Abstract
We studied the population genetics of Mycobacterium kansasii isolates from the United States by PCR restriction enzyme analysis (PRA) of the 441-bp Telenti fragment of the hsp-65 gene and pulsed-field gel electrophoresis (PFGE) of genomic DNA with the restriction endonucleases AseI, DraI, and XbaI, and we compared the patterns to those previously reported from France and Japan. By PRA, 78 of 81 clinical isolates (96%) from the United States belonged to subspecies I. With PFGE, 28 AseI patterns, 32 DraI patterns, and 35 XbaI patterns were produced. PFGE showed marked clonality of the U.S. isolates, with differences between genotypes involving only one or two bands. Isolates within Texas showed lower pattern diversity than those from different states. With DraI, 31 of 71 isolates (44%) had the same common PFGE pattern, which matched the predominant pattern in France (pattern Ia), determined by Picardeau et al. (M. Picardeau, G. Prod'hom, L. Raskine, M. P. LePennec, and V. Vincent, J. Clin. Microbiol. 35:25-32, 1997), and in Japan (type M), determined by Iinuma et al. (Y. Iinuma, S. Ichiyama, Y. Hasegawa, K. Shimokata, S. Kawahara, and T. Matsushima, J. Clin. Microbiol. 35:596-599, 1997). With AseI, 42% of isolates produced a common pattern indistinguishable from the common pattern seen in French isolates (Ia) and with only one band difference from the common pattern (type M) in Japan. This study demonstrates that subspecies I is the predominant subspecies of M. kansasii among clinical isolates in the United States, as it is in Europe and Japan, and that genotype I is highly clonal worldwide, with the same major genotype responsible for human infection. The fact that a single clone of M. kansasii is responsible for most cases of human disease suggests that specific virulence factors may be associated with this specific genotype.
Mycobacterium kansasii, a nontuberculous mycobacterium, is an opportunistic pathogen of human diseases. It may produce pulmonary or disseminated infections in human immunodeficiency virus (HIV)-infected patients. It is best known as a cause of fibrocavitary lung disease in non-HIV-infected patients, although it has also produced noncavitary nodular disease in the setting of bronchiectasis (3-5, 7, 9-11, 14). M. kansasii is rarely recovered from dust, soil, or natural water supplies and is recovered almost exclusively from municipal tap water, which is considered to be its major environmental reservoir. In Texas M. kansasii disease is an urban disease (1), and the organism has been recovered from piped water systems in areas where M. kansasii is prevalent (16). As with other nontuberculous mycobacteria, human infections are believed to be acquired from the environment.
Over the past 10 years, molecular typing methods have been applied to the study of M. kansasii. Ross et al. published one of the earliest studies in 1992, when they characterized isolates from Australia, Belgium, Japan, South Africa, and Switzerland (15). They found that 20 of 105 isolates were pMK1-9 probe negative and that 19 of the 20 probe-negative isolates had a partial 16S rRNA gene sequence different from that of M. kansasii, suggesting the existence of a molecular subspecies or even possibly a new species. Using molecular-based techniques for M. kansasii identification and strain typing, including restriction fragment length polymorphism analysis with the major polymorphic tandem repeat probe and IS1652, pulsed-field gel electrophoresis (PFGE), and PCR restriction analysis (PRA) of the hsp-65 gene, Picardeau et al. (13) studied 38 clinical M. kansasii strains and 24 M. kansasii strains from water samples, all isolated in France. They found that the clinical and environmental isolates of M. kansasii actually belonged to five different M. kansasii subspecies, with genotype II corresponding to the subspecies proposed by Ross et al. (15). Alcaide et al. (2) conducted a study by genotypic analysis of 276 M. kansasii isolates from various geographic areas within Europe. They identified the same five PRA subtypes as Picardeau et al. did, and they found that genotype I represented the most common isolates from humans. Iinuma et al. typed 84 M. kansasii isolates from patients with bronchopulmonary infections in Japan by PFGE with VspI digestion, and 21 distinctive PFGE types were identified (6). PRA genotyping was not performed in that study.
