Abstract
Cholesterol influences ion-channel function, distribution and clustering in the membrane, endocytosis, and exocytic sorting of the nicotinic acetylcholine receptor (AChR). We report the occurrence of a cholesterol recognition motif, here coined “CARC”, in the transmembrane regions of AChR subunits that bear extensive contact with the surrounding lipid, and are thus optimally suited to convey cholesterol-mediated signaling from the latter. Three cholesterol molecules could be docked on the transmembrane segments of each AChR subunit, rendering a total of 15 cholesterol molecules per AChR molecule. The CARC motifs contribute each with an energy of interaction between 35 and 52 kJ.mol−1, adding up to a total of about 200 kJ.mol−1 per receptor molecule, i.e. ∼40% of the lipid solvation free energy/ AChR molecule. The CARC motif is remarkably conserved along the phylogenetic scale, from prokaryotes to human, suggesting that it could be responsible for some of the above structural/functional properties of the AChR.
Within the superfamily of ligand-gated ion channels, the Cys-loop family comprises several evolutionarily related neurotransmitter receptor proteins, coded by a few hundred genes which function as transducers of various chemical signals. The family includes the nicotinic acetylcholine receptor (AChR), the subtype 3 of the 5-hydroxytryptamine (serotonin, 5-HT3) receptor, and the glycine and gamma aminobutyric acid type A (GABAA) receptors. Of these, the AChR, a cation selective channel, is one of the best characterized members1. Structurally, AChRs are transmembrane (TM) proteins predominantly localized at the postsynaptic membrane of chemically excitable synapses, although some of these macromolecules are also found in the presynaptic nerve terminals or in neuronal somas. Extensive experimental evidence supports the view that cholesterol, a very abundant component of the postsynaptic membrane, modulates the functional properties and distribution of the AChR protein2,3. The mechanisms by which cholesterol-AChR interactions translate into the observed changes in the receptor's ligand binding affinity4 or ion channel properties5 are still not fully understood.
Attempts to identify cholesterol binding sites on the AChR have mainly relied on the use of photoaffinity labeling techniques. Initial experiments using chemically reactive cholesterol analogs led to the identification of the individual AChR subunits targeted by these probes6,7. Subsequent studies with photoactivatable sterols, which are not necessarily bona fide functional cholesterol substitutes, have provided further insight into putative regions of the AChR involved in interactions with cholesterol; the label was found in peptides that contain almost exclusively the α-TM4, α-TM1 and γ-TM4 membrane spanning segments8,9. More recent photoaffinity labeling studies using azido-cholesterol analogs led to the identification of putative cholesterol-AChR interaction sites on the TM segments TM4, TM3, and TM1 of each subunit, with TM4 having the greatest interaction with membrane cholesterol, and in all cases fully overlapping the lipid-protein interface of the AChR10.
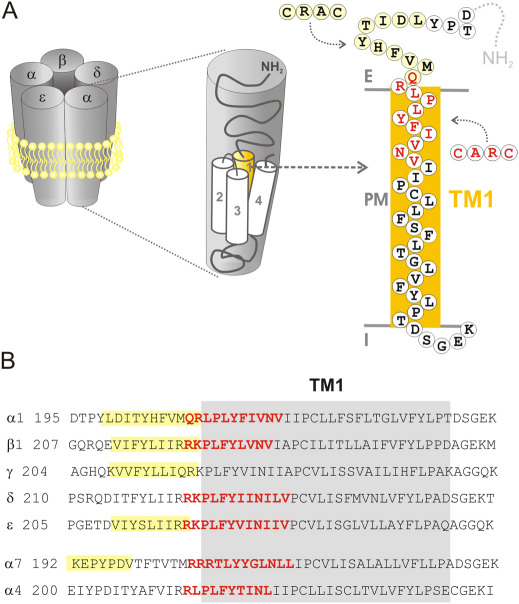
It has been reported that many proteins that interact with cholesterol possess an amino acid consensus sequence termed “CRAC” (cholesterol recognition/interaction amino acid consensus) in their juxtamembrane region, with the pattern –L/V–(X)(1–5)–Y–(X)(1–5)–R/K–, where (X)(1–5) represents between one and five residues of any amino acid11,12,13,14. As a consequence of such a broad definition its occurrence is very high in many proteomes, so that the mere presence of CRAC does not indicate the occurrence of specific protein-cholesterol interactions13. The CRAC sequence has been suggested to relate to the propensity of membrane proteins to be incorporated into cholesterol-rich lipid domains14 and also to promote the segregation of cholesterol into lipid domains15. Given the profound modulatory effects of cholesterol on the structural and functional properties of the AChR we undertook a search for this region in AChR amino acid sequences. Indeed, the sequence was found to be present in virtually all AChR muscle-type AChR subunits in a region immediately adjacent to the TM1 transmembrane domain (Fig. 1). The CRAC domain, however, was found in segments of the AChR polypeptide chain that stretch out of the membrane bilayer, and hence are probably not the energetically most favorable partners for cholesterol in a hydrophobic membrane environment.
