a positive control.
The pag1-associated small RNA samples from pag1 transfected cells at different time points were isolated using a mirVanaTM miRNA isolation kit (Ambion) according to the manufacturer's instructions. For northern blot analysis of small RNAs, 1 µg aliquots of each small RNA fraction, as well as radio-labeled Decade Markers (Ambion), were fractionated in 15% denaturing polyacrylamide gel electrophoresis (PAGE) gels (acrylamide∶bis ratio, 19∶1) containing 8 M urea in 0.5 × TBE buffer. The gels were soaked briefly in 0.2 µg/ml ethidium bromide in TBE to allow visualization of the RNA using a UV transilluminator (Bio-Rad, Hercules, CA). RNAs were transferred by electroblotting to a Hybond-N+ nylon transfer membrane (GE Healthcare) and UV cross-linked to the membrane. PAGE-purified DNA oligonucleotides (Integrated DNA Technologies) with the reverse complementary sequence to either candidate miRNAs or let-7a, were end labeled with [32P]ATP (MP Biomedicals, Irvine, CA) to high specific activity. The labeled probes were purified using the mirVana Probe & Marker kit (Ambion) according to the manufacturers' protocols. Hybridizations and washes were carried out using the ULTRAhyb-Oligo hybridization buffer according to the manufacturer's directions (Ambion).
Mutant miRNA experiments
2 × 105 cells/well were seeded in a 24-well culture plate and then transfected the plasmid pKShHI (0.5µg) with hv-miR-246, hv-miR-246m, hv-miR-2959 and hv-miR-2959m (50nM each), separately, using Lipofectamine RNAiMAX (Invitrogen). Transfected cells were harvested at 12 hours post-transfection (hpt) for northern analysis using a hhi1 specific probe. hv-miR-246m and hv-miR-2959m are three base mutants created in the perfect pairing regions of hv-miR-246 and hv-miR-2959, separately, to abolish the function of these miRNAs. All miRNAs duplex were synthesized by MDbio Inc. The miRNAs or mutant miRNAs used in this study are as follows:
hv-miR-246 (5′-AGGCUAAGCCCAGCUAAUGAGGCGAG-3′)
hv-miR-246m (5′-AGACAAACCCCAGCUAAUGAGGCGAG-3′)
hv-miR-2959 (5′-CGAGAACGGUUAAUUGCAAUUGCAUC-3′)
hv-miR-2959m (5′-CGAGAAUGAUCAAUUGCAAUUGCAUC-3′)
miRNA transfection upon HzNV-1 infection
For hhi1 and pag1 miRNA transfected experiments, all miRNAs were synthesized by MDbio Inc. Sf-21 cells (4 × 104/well) in 96-well plates were transfected with 50 nM of the miRNA using Lipofectam RNAiMAX (Invitrogen). Cells were transfected with pag1 miRNAs; then, at 4 hpt, the cells were infected with HzNV-1 virus (MOI = 1), and the number of latently infected colonies was again calculated at 12 dpi4. Several primers were designed for the RT-PCR amplification of hhi1, pag1 and actin as follows:
hhi1-1F: 5′-CGATATGAACATTAACGATGACGATC-3′;
hhi1-1R: 5′-AAACGGATGCAAAATGGACTCAA-3′;
pag1-F: 5′-ACGGGAATTCAGTGTCGAGGACTT-3′
pag1-R:5′-CATGTCTAGAACCCTACCTACCT-3′
actin-F: 5′-CGTGATGGTGGGCATGGGTCAG-3′;
actin-R:5′-CTAATGTCACGCACGTATTCC-3′.
RNA interference
For pag1 knockdown experiments, all siRNAs were predicted and synthesized by MDbio Inc. The siRNAs used in this study were pag1 siRNA (5′- GGAGUAGGUAGCAUAGAUG-3′) and egfp siRNA (5′- GGCGAUGCCACCUACGGCAAG-3′ ). Sf-21 cells (4 × 104) in 96-well plates were transfected with 50 nM of the siRNA using Silencer siRNA transfection kit II (Applied Biosystems). Cells were transfected with designed siRNAs and, at 4 hpt, the cells were infected with HzNV-1 virus (MOI = 1). The number of latently infected colonies was calculated at 12 dpi4. Primers designed for the detection of hhi1 and pag1 by RT-PCR are listed as following:
hhi1-1F: 5′-CGATATGAACATTAACGATGACGATC-3′
hhi1-1R: 5′-AAACGGATGCAAAATGGACTCAA-3′
pag1-F: 5′-ACGGGAATTCAGTGTCGAGGACTT-3′
pag1-R:5′-CATGTCTAGAACCCTACCTACCT-3′
Construction of pag1-null HzNV-1
A fragment containing egfp gene flanked by EcoRV sites was inserted down-stream of the hsp 70 promoter (p-hsp) of plasmid pBShsp70, generating plasmid pBShE. Fragments containing −50 to −300 nt and +300 to +550 nt of pag1, respectively, were obtained by PCR and subsequently cloned into the upstream and downstream, respectively, of hsp-egfp of pBShE, yielding pBSphEp. pBSphEp and HzNV-1 viral DNA were co-transfected into Sf-21 cells to generate pag1-null HzNV-1 virus. Several primers were designed for the PCR amplification of pag1 upstream and downstream fragments and they are listed as following:
pag1UF: 5′-ATTGAGCTCCCGAGACTGCACTTTTTAAA-3′
pag1UR: 5′-ATTCCGCCGTAGTAGGTGTGCACGTTTT 3′
pag1DF: 5′-ATTATCGATGTGTATTTATAGATGCCCGA-3′
pag1DR: 5′-ATTATCGATAATACGCAGGGTGGACTCTT-3′
Author Contributions
Guarantors of integrity of entire study, study concepts, and manuscript preparation, Y.C.C.; study design, data acquisition/analysis, literature research and manuscript preparation, Y.L.W.; computational prediction, P.W.C.H.; data acquisition/analysis, manuscript editing, and revision, C.P.W., C.Y.Y.L. and E.C.W. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Drs. Chang-Chi Lin, Wen Chang, Bryan R. Cullen, and May-Ru Chen for valuable discussions and suggestions; and Ms. Miranda Loney and Dr. Harry Wilson for revision. This research is funded by grants 098-2811-B-001-025, 098-2811-B-001-036, and 98-2321-B-001-031-MY3 from the National Science Council, and 94S-1303 from Academia Sinica, Taiwan.