In the United States, M. kansasii is one of the most frequently isolated pathogenic nontuberculous mycobacteria, second only to Mycobacterium avium complex in many areas (1, 3, 10, 16). No previous studies, however, have provided a molecular analysis of M. kansasii isolates from the United States similar to those of isolates from Europe and Japan. In the present study we analyzed clinical isolates of M. kansasii from Texas and elsewhere in the United States by PRA of the hsp-65 gene and PFGE and compared the results to those from Europe and Japan.
(This study was presented at the 101st General Meeting of the American Society for Microbiology, Orlando, Fla., 20-24 May 2001 [Y. Zhang, L. B. Mann, R. W. Wilson, B. A. Brown-Elliott, V. Vincent, and R. J. Wallace, Jr., Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. U-14, p. 700].)
MATERIALS AND METHODS
Strains.
Eighty-one clinical isolates from the United States, collected between 1982 and 1999 from 69 patients with bronchopulmonary infection, were studied with PFGE and PRA. Isolates either came from patients treated at the University of Texas Health Center at Tyler or had been submitted to the Mycobacterium/Nocardia Laboratory for susceptibility testing. Ten patients (14.5%) were known to be HIV positive, and 48 of 81 isolates had been referred for treatment failure associated with rifampin resistance. Forty-four patients (63.8%) were from 15 Texas cities. The remaining 25 patients (36.2%) were from 18 states other than Texas (Table 1). Only single isolates from each of 62 patients were studied, while the remaining 7 patients provided multiple isolates. All of the isolates were identified as M. kansasii on the basis of conventional biochemical tests (8) and PRA. Isolates had been stored at −70°C in tryptic soy broth with 15% glycerol following isolation.
TABLE 1.
Locations of 69 patients infected with M. Kansasii
| City and/or state (no. of patients) | No. of cities or states | No. of patients (% of total) | Number of PFGE patternsa |
|---|---|---|---|
| Texas | |||
| Dallas (16), Houston (6), Austin (6), Fort Worth (5), Amarillo (1), Arlington (1), Brownsville (1), Corpus Christi (1), Galveston (1), Harlingen (1), Longview (1), Lubbock (1), Mexia (1), Paris (1), San Antonio (1) | 15 | 44 (63.8) | 33 |
| Other states | |||
| California (3), Alabama (2), Florida (2), Missouri (2), Pennsylvania (2), Tennessee (2), Arkansas (1), Arizona (1), Colorado (1), Connecticut (1), Kansas (1), Louisiana (1), Maryland (1), Minnesota (1), Mississippi (1), North Carolina (1), North Dakota (1), Oklahoma (1) | 18 | 25 (36.2) | 24 |
PFGE patterns with combination of AseI, DraI, and XbaI.
Control strains included 2 reference strains, ATCC 12478 and ATCC 35775, obtained from the American Type Culture Collection (ATCC) (Manassas, Va.); 21 M. kansasii isolates from France, including five isolates in genotype I, six in genotype II, three in genotype III, four in genotype IV, and three in genotype V, that were part of the study by Picardeau et al. (13); and 16 M. kansasii isolates from Japan, including six isolates with PFGE type B, four with type M, and one each with types C, E, F, G, J, and L, that were included in the study by Iinuma et al. (6).
PRA.
The Telenti fragment (441 bp) of the 65-kDa heat shock protein gene (hsp) was amplified from cell supernatants by PCR with primers TB11 and TB12 and subjected to restriction enzyme analysis with BstEII and HaeIII as recommended by Telenti et al. (19) and as performed previously (17). Band patterns were characterized as described by Picardeau et al. (13) and Alcaide et al. (2).
PFGE.
Patterns of large restriction fragments of genomic DNA were obtained by PFGE as described previously (21). Organisms in low-melting-point agarose plugs were lysed with lysozyme, sodium dodecyl sulfate, and proteinase K. Genomic DNA contained in the plugs was digested with restriction endonucleases DraI, AseI, and XbaI (each enzyme was used alone) and separated by PFGE with a CHEF Mapper system (Bio-Rad Laboratories, Richmond, Calif.). The gel photos were scanned and analyzed by using Advanced Quantifier 1-D Match (Bio Image, Ann Arbor, Mich.).
To simplify the analysis of XbaI patterns, because of their large number of bands, only the bands larger than 97.0 kb were counted. The fragment resolution on electrophoresis gels became optimal by doing so. Generally, differences should also be seen among the larger fragments if differences were found in smaller fragments. Thus, counting only XbaI fragments larger than 97.0 kb did not reduce the strain discrimination ability.
In this study, patterns with one or more PFGE band differences were considered different; otherwise they were considered indistinguishable. Isolates were considered clonal if they exhibited six or fewer band differences as defined by Tenover et al. for outbreak strains (20). They were considered unrelated (nonclonal) if they exhibited seven or more band differences.
Statistics.
The chi-square test was used for comparison of PFGE patterns produced by Texas and non-Texas isolates, with a P value of <0.05 indicating significance.
RESULTS
PRA.
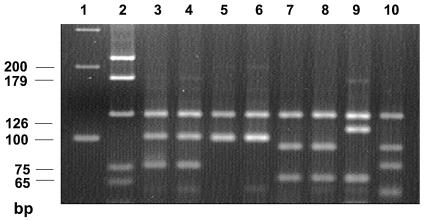
Restriction enzyme analysis of the hsp-65 gene sequence showed that all 81 clinical U.S. isolates and the two ATCC reference strains matched one of the five recognized subspecies patterns of M. kansasii (13). Seventy-eight of 81 clinical isolates (96.3%) had indistinguishable patterns (BstEII bands at 240 and 210 bp and HaeIII bands at 125, 100, and 80 bp), which matched genotype I described by Picardeau et al. (13) and Alcaide et al. (2). The two reference strains (ATCC 12478 and ATCC 35775) also exhibited the genotype I pattern. Of the remaining three U.S. clinical isolates, two belonged to genotype III and one belonged to genotype II. The identity of the PRA pattern was confirmed by electrophoresis of the U.S. isolates and the reference strains of each genotype from France together on the same gel. Examples of the five genotypes are shown in Fig. 1. Eight M. kansasii isolates from Japan, including one each of PFGE types B, C, E, F, G, J, L, and M (6), were also tested. All eight isolates belonged to genotype I.
FIG. 1.
Examples of the five PRA patterns (genotypes) seen with HaeIII digestion of the hsp-65 gene sequence (441 bp) of M. kansasii as described by Picardeau et al. (13). Lanes 1 and 2, 100-bp and pGem markers, respectively. Lanes 3 and 4, genotype 1 (M. kansasii 404 and 340, respectively). Lanes 5 and 6, genotype 2 (M. kansasii 234 and 335, respectively). Lanes 7 and 8, genotype 3 (M. kansasii 241 and 343, respectively). Lane 9, genotype 4 (M. kansasii 339). Lane 10, genotype 5 (M. kansasii 334).
PFGE.
Restriction endonucleases DraI, AseI, and XbaI were used to generate restriction patterns, which were then compared by PFGE. With 69 clinical isolates (one isolate per patient) and the two ATCC reference strains, 28 AseI patterns, 32 DraI patterns, and 35 XbaI patterns were produced (Table 2).
TABLE 2.
PFGE patterns and isolate distribution
| Restriction endonuclease and pattern(s)a,b | No. of isolatesc |
|---|---|
| AseI | |
| Aa | 30 (42.3) |
| Ac | 6 (8.5) |
| Ag | 6 (8.5) |
| Ad | 3 (4.2) |
| Ab, An | 2d (2.8) |
| Other (22 different patterns) | 1d (1.4) |
| DraI | |
| Da | 31 43.7) |
| Dd | 5 (7.0) |
| Dh | 4 (5.6) |
| Dm, Dw | 2d (2.8) |
| Other (27 different patterns) | 1d (1.4) |
| XbaI | |
| Xa | 23 (32.4) |
| X1 | 4 (5.6) |
| Xt | 4 (5.6) |
| Xb, Xf, Xd, Xi, Xk, Xr, Xs, Xdd | 2d (2.8) |
| Other (24 different patterns) | 1d (1.4) |
Restriction patterns were generated for 71 isolates.
The total numbers of patterns observed for the restriction endonuclease were as follows: for AseI, 28 patterns; for DraI, 32 patterns; and for XbaI, 35 patterns.
Values in parentheses are percentages.
Number of isolates with each different pattern.
With DraI digestion, 31 of 71 isolates (43.7%) had the same pattern (pattern Da), while another 13 isolates belonged to four other clusters. Thus, only 27 isolates (38.0%) had unique patterns. Compared with the most common pattern (pattern Da), 33 isolates with 24 patterns generated one to three band differences, and 4 isolates with 4 patterns generated four to six band differences. Thus, 68 of 71 isolates (95.8%) were clonal. Comparison of our major DraI pattern (pattern Da) lane by lane on the same electrophoresis gel with the predominant pattern seen in France (genotype Ia) by Picardeau et al. (13) and the pattern of the major M. kansasii group with PFGE VspI pattern M found in Japanese patients by Iinuma et al. (6) showed they were indistinguishable. Type strain ATCC 35775 also showed the same major DraI pattern, and another type strain, ATCC 12478, showed only one band difference.
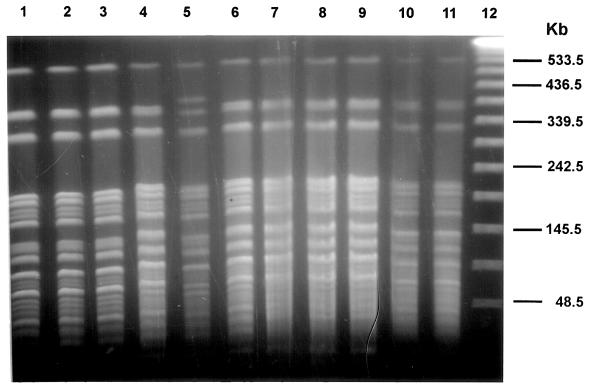
With AseI, 30 of 71 isolates (42.3%) generated a common pattern (pattern Aa), another 19 isolates were in five other clusters, and only 22 isolates (31.0%) had unique patterns. Compared with the most common pattern (pattern Aa), 25 isolates with 14 patterns generated one to three band differences, and eight isolates with five patterns generated four to six band differences. Thus, 63 of 71 isolates (88.7%) were clonal. Lane-by-lane comparison in the same gel showed that our common AseI pattern (Aa) was indistinguishable from the patterns produced by the major group of French isolates (genotype Ia) (13) and had only one band difference with the most frequently observed pattern (type M) in Japan (6) (Fig. 2). Figure 2 also showed that isolates with the same DraI pattern may produce differences with AseI digestion (Fig. 2, lanes 4 to 6).
FIG. 2.
Comparison of PFGE patterns of major M. kansasii groups seen in France, the United States, and Japan, with AseI digestion. Lanes: 1 to 3, French isolates (W47, C5, and W45) with major pattern Ia (13); 4 to 6, isolates in the United States (M. kansasii 88, 223, and 224) with major pattern Da; 7 to 9, Japanese isolates (65190, 95265, and 95315) of major group with VspI pattern M (6); 10, ATCC 35775; 11, ATCC 12478; 12, lambda DNA standard.
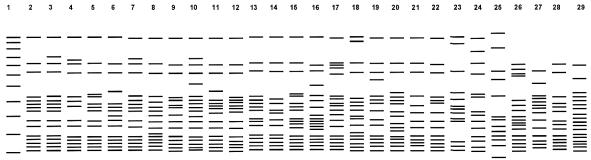
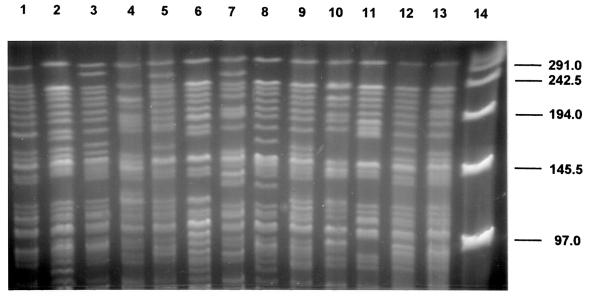
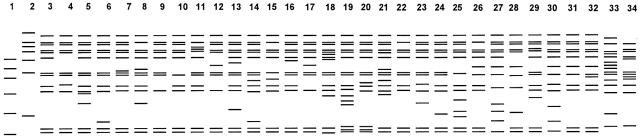
With XbaI, results similar to those with DraI and AseI were obtained (Table 2). Thus, with each of the three restriction enzymes, 32.4 to 43.7% of the isolates fell into a single cluster showing an indistinguishable pattern, with 88.7 to 95.8% of the isolates considered clonal. Most PFGE patterns were highly similar; differences between patterns often involved only one or two bands (Fig. 3 to 5). Some isolates showed indistinguishable patterns with one enzyme but different patterns with another enzyme. These isolates were considered different. Thus, with different combinations of the three enzyme patterns, the strain differentiation was improved. By combining the AseI, DraI, and XbaI patterns, 57 different patterns were identified within 71 M. kansasii isolates. The use of XbaI helped to further subdivide isolates, but the gel patterns were difficult to analyze because of the large number of bands. Counting only the bands larger than 97.0 kb on electrophoresis gels simplified the analysis of XbaI patterns.
FIG. 3.
Schematic representation of PFGE patterns with AseI digestion. Lane 1, lambda DNA standards; lane 2, common pattern Aa; all other lanes, clinical isolates with different patterns.
FIG. 5.
PFGE patterns of M. kansasii isolates with XbaI digestion. Lanes 1 through 11, clinical U.S. isolates; lane 12, ATCC 12478; lane 13, ATCC 35775; lane 14, lambda DNA standard.
Even with different combinations of restriction enzyme digestion patterns, there were still 20 isolates in six clusters each having indistinguishable patterns. Forty-four isolates from Texas showed 33 patterns, and 25 isolates from other states had 24 patterns (Table 1).
Of the 44 clinical isolates found in Texas, 26 (59.1%) had the common DraI pattern (pattern Da), while only 4 of 25 isolates (16.0%) found in other states had this pattern (P < 0.005). A higher percentage of Texas isolates had the common DraI pattern. However, 18 of 44 Texas isolates (40.9%) and 10 of 25 non-Texas isolates (40.0%) showed the common AseI pattern (pattern Aa) (P > 0.75). Our result also showed that 44 isolates from Texas had 33 patterns and 25 isolates from other states had 24 patterns (Table 1) (P > 0.05). Isolates within a single state (Texas) were more closely related than those from different states. However, the geographic sources of the isolates with the same common patterns were not actually limited in a single state.
To determine if PFGE patterns over time were stable, 19 isolates from seven patients were chosen. Each patient had two to six isolates, with the time between specimen recovery ranging from 1 to 28.5 months. The results showed that all of the isolates from the same patients over time gave indistinguishable PFGE patterns, with no changes in any visible band.
DISCUSSION
In recent years a number of molecular-based techniques for strain identification and comparison have been developed. These techniques have greatly facilitated investigation of M. kansasii identification. Picardeau et al. (13) used five fingerprinting methods, including RFLP analysis with the major polymorphic tandem repeat probe and the IS1652 probe, PFGE, amplified fragment length polymorphism analysis, and PRA of the hsp-65 gene. All of these methods showed excellent typeability and reproducibility. The five methods revealed independent polymorphisms. Nevertheless, the genotypes indicated by each method were in agreement. PRA defined five genotypes, while PFGE allowed recognition of subgroups within the genotypes.
Picardeau et al. (13) studied 63 M. kansasii isolates, including 38 clinical strains and 24 water isolates (all recovered in France) and 1 reference strain. The most common of the five genotypes, genotype I, included 25 of 63 strains studied (39.7%). Among clinical isolates only, 16 of 38 (42.1%) belonged to genotype I. Alcaide et al. (2) found the same five genotypes among 276 M. kansasii isolates from all over Europe. Genotype I was present among 109 of 163 clinical isolates (66.9%). A recent Swiss national survey (18) and studies of isolates from Bilbao, Spain (5a), found similar results. In the present study, 78 of 81 isolates (96.3%) recovered from the United States had an indistinguishable PRA pattern which matched PRA genotype I of Picardeau et al. (13) and Alcaide et al. (2). The isolates with the eight most common PFGE types from Japan (6) belonged to genotype I as well. Thus, genotype I is the predominant subspecies of M. kansasii among clinical isolates worldwide in all studies to date. Other genotypes are prevalent in Europe and were identified much less commonly in the present study in the United States.
Using PFGE and the restriction enzyme DraI, Picardeau et al. (13) found the most prevalent subgroup to be subgroup Ia, which was found in genotype I. This subgroup consisted of 17 clinical isolates and 1 water isolate, and represented 28.6% of the strains studied. Iinuma et al. (6) used restriction enzyme VspI and found that 22 of 84 clinical isolates (26.2%) and ATCC 12478 produced their most frequently observed PFGE pattern (type M) in Japan. In our PFGE study, both DraI and AseI (an isoschizomer of VspI) were used. The most prevalent PFGE pattern with DraI was pattern Da, and it comprised 31 of 71 strains (43.7%). The most common AseI pattern was pattern Aa, which was seen in 30 of 71 (42.3%) of the strains. Lane-by-lane comparison on the same electrophoresis gel showed that the isolates with common PFGE patterns found in the United States (pattern Da), France (pattern Ia), and Japan (type M) gave the same patterns with DraI. The comparison also showed that with AseI our common pattern (pattern Aa) was indistinguishable from the pattern produced by the major group of French isolates (subgroup Ia) and had only one band difference with the most frequently observed pattern (type M) in Japan. Thus, a single common PFGE genotype with DraI or VspI, representing approximately 30% of studied strains, has been observed in all PFGE studies of M. kansasii to date.
In our study, the isolates which had the common DraI pattern (pattern Da) were not in entire accordance with the isolates which produced the common AseI pattern (pattern Aa). As shown in Fig. 2, isolates with the same DraI pattern may exhibit minor band differences with AseI digestion. Of the 31 isolates with the common DraI pattern (pattern Da), only 17 had both the common DraI pattern (pattern Da) and the common AseI pattern (pattern Aa). The geographic sources of the 17 isolates were mainly in Texas. Only 3 of the 17 isolates were from states other than Texas (one from Arkansas, one from Alabama, and one from North Dakota). When the XbaI results were combined with the DraI and AseI results, 7 of the 17 isolates were still indistinguishable. The seven isolates were from three states (five from Texas, one from Arizona, and one from North Dakota) and were discovered in 1987 to 1997. There was no evidence of an epidemiologic relationship between these seven isolates, considering the sources and the years that the isolates were discovered.
Iinuma et al. (6) found that the largest group (type M) within 85 strains tested by PFGE with VspI contained 22 clinical isolates. These isolates were collected from four different districts in Japan. In the study by Alcaide et al. (2), 11 of 51 genotype I clinical isolates from Spain (21.5%) showed a PFGE pattern indistinguishable from those of both type strains and one of M. kansasii genotype I isolates from Switzerland. Iinuma et al. (6) analyzed the 16S rRNA gene sequences of six strains included in their study and verified that four strains, including one with a type M pattern, had sequences identical to that of an M. kansasii genotype isolated principally in Europe (presumably subspecies type I). These findings were quite similar to the results in the present study.
Using an M. kansasii-specific DNA probe (pMK1-9), Ross et al. (15) tested 105 M. kansasii isolates from Australia, Belgium, Japan, South Africa, and Switzerland. Eighty-five probe-positive strains shared most bands in common, with the only very minor variations occurring in the highest-molecular-weight band. This indicated that within the typical M. kansasii group, genetic variability is low. Picardeau et al. (13) found that within each of the five fingerprinting genotype PFGE patterns seen in French isolates, there was a relatively low degree of genotypic divergence. In our study, the PFGE patterns showed that approximately 90% of the isolates collected from 19 states were clonal. Differences between PFGE patterns often involved only one or two bands. Even with different combinations of restriction digestion patterns, there were still 20 isolates in six clusters having indistinguishable patterns. All of these findings indicated that M. kansasii isolates had a marked clonal structure, especially in the most prevalent genotype found in humans, regardless of the geographic sources of the isolates worldwide.
Tenover et al. (20) proposed PFGE criteria for interpreting the relatedness of isolates epidemiologically involved in outbreaks. By their definition, if two or three fragment differences compared with the outbreak pattern are found, the isolate is probably part of the outbreak. If four to six fragment differences are found, the isolate is possibly part of the outbreak. The isolate is considered different only if more than six band differences are seen (presumably with any given enzyme). Obviously, however, these definitions could not be used for investigating outbreaks involving M. kansasii, which has a very tight clonal structure within each subspecies. Isolates belonging to unusual subspecies or with unique PFGE patterns would be relatively easily identified as different by the definition of Tenover et al. Isolates belonging to subspecies I would be difficult to compare.
A key component of studying organism relatedness is the stability of the organisms in individual patients or outbreaks. Iinuma et al. (6) tested additional strains isolated at different times from two patients. The VspI PFGE patterns of each of two strains from the two patients remained unchanged. Our results also showed that multiple isolates from the same patients over time gave indistinguishable PFGE patterns. However, additional studies are needed to establish this observation.
This is the first subspecies molecular characterization of M. kansasii isolates from the United States. The study demonstrates that genotype I is the predominant subspecies, as it is in Europe. PFGE with DraI and AseI showed marked clonality of the clinical isolates, with patterns indistinguishable from or close to those from Europe and Japan. Epidemiological studies of strain relatedness of M. kansasii in suspected outbreaks will be difficult given the higher pattern similarity of most clinical isolates by PFGE, a finding similar to that for other nontuberculous mycobacteria such as Mycobacterium celatum type I and Mycobacterium xenopi.
The majority of U.S. isolates chosen for this study were rifampin resistant. Most had detailed histories that have previously been published (22) and that clearly established them as definite pathogens. Such a selection factor could have biased the molecular genotypes observed if one genotype more than another tends to be associated with rifampin resistance. We have no evidence of such a genotype difference, and the same genotype with each restriction enzyme was the predominant one with both rifampin-resistant and -susceptible isolates (data not shown).
The finding of a single clone of M. kansasii being responsible for most cases of human disease is surprising for an environmental species. Studies of most environmental species, such as M. avium complex, have shown almost as many PFGE patterns as there are isolates (12, 23). A few species, such as M. xenopi, appear to have limited genetic diversity like M. kansasii, however, suggesting that specific factors may select for one or more specific genotypes. These could be colonization factors within piped water systems or, more likely, could be related to one or more virulence factors present in this clone of organisms. This is supported by the finding that five subspecies and multiple organism clones are present in tap water (2, 13) but that only one subspecies and one clone are responsible for up to 90% of clinical disease. It is also possible that M. kansasii is a relatively recently derived species or that the time for genetic change is very slow. The presence of one or more virulence factors in this clone seems the most likely explanation, although little has been learned about these factors.
FIG. 4.
Schematic representation of PFGE patterns with DraI digestion. Lane 1, lambda DNA standards; lane 2, yeast DNA standards; lane 3, common pattern Da; lane 23, ATCC 12478; all other lanes, clinical isolates with different patterns.
REFERENCES
- 1.Ahn, C. H., J. R. Lowell, G. D. Onstad, E. H. Shuford, and G. A. Hurst. 1979. A demographic study of disease due to Mycobacterium kansasii or M intracellulare-avium in Texas. Chest 75:120-125. [DOI] [PubMed] [Google Scholar]
- 2.Alcaide, F., I. Richter, C. Bernasconi, B. Springer, C. Hagenau, R. Schulze-Röbbecke, E. Toptoli, R. Martin, E. C. Böttger, and A. Telenti. 1997. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J. Clin. Microbiol. 35:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Block, K. C., L. Zwerling, M. J. Pletcher, J. A. Hahn, J. L. Gerberding, S. M. Ostroff, D. J. Vugia, and A. L. Reingold. 1998. Incidence and clinical implications of isolation of Mycobacterium kansasii: results of a 5-year, population-based study. Ann. Intern. Med. 129:698-704. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter, J. L., and J. M. Parks. 1991. Mycobacterium kansasii infections in patients positive for human immunodeficiency virus. Rev. Infect. Dis. 13:789-796. [DOI] [PubMed] [Google Scholar]
- 5.Corbett, E. L., M. Hay, G. J. Churchyard, P. Herselman, T. Clayton, B. G. Williams, R. Hayes, D. Mulder, and K. M. De Cock. 1999. Mycobacterium kansasii and M. scrofulaceum isolates from HIV-negative South African gold miners: incidence, clinical significance and radiology. Int. J. Tuberc. Lung Dis. 3:501-507. [PubMed] [Google Scholar]
- 5a.Gaafar, A., M. J. Unzaga, R. Cisterna, F. E. Clavo, E. Urra, R. Ayarza, and G. Martín. 2003. Evaluation of a modified single-enzyme amplified-fragment length polymorphism technique for fingerprinting and differentiating of Mycobacterium kansasii type I isolates. J. Clin. Microbiol. 41:3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iinuma, Y., S. Ichiyama, Y. Hasegawa, K. Shimokata, S. Kawahara, and T. Matsushima. 1997. Large-restriction-fragment analysis of Mycobacterium kansasii genomic DNA and its application in molecular typing. J. Clin. Microbiol. 35:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobson, K. L., R Teira, H. I. Libshitz, I. Raad, K. V. I. Rolston, J. Tarrand, and E. Whimbey. 2000. Mycobacterium kansasii infection in patients with cancer. Clin. Infect. Dis. 30:965-969. [DOI] [PubMed] [Google Scholar]
- 8.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control and Prevention, Atlanta, Ga.
- 9.Klein, J. L., E. L. Corbett, P. M. Slade, R. F. Miller, and R. J. Coker. 1998. Mycobacterium kansasii and human immunodeficiency virus co-infection in London. J. Infect. 37:252-259. [DOI] [PubMed] [Google Scholar]
- 10.Lillo, M., S. Orengo, P. Cernoch, and R. L. Harris. 1990. Pulmonary and disseminated infection due to Mycobacterium kansasii: a decade of experience. Rev. Infect. Dis. 12:760-767. [DOI] [PubMed] [Google Scholar]
- 11.Lortholary, O., F. Deniel, P. Boudon, M. P. Le Pennec, M. Mathieu, M. Soilleux, C. Le Pendeven, P. Loiseau, V. Vincent, D. Valeyre, et al. 1998. Mycobacterium kansasii infection in a Paris suburb: comparison of disease presentation and outcome according to human immunodeficiency virus status. Int. J. Tuberc. Lung Dis. 3:68-73. [PubMed] [Google Scholar]
- 12.Mazurek, G. H., S. Hartman, Y. Zhang, B. A. Brown, J. S. R. Hector, D. Murphy, and R. J. Wallace, Jr. 1993. Large DNA restriction fragment polymorphism in the Mycobacterium avium-M. intracellulare complex: a potential epidemiologic tool. J. Clin. Microbiol. 31:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picardeau, M., G. Prod'hom, L. Raskine, M. P. LePennec, and V. Vincent. 1997. Genotypic characterization of five subspecies of Mycobacterium kansasii. J. Clin. Microbiol. 35:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pintado, V., E. Dómez-Mampaso, P. Martin-Dávila, J. Cobo, E. Navas, C. Quereda, J. Fortúm, and A. Guerrero. 1999. Mycobacterium kansasii infection in patients infected with the human immunodeficiency virus. Eur. J. Clin. Microbiol. Infect. Dis. 18:582-586. [DOI] [PubMed] [Google Scholar]
- 15.Ross, B. C., K. Jackson, M. Yang, A. Sievers, and B. Dwyer. 1992. Identification of a genetically distinct subspecies of. Mycobacterium kansasii. J. Clin. Microbiol. 30:2930-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steadham, J. E. 1980. High-catalase strains of Mycobacterium kansasii isolated from water in Texas. J. Clin. Microbiol. 11:496-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taillard, C., G. Greub, R. Weber, G. E. Pfyffer, T. Bodmer, S. Zimmerli, R. Frei, S. Bassetti, P Rohner, J. Piffaretti, E. Bernasconi, J. Bille, A. Telenti, and G. Prod'hom. 2003. Clinical implication of Mycobacterium kansasii species heterogeneity: Swiss national survey. J. Clin. Microbiol. 41:1240-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produces by pulsed-field electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace, R. J., Jr., Y. Zhang, B. A. Brown, V. Fraser, G. H. Mazurek, and S. Maloney. 1993. DNA large restriction fragment pattern of sporadic and epidemic nosocomial strains of Mycobacterium chelonae and Mycobacterium abscessus. J. Clin. Microbiol. 31:2697-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace, R. J., Jr., D. Dunbar, B. A. Brown, G. Onyi, R. Dunlap, C. H. Ahn, and D. T. Murphy. 1994. Rifampin-resistant Mycobacterium kansasii. J. Infect. Dis. 18:736-743. [DOI] [PubMed] [Google Scholar]
- 23.Wallace, R. J., Jr., Y. Zhang, B. A. Brown, D. Dawson, D. T. Murphy, R. Wilson, and D. Griffith. 1998. Polyclonal Mycobacterium avium complex infections in patients with nodular bronchiectasis. Am. J. Respir. Crit. Care Med. 158:1235-1244. [DOI] [PubMed] [Google Scholar]