Figure 1. (A) Scheme depicting the whole AChR protein molecule inserted in the plasma membrane (left), an individual subunit of the pentamer, showing the four (TM1 to TM4) transmembrane segments (middle), and the amino acid sequence of a single TM domain, corresponding to the human muscle TM1 (right).
All subunits possess a relatively large extracellular amino terminal domain, four TM segments (TM1 to TM4), a large intracellular loop between TM3 and TM4, and a short extracellular C-terminus portion. The CRAC and CARC regions are indicated in the scheme. (B) Alignment of the TM1 (highlighted in gray) amino acid sequences of human muscle and neuronal (α7 and α4) AChR subunits. The cholesterol recognition/interaction amino acid consensus (CRAC) motif [−L/V−(X)(1−5)−Y−(X)(1−5)−R/K−] is highlighted in yellow. CARC, the inverted CRAC sequence, ([−R/K−(X)(1−5)−Y−(X)(1−5)−L/V−]) is indicated by bold red letters. See text for details.
In the course of our analysis, more detailed inspection of the AChR sequences disclosed the occurrence of a new potential cholesterol-recognizing domain with a motif similar to the CRAC sequence but oriented in the opposite direction along the polypeptide chain, constituting an “inverted CRAC” domain, for which reason we coined it “CARC” sequence (Fig. 1). In contrast to CRAC, a search in the AChR family revealed that the newly found CARC motif is totally embedded in the membrane bilayer region and thus appears more suitable for interaction with membrane-bound cholesterol than CRAC. Furthermore, the CARC motif is found to be evolutionarily conserved among members of the AChR family, from prokaryotes to Homo sapiens, and is also present in other members of the Cys-loop family and its bacterial homologs as well as in members of the important family of G-protein coupled receptors.
Results
Initial studies in search of the so-called “CRAC” domain in AChR amino acid sequences, postulated to interact with cholesterol13,14, revealed its presence in almost all subunits of human muscle-type AChR in a region immediately adjacent to TM1 and in mouse TM1 (Fig. 1 and Supplementary Fig. S1 online). The CRAC sequence, which is present in proteins such as caveolin-113,16, was found to occur in the extramembranous amino-terminal region of the AChR but was conspicuously absent from some subunits such as γ in the Torpedo AChR, δ in human muscle AChR, or the α4 subunit in human neuronal AChR (Fig. 1 and Supplementary Fig. S1 online). The CRAC sequence spans amino acids in the TM region and beyond, extending into the extramembranous domain of the AChR.
A closer look at the AChR sequences disclosed that immediately adjacent to the CRAC sequence, a mirror or “inverted” sequence follows within the membrane-embedded region of the AChR, which we refer to as the "CARC" motif. Indeed, the CARC sequence is found in the TM1 segments of human and mouse muscle AChR subunits as well as in Torpedo sp. (with the exception of the γ subunit for the three above-mentioned species) and in neuronal α7 and α4 AChR subunits (Fig. 1 and Supplementary Fig. S1 online). CARC is also found in other members of the Cys-loop family (Supplementary Fig. S1 online). Interestingly, the CARC domain is present in the TM1 of two prokaryote pentameric channel homologs of the AChR, the proteins ELIC and GLIC (Supplementary Fig. S1 online), found in Erwinia chrysanthemi and Gloeobacter violaceus, respectively17,18.
It is also interesting to note that the CARC sequence satisfies several other criteria to qualify as a cholesterol binding motif: i) CARC is present in TM1, a receptor region firmly established as a membrane-spanning domain and, more importantly, to interact with membrane lipids19; ii) there appears to be a good correlation between the amino acids in the CARC motifs in the AChR subunits and some of the residues in TM1 recently postulated by Brannigan et al.20 to be involved in AChR-cholesterol interactions. iii) In addition, the Asp residue at the N-terminal region of the βTM4 segment, postulated by Hamouda et al.10 to interact with cholesterol, is immediately adjacent to the CARC segment.
In order to further define whether the proposed CARC domain is an energetically plausible cholesterol-binding motif we employed molecular modeling techniques to analyze possible interactions between cholesterol and the AChR, using the molecular coordinates of the Torpedo AChR (PDB 2bg9 ref. 21) as a template for the human receptor.
Optimal fit of cholesterol on the CARC domain of α1TM1
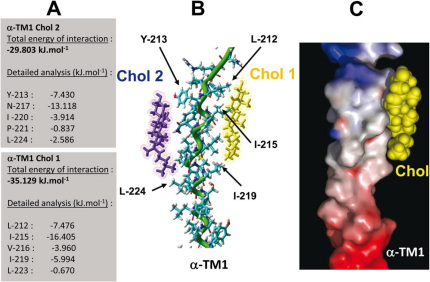
The CARC domain of human α1TM1 is defined as 209RLPLYFIV216 (the underlined amino acids belonging to the TM domain). The definition of the motif is based on a triad of basic (R or K), aromatic (Y), and aliphatic (L or V) residues. Two distinct cholesterol molecules (Chol 1 and Chol 2) fit onto this motif. Because of its charged characteristics, the basic residue (R-209) is located outside the TM domain (see Supplementary Fig. S2 online). Its side chain is oriented towards the same side as the aromatic side chain of Y-213. Although the -OH group of cholesterol could theoretically interact with any of these polar side chains, obviously it can only interact with one. An interaction with R-209 is unlikely because once the -OH group of cholesterol is bound to the guanidinium cation, there is no other available polar group in the cholesterol molecule to accommodate the -OH group of Y-213. Thus an interaction between the -OH group of cholesterol (i.e. Chol 2) and Y-213 is far more likely (Fig. 2). The energy of interaction between Chol 2 and Y-213 was found to be −7.430 kJ.mol−1. There is no other contact between Chol 2 and any other amino acid residue of the CARC domain, yet the fit between Chol 2 and TM1 is quite good, with a total energy of interaction of −29.803 kJ.mol−1. However, a second cholesterol molecule (Chol 1) could be positioned on the opposite face of TM1, establishing several tight contacts with CARC amino acid residues: L-212 (−7.476 kJ.mol−1), I-215 (−16.405 kJ.mol−1), and V-216 (−3.960 kJ.mol−1). The total energy of interaction between Chol 1 and TM1 is −35.129 kJ.mol−1, a significantly better fit than the −23.318 kJ.mol−1 value calculated from the model obtained in ref. 20, which also places a cholesterol molecule in TM1. It should be noted that the best fit has been found for the α face of Chol 1 and the β face of Chol 222. If one tries the symmetric orientations (i.e. β face for Chol 1 and α face for Chol 2) the energy of interaction decreases in both cases, indicating a worse fit for TM1.
Figure 2. Docking of two cholesterol molecules on human AChR α-TM1.
Two cholesterol molecules (Chol 1 and Chol 2) were docked on the αTM1 domain of the human AChR as described in Materials and Methods. A detailed analysis of the energy of interaction of each cholesterol-TM1 complex is shown on the left panel (A). A tube rendering of the model is shown in (B). The α-helix of the TM domain is colored in green. The apolar part of Chol 1 (in yellow) interacts with Ile212, Ile215, and Ile219 (van der Waals interactions), whereas its polar part (the OH group) is located at the apolar-polar interface of the membrane where it can interact with water molecules. The OH group of Chol 2 (in violet) forms an H-bond with the phenolic OH of Tyr213. The surface complementarity between the α-TM1 domain and Chol 2 (in yellow) is illustrated by a surface view of the complex (C). The electrostatic surface of the α-TM1 domain is rendered in blue for positive, red for negative, and grey for neutral regions, respectively.
The obvious question arises: can the whole AChR protein accommodate both Chol 1 and Chol 2? To answer this question, the calculated ternary complex [Chol1-TM1-Chol2] was modeled onto the human AChR structure derived from 2bg9. A clear steric clash became apparent between Chol 2 and the βTM2 domain. This suggests that TM1 can accommodate only one cholesterol molecule, Chol 1. Accommodating a second cholesterol molecule, Chol 2, produces a serious and unacceptable distortion of the protein.
Unfavorable docking of cholesterol on α1TM2
TM2 is the channel-lining, innermost ring of TM segments of the AChR. It does not possess a CARC domain. Two cholesterol molecules could be docked onto the TM2 transmembrane domain (Supplementary Fig. S3 online): one on the extracellular-facing leaflet of the bilayer (Chol 1), and the other on the intracellular leaflet (Chol 2) of the plasma membrane. However, the fit between TM2 and Chol 1 is significantly weaker than with other TM segments (−17.139 kJ.mol−1). In contrast, the fit with Chol 2 is significantly better (−31.695 kJ.mol−1), essentially because of the possibility of an H-bond between the -OH group of Chol 2 and the -OH group in the side chain of T-244 (overall the energy of interaction between Chol 2 and T-244 is −10.074 kJ.mol−1, i.e. one third of the total energy of interaction. See Discussion for more details). In spite of the theoretical possibility of establishing Chol-TM2 contacts, none of the two cholesterol molecules are likely to interact with the intact AChR protein, since both overlap with the α1TM1 domain.
Docking of cholesterol on α1TM3
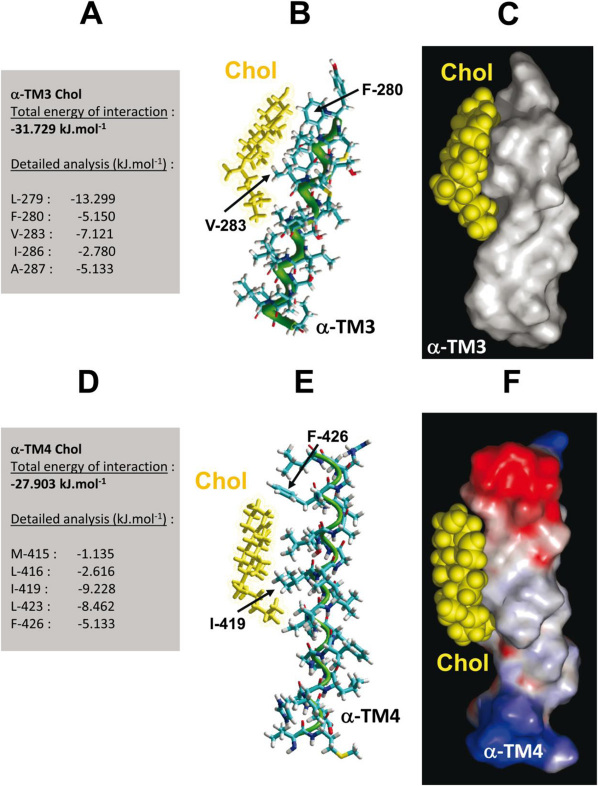
There is no strict CARC domain in TM3, unless the domain definition permits replacement of a Y residue by an F residue. Indeed, the TM3 domain displays a 276KYMLFTMV283 “CARC-like” sequence (the “CARC-like” motif is operationally defined as follows: [R/K]–[X](1–5)–[F]–[X](1–5)–[L/V]). Cholesterol could be docked onto this motif, with a total energy of interaction of −31.729 kJ.mol−1. Most of the interaction involves amino acid residues belonging to this CARC-like domain (Fig. 3). Moreover, the Chol-TM3 complex is fully consistent with the AChR structure.
Figure 3. Docking of cholesterol on α-TM3 and α-TM4 domains of the human AChR.
The predicted interaction between cholesterol and α-TM3 relies exclusively on van der Waals interactions involving aliphatic residues (Leu279, Val283) and the aromatic Phe280 (A). The geometry of the iso-octyl chain of cholesterol is consistent with a bend that is necessary to adapt the form of the lipid molecule to the crevices of the α-TM3 domain as shown in both the tube (B) and surface renderings (C). In the α-TM4/cholesterol complex, cholesterol is not only bent but also twisted on its main axis, allowing an optimal fit for α-TM4 (E, F). As for α-TM3, this complex is chiefly stabilized by van der Waals interactions involving both aliphatic (Ile419, Leu423) and aromatic (Phe426) residues (D).
Docking of cholesterol on α1TM4
An energetically favorable fit was also found between TM4 and cholesterol (−27.903 kJ.mol−1). The sequence interacting with cholesterol lies between amino acid residues M-415 and F-426, resembling a “CRAC-like” motif 423LAVFAGR429 (Fig. 3), i.e. the CRAC motif with an F instead of a Y residue in the sequence: [L/V]–[X](1–5)–[F]–[X](1–5)–[R/K]. This fit is also consistent with the AChR structure.
Docking of cholesterol molecules to the whole AChR and energetics of cholesterol-AChR interactions
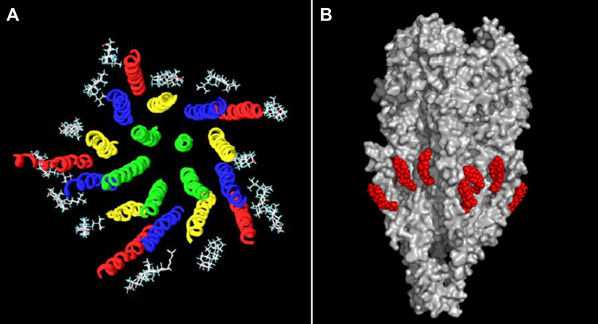
As shown in Fig. 4, a total of fifteen cholesterol molecules could be accommodated around the belt of the AChR macromolecule, in accordance with early work by Narayanaswami and McNamee23 and Mantipragada et al.24. This molecular arrangement does not produce distortions of the whole AChR molecular structure. The free energy of interaction between cholesterol molecules and the AChR is about −510/−530 kJ.mol−1, i.e. more than −100 kJ.mol−1 per subunit. The particularly good fit between the “CARC-like” γTM4 domain from human AChR (428RVCFLAML435) and cholesterol is noteworthy, with an energy of interaction of about −60 kJ.mol−1, i.e. ∼60% of the total energy of interaction of the entire γ subunit, which exhibits the highest affinity for cholesterol (cf. Table 1).
Figure 4. Docking of 15 cholesterol molecules onto the whole human AChR.
(A) End-on view ribbon rendering of the human AChR (α2βδε) with 15 cholesterol molecules (grey), in contact with the outer ring29, made up of TM4 segments (red), and middle ring, made up up TM1 (yellow) and TM3 (blue) transmembrane segments. Note that the inner ring, which lines the walls of the ion channel (TM2 domains, green), does not establish contact with cholesterol molecules. (B) Side view of the human AChR (in grey) with energy-minimized cholesterol molecules (red) docked on the surface of its transmembrane region.
Table 1. Energetics of interaction of cholesterol and the CARC, CARC-like and CRAC-like motifs in the transmembrane domains of human muscle-type AChR.
| AChR TM domain | Energy of interaction (kJ.mol−1) |
|---|---|
| αTM1 | −35.129 |
| αTM3 | −31.729 |
| αTM4 | −27.903 |
| Total α subunit | −94.761 |
| βTM1 | −52.332 |
| βTM3 | −20.808 |
| βTM4 | −26.241 |
| Total β subunit | −99.453 |
| γTM1 | −30.542 |
| γTM3 | −37.066 |
| γTM4 | −59.961 |
| Total γ subunit | −127.569 |
| δTM1 | −46.184 |
| δTM3 | −33.083 |
| δTM4 | −29.197 |
| Total δ subunit | −108.464 |
| εTM1 | −44.438 |
| εTM3 | −24.421 |
| εTM4 | −44.050 |
| Total ε subunit | −112.909 |
| Embryonic AChR1 (α2βγδ) | −525.008 |
| Adult AChR1 (α2βεδ) | −510.348 |
1The stoichiometric contribution of two α subunits is taken into account in the estimation of the total energy of interaction of the AChR pentamer with cholesterol molecules.
Presence of CARC domains in G protein-coupled receptors (GPCRs)
Is the CARC motif exclusive to the Cys-loop receptor superfamily? To address this question, we investigated its presence in another very important group of membrane proteins, the G-protein coupled receptors (GPCRs), which are documented to interact with cholesterol in lipid microdomains25. Indeed, the CARC domain is conspicuously present in the GPCR superfamily (Supplementary Fig. S4 online). We further narrowed down the search to GPCRs involved in Alzheimer's disease25,26, because of the purported link between neuronal AChRs and cholesterol in this pathology27. As shown in Supplementary Fig. S4B (online), the CARC and CRAC domains are present in GPCRs involved in Alzheimer's disease. In order to quantify the degree of interaction between cholesterol and the CARC domain in GPCRs we chose a TM domain of the somatostatin receptor (Supplementary Fig. S5 online) known to interact with cholesterol28. Interestingly, this GPCR was found to contain CARC and CRAC sequences in the same TM segment (Figs. S4B–S5). As shown in Supplementary Fig. S5 (online), both CARC and CRAC domains bind cholesterol with high affinity, the interaction with CARC being 23% stronger.
Discussion
In the present work we show the existence of CARC, CARC-like and CRAC-like amino acid consensus patterns in the TM segments of the AChR that are ideally suited to interact with cholesterol molecules present in the vicinal lipid microenvironment of the macromolecule2,3. These interaction sites are in full accordance with published experimental data2,29. A more recent proposal20 is based on the observation of "gaps" in the electron density maps of the AChR cryoelectron microscopy images of Unwin and colleagues21: up to 5 cholesterol molecules could be accommodated into these deeply buried inter-subunit "holes", with a total of 15 cholesterol molecules per receptor molecule. Both Hamouda et al.10 and our laboratory24 had calculated the same number of cholesterol molecules from independent photolabeling and electron spin resonance experimental data, respectively, but unlike the deeply buried cholesterols postulated from in silico calculations in the more recent proposal20, all the cholesterol molecules readily exchange with bulk lipids10,24 (Supplementary Fig. S6 online).
Three cholesterol molecules could be docked on the transmembrane segments TM1, TM3 and TM4 of each of the five human AChR subunits, with energies of interaction per subunit ranging between −90 and −130 kJ.mol−1 (Table 1). The highest energetic contribution stemmed from the CARC domains in the TM1 segments, each with an energy of interaction between 35 and 52 kJ.mol−1, adding up to a total of about 200 kJ.mol−1 per AChR molecule, that is, ca. 40% of the total lipid solvation free energies for the whole AChR molecule30.
The best fit was found for the CARC-like motif present in the human AChR γTM4 segment (Table 1). The latter constitutes an exceptional case, in as much as it supplies an energy of interaction (−60 kJ.mol−1) amounting to roughly half the total energy contributed by the other TM segments in the γ subunit, which incidentally provides the highest energetic contribution of all subunits (Table 1). The AChR γTM4 segment exhibits other interesting features: in a pure phospholipid (phosphatidyl-oleoyl phosphatidylcholine) system in the ld phase, γTM4 was found to occur as an isolated peptide, matching the hydrophobic region of the bilayer; in contrast, if the same phospholipid system contains cholesterol (and is thus in the lo phase) γTM4 maximizes homophilic hydrophobic interactions and forms bundles with other γTM4s31.
The CARC domain identified in TM1 exhibits a comparable or even higher energy of interaction than the values found for lipophilic xenobiotics such as general anesthetics in the AChR32. The energetics of interaction between CARC in TM1 and cholesterol do not involve any H-bond, but rather van der Waals interactions among cholesterol and non-polar side chains in TM1. Several other fits were tested between cholesterol and TM domains, but most of them resulted in steric clashes, implying that the AChR protein could not accommodate more cholesterol molecules without significant structural distortions. A recent study employing molecular dynamics simulations disclosed interactions between cholesterol and the TM1 and TM4 segments of the neuronal (α4β2) subtype of AChR33. It is worth noting that the amino acids identified by these authors fit very well with the CARC consensus motif disclosed in the present work.
The TM3-cholesterol complex involves a “CARC-like” domain in which the aromatic Tyr residue is replaced by Phe, another aromatic residue. A similarly “F/Y” modified “CRAC-like” domain is also involved in the interaction between cholesterol and TM4. As mentioned above, the best fit in the present study was obtained with the CARC motif present in TM1. Overall, the energy of interaction between cholesterol and individual amino acid residues in human AChR TM domains are compatible with previous values calculated for an unrelated cholesterol binding site34. For instance, our values of −9.228 kJ.mol−1 for I-419 in TM4 and −16.405 kJ.mol−1 for I-215 in TM1 are consistent with the −10.032 and −10.868 kJ.mol−1 calculated for I-167 and I-203 respectively in the Osh4 protein.
The molecular mechanisms underlying the physical interaction between cholesterol and CARC/CARC-like domains warrant further experimental study. Inspection of the specific amino acid requirements of the CARC/CARC-like consensus motif already reveals several interesting features. The motif is based on a sequence of amino acids which includes, from the N-term to C-term endings, a basic (K/R), an aromatic (Y/F), and a branched aliphatic residue (L/V). However, this does not imply that all these residues interact with cholesterol. Indeed, there is no evidence in our modeling studies of a direct interaction between the basic residue of CARC and cholesterol. Instead, we assume that the presence of K/R ensures that the CARC motif is correctly positioned at the polar-apolar interface of a TM domain, just where cholesterol is supposed to be. This is due to the ‘snorkeling' effect35 which is attributable to the burial of the long and flexible side chain of Lys (or Arg) in the hydrophobic region of the membrane, whereas the cationic ε-NH3+ group ‘breathes' at the more polar interface region36. As a consequence of this, if Lys and Arg residues are snorkeling, they can be present in a TM domain and be followed by several hydrophobic residues which also reside in the membrane. In addition, since arginine and lysine residues are more frequently found in the N-terminal than in the C-terminal third of a TM domain, snorkeling is an asymmetric phenomenon36. The specific topology of the CARC sequence (K/R…Y/F…L/V, from the N-terminus to the C-terminus) implies that the key residues following K/R in the motif (i.e. Y/F and L/V) actually belong to a TM domain. Therefore, even if there is no direct interaction between cholesterol and K/R, the basic amino acid in the first position is critical for identifying a functional CARC motif belonging to a TM domain. Incidentally, this also explains why the CRAC motif, which has an inverse topology (L/V…Y… K/R, from the N-terminus to the C-terminus), does not always belong to a TM domain14. Moreover, the Tyr residue (an absolute requirement in the case of CRAC) can be replaced by Phe in the CARC motif. Since CRAC is a juxtamembrane motif, the phenol group of Tyr is required to form a H-bond with the OH group of cholesterol14, and this would not be possible with Phe. In the case of CARC, the interaction between the aromatic amino acid and cholesterol occurs in the apolar region of the membrane, far from the OH group of cholesterol. Thus the interaction with cholesterol is mediated by the stacking between the aromatic ring of the amino acid (either Y or F) and one of the cycles of cholesterol. The energy of interaction between the aromatic amino acids of AChR TM3 and TM4 domains is about −5 kJ/mol, which is fully consistent with this type of interaction. Finally, the requirement for L/V is justified by the need to accommodate the crevices and asperities of the cholesterol molecule22 and there are numerous van der Waals contacts between these residues and cholesterol in the TM domains of the AChR (Figs. 2–4).
The newly discovered consensus cholesterol-recognition motif, CARC, covers a wide evolutionary span, ranging from the fairly “recent” Torpedo sp. to Homo sapiens and even to the bacterial pentameric channels, structural17,18,37,38 homologs of the AChR recently found in prokaryotes, i.e. the cyanobacterium Gloebacter violaceous and its orthologue from Erwinia chrysanthemi. Even though the presence of cholesterol in the plasma membrane is an almost exclusive attribute of eukaryotes, there are a few examples of the presence of sterols in bacteria39,40. Cyanobacteria possess hopanoids, which are structurally and functionally similar to sterols40. Thus, the preservation of the CARC motif through such a wide span in the evolutionary scale -from prokaryotes to humans- could indicate that this domain has an important structural and/or functional role. In support of this hypothesis is the extensive experimental work showing that mutations in amino acids in the transmembrane regions of the AChR alter channel gating (see review in41). Interestingly, some of these functionally relevant mutations are very close to or within CARC/CARC-like domains. Another possibility is that this motif initially had no functional properties in prokaryotic AChR-like proteins, but upon appearance of cholesterol in the course of phylogeny it acquired a role in higher organisms for transducing regulatory signals from the adjacent plasma membrane lipid moiety to the protein moiety. We would like to entertain the idea that the CARC domain present in the AChR and its bacterial homologues is involved in some of the observed effects of cholesterol on AChR function and distribution (see review in41). Future mutational and biophysical studies should be oriented to determining the functional relevance of the CARC motif in the AChR. Finally, we have shown that the CARC motif is also present in the TM domains of the important group of GPCRs (Supplementary Figs. S4 and S5 online). Interestingly, both GPCRs and AChR are thought to play a role in the pathology of Alzheimer's disease25,26,27. Altogether, these data provide an interesting link between Alzheimer's disease, high cholesterol, and neurotransmitter receptors including AChR and GPCRs displaying CARC motifs in their TM domains.
Methods
The definition of the motif is based on a triad of basic (R or K), aromatic (Y), and aliphatic (L or V) residues along the primary sequence. Search for the presence and localization of the consensus sequence for the "CRAC" motif, [L/V]–[X](1–5)–[Y]–[X](1–5)–[R/K]11,12,13,15 or the "CARC" motif introduced in the present work, [R/K]–[X](1–5)–[Y]–[X](1–5)–[L/V], as well as the “CARC-like” motif [R/K]–[X](1–5)–[F]–[X](1–5)–[L/V] and the corresponding “CRAC-like” motif [L/V]–[X](1–5)–[F]–[X](1–5)–[R/K], was carried out on the AChR subunits and other Cys-loop receptor sequences using the Fuzzpro application of the Jemboss software (European Molecular Biology Open Software Suite, EMBOSS). The same procedure was also applied to analyze the sequences of the large family of G-protein coupled receptors (GPCRs).
To analyze the interaction between cholesterol and the TM domains of the AChR we applied a modeling procedure derived from the fragment-based approach42. The TM domains of the human AChR protein (Swiss-Prot entries P02708, P11230, P07510, Q07001, and Q04844 respectively for the α,β,γ,δ, and ε subunits) were generated by structural homology modeling with the Swiss-Pdb modeling software, using the molecular coordinates of the Torpedo AChR (PDB No. 2bg9, ref. 21) as a template. Cholesterol-TM complexes were then modeled with Hyperchem 8 (Hypercube Inc. Gainesville, FL). In each case, one cholesterol molecule was initially positioned in the vicinity of the TM domain to undertake the manual search for geometric fit. Geometry optimization of each cholesterol-TM complex was first achieved using the unconstrained optimization rendered by the Polak-Ribière conjugate gradient algorithm43. Molecular dynamics simulations were then performed in vacuo for 10 ns with the Bio+ (CHARMM) force field of the Hyperchem 8 software suite34. The ligand (cholesterol)-protein energy of interaction was determined with the Molegro Molecular Viewer. Briefly, the piece-wise linear potential method was applied44. The method takes into account two different sets of parameters: one set for approximating the steric (van der Waals) term between atoms and another set for stronger potentials (hydrogen bonds). Several initial conditions were tested (with distinct orientations of cholesterol with respect to the given TM domain) to select those cholesterol-TM complexes having the highest energy of interaction. Individual cholesterol-TM complexes were then superimposed step-by-step with the structure of the AChR protein using the fit menu of the Swiss-Pdb program. This allowed us to identify any potential steric clash induced by the presence of cholesterol molecules bound to the receptor. Fine conformational tuning was performed with Hyperchem 8 to generate the final model of the human AChR protein complexed with 15 cholesterol molecules.
Author Contributions
C.J.B., J.F. and F.J.B. designed research, performed research, analyzed data and wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported in part by grants from the Ministry of Science and Technology of Argentina (MINCyT), the Scientific Research Council of Argentina (CONICET); Philip Morris USA Inc. and Philip Morris International to F.J.B. Thanks are due to Dr. M. Palmer, Dept. Chemistry, Univ. Waterloo, Ontario, Canada, for help with the use of the Fuzzpro application of the European Molecular Biology Open Software Suite, EMBOSS.
References
- Taly A., Corringer P. J., Guedin D., Lestage P., & Changeux J. P. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat. Rev Drug Discov. 8, 733–750 (2009). [DOI] [PubMed] [Google Scholar]
- Barrantes F. J. Structural basis for lipid modulation of nicotinic acetylcholine receptor function. Brain Res. Brain Res. Rev. 47, 71–95 (2004). [DOI] [PubMed] [Google Scholar]
- Barrantes F. J. Cholesterol effects on nicotinic acetylcholine receptor: cellular aspects. Subcell. Biochem. 51, 467–487 (2010). [DOI] [PubMed] [Google Scholar]
- Criado M., Eibl H., & Barrantes F. J. Functional properties of the acetylcholine receptor incorporated in model lipid membranes. Differential effects of chain length and head group of phospholipids on receptor affinity states and receptor-mediated ion translocation. J Biol. Chem. 259, 9188–9198 (1984). [PubMed] [Google Scholar]
- Borroni V. et al.. Cholesterol depletion activates rapid internalization of submicron-sized acetylcholine receptor domains at the cell membrane. Mol. Membr. Biol. 24, 1–15 (2007). [DOI] [PubMed] [Google Scholar]
- Fernandez A. M., Fernandez-Ballester G., Ferragut J. A., & Gonzalez-Ros J. M. Labeling of the nicotinic acetylcholine receptor by a photoactivatable steroid probe: effects of cholesterol and cholinergic ligands. Biochim. Biophys. Acta 1149, 135–144 (1993). [DOI] [PubMed] [Google Scholar]
- Middlemas D. S. & Raftery M. A. Identification of subunits of acetylcholine receptor that interact with a cholesterol photoaffinity probe. Biochemistry 26, 1219–1223 (1987). [DOI] [PubMed] [Google Scholar]
- Blanton M. P., Xie Y., Dangott L. J., & Cohen J. B. The steroid promegestone is a noncompetitive antagonist of the Torpedo nicotinic acetylcholine receptor that interacts with the lipid-protein interface. Mol. Pharmacol. 55, 269–278 (1999). [DOI] [PubMed] [Google Scholar]
- Corbin J., Wang H. H., & Blanton M. P. Identifying the cholesterol binding domain in the nicotinic acetylcholine receptor with [125I]azido-cholesterol. Biochim. Biophys. Acta 1414, 65–74 (1998). [DOI] [PubMed] [Google Scholar]
- Hamouda A. K., Chiara D. C., Sauls D., Cohen J. B., & Blanton M. P. Cholesterol interacts with transmembrane alpha-helices M1, M3, and M4 of the Torpedo nicotinic acetylcholine receptor: photolabeling studies using [3H]Azicholesterol. Biochemistry 45, 976–986 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 139, 4991–4997 (1998). [DOI] [PubMed] [Google Scholar]
- Li H., Yao Z., Degenhardt B., Teper G., & Papadopoulos V. Cholesterol binding at the cholesterol recognition/ interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc. Natl. Acad. Sci. U. S. A 98, 1267–1272 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R. M. Proteins and cholesterol-rich domains. Biochim. Biophys. Acta 1778, 1576–1582 (2008). [DOI] [PubMed] [Google Scholar]
- Epand R. M., Thomas A., Brasseur R., & Epand R. F. Cholesterol interaction with proteins that partition into membrane domains: an overview. Subcell. Biochem. 51, 253–278 (2010). [DOI] [PubMed] [Google Scholar]
- Epand R. F., Sayer B. G., & Epand R. M. The tryptophan-rich region of HIV gp41 and the promotion of cholesterol-rich domains. Biochemistry 44, 5525–5531 (2005). [DOI] [PubMed] [Google Scholar]
- Epand R. M. Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res. 45, 279–294 (2006). [DOI] [PubMed] [Google Scholar]
- Hilf R. J. & Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 (2009). [DOI] [PubMed] [Google Scholar]
- Bocquet N. et al.. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 (2009). [DOI] [PubMed] [Google Scholar]
- Barrantes F. J. Lipid matters: nicotinic acetylcholine receptor-lipid interactions (Review). Mol. Membr. Biol. 19, 277–284 (2002). [DOI] [PubMed] [Google Scholar]
- Brannigan G., Henin J., Law R., Eckenhoff R., & Klein M. L. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. U. S. A 105, 14418–14423 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J. Mol. Biol. 346, 967–989 (2005). [DOI] [PubMed] [Google Scholar]
- Fantini J. & Barrantes F. J. Sphingolipid/cholesterol regulation of neurotransmitter receptor conformation and function. Biochim. Biophys. Acta 1788, 2345–2361 (2009). [DOI] [PubMed] [Google Scholar]
- Narayanaswami V. & McNamee M. G. Protein-lipid interactions and Torpedo californica nicotinic acetylcholine receptor function. 2. Membrane fluidity and ligand-mediated alteration in the accessibility of gamma subunit cysteine residues to cholesterol. Biochemistry 32, 12420–12427 (1993). [DOI] [PubMed] [Google Scholar]
- Mantipragada S. B. et al.. Lipid-protein interactions and effect of local anesthetics in acetylcholine receptor-rich membranes from Torpedo marmorata electric organ. Biochemistry 42, 9167–9175 (2003). [DOI] [PubMed] [Google Scholar]
- Maudsley S., Martin B., & Luttrell L. M. G protein-coupled receptor signaling complexity in neuronal tissue: implications for novel therapeutics. Curr. Alzheimer Res. 4, 3–19 (2007). [DOI] [PubMed] [Google Scholar]
- Thathiah A. & De S. B. The role of G protein-coupled receptors in the pathology of Alzheimer's disease. Nat. Rev. Neurosci. 12, 73–87 (2011). [DOI] [PubMed] [Google Scholar]
- Barrantes F. J., Borroni V., & Valles S. Neuronal nicotinic acetylcholine receptor-cholesterol crosstalk in Alzheimer's disease. FEBS Lett. 584, 1856–1863 (2010). [DOI] [PubMed] [Google Scholar]
- Reversi A., Rimoldi V., Brambillasca S., & Chini B. Effects of cholesterol manipulation on the signaling of the human oxytocin receptor. Am. J. Physiol Regul. Integr. Comp Physiol 291, R861–R869 (2006). [DOI] [PubMed] [Google Scholar]
- Barrantes F. J. Modulation of nicotinic acetylcholine receptor function through the outer and middle rings of transmembrane domains. Curr. Opin. Drug Discov. Devel. 6, 620–632 (2003). [PubMed] [Google Scholar]
- Sandermann H. Jr High free energy of lipid/protein interaction in biological membranes. FEBS Lett. 514, 340–342 (2002). [DOI] [PubMed] [Google Scholar]
- De Almeida R. F. et al.. Cholesterol modulates the organization of the gammaM4 transmembrane domain of the muscle nicotinic acetylcholine receptor. Biophys. J 86, 2261–2272 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher S., Altschuh J., & Sandermann H. Jr The lipid/