References
- Burand J. P., Stiles B. & Wood H. A. Structural and Intracellular Proteins of the Nonoccluded Baculovirus Hz-1. J. Virol. 46, 137–142 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. S., Hedberg M. & Kawanishi C. Y. Characterization of the DNA of a Nonoccluded Baculovirus, Hz-1V. J. Virol. 43, 174–181 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y. C. et al.. Differential expression of Hz-1 baculovirus genes during productive and persistent viral infections. J. Virol. 66, 1442–1448 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y. C. et al.. A 2.9-kilobase noncoding nuclear RNA functions in the establishment of persistent Hz-1 viral infection. J. Virol. 72, 2233–2245 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. H. et al.. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76, 9024–9034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D. C. et al.. Induction of a nonoccluded baculovirus persistently infecting Heliothis zea cells by Heliothis armigera and Trichoplusia ni nuclear polyhedrosis viruses. Virology 112, 174–189 (1981). [DOI] [PubMed] [Google Scholar]
- Wang Y., Kleespies R. G., Huger A. M. & Jehle J. A. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J. Virol. 81, 5395–5406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., van Oers M. M., Crawford A. M., Vlak J. M. & Jehle J. A. Genomic analysis of Oryctes rhinoceros virus reveals genetic relatedness to Heliothis zea virus 1. Arch. Virol. 152, 519–531 (2007). [DOI] [PubMed] [Google Scholar]
- Granados R. R. Early events in the infection of Hiliothis zea midgut cells by a baculovirus. Virology 90, 170–174 (1978). [DOI] [PubMed] [Google Scholar]
- McIntosh A. H. & Ignoffo C. M. Establishment of a persistent baculovirus infection in a lepidopteran cell line. J. Invertebr. Pathol. 8, 395–403 (1981). [Google Scholar]
- Ralston A. L., Huang Y. S. & Kawanishi C. Y. Cell culture studies with the IMC-Hz-1 nonoccluded virus. Virology 115, 33–44 (1981). [DOI] [PubMed] [Google Scholar]
- Podgwaite J. D. & Mazzone H. M. Latency of insect viruses. Adv. Virus Res. 31, 293–320 (1986). [DOI] [PubMed] [Google Scholar]
- Wood H. A. & Burand J. P. Persistent and productive infections with the Hz-1 baculovirus. Curr. Top. Microbiol. Immunol. 131, 119–133 (1986). [DOI] [PubMed] [Google Scholar]
- Lin C. L. et al.. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J. Virol. 73, 128–139 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. L. et al.. Cooperation of ie1 and p35 genes in the activation of baculovirus AcMNPV and HzNV-1 promoters. Virus Res. 135, 247–254 (2008). [DOI] [PubMed] [Google Scholar]
- Wu Y. L. et al.. The early gene hhi1 reactivates Heliothis zea nudivirus 1 in latently infected cells. J. Virol. 84, 1057–1065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach J. L. et al.. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454, 780–783 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. et al.. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D. et al.. Real-time PCR quantification of precursor and mature microRNA. Methods 44, 31–38 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M. E. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 8, 843–852 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. L. et al.. HzNV-1 Viral Gene hhi1 Induces Apoptosis which is Blocked by Hz-iap2 and a Non-coding Gene pag1. J. Virol. 85, 6856–6866 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C. et al.. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 38, 789–802 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent J. R. & Fraser N. W. The cellular response to herpes simplex virus type 1 (HSV-1) during latency and reactivation. J. Neurovirol. 11, 376–383 (2005). [DOI] [PubMed] [Google Scholar]
- Perng G. C. & Jones C. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip. Perspect. Infect. Dis. 2010, 262415 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach J. L. et al.. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 84, 1189–1192 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. et al.. Two small RNAs encoded within the first 1.5 kilobases of the herpes simplex virus type 1 latency-associated transcript can inhibit productive infection and cooperate to inhibit apoptosis. J. Virol. 83, 9131–9139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly D. R., Miller L. K. and Luckow V. A. Baculovirus Expression Vectors: A Laboratory Manual. 109–123 (Oxford University Press, 1994). [Google Scholar]
- Wu C. P., Wang J. Y., Huang T. Y., Lo H. R. & Chao Y. C. Identification of baculoviral factors required for the activation of enhancer-like polyhedrin upstream (pu) sequence. Virus Res. 138, 7–16 (2008). [DOI] [PubMed] [Google Scholar]
- Lo H. R. & Chao Y. C. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 20, 354–360 (2004). [DOI] [PubMed] [Google Scholar]
- Ingolia T. D., Craig E. A. & McCarthy B. J. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell 21, 669–679 (1980). [DOI] [PubMed] [Google Scholar]
- Li S. C., Shiau C. K. & Lin W. C. Vir-Mir db: prediction of viral microRNA candidate hairpins. Nucleic Acids Res. 36, D184–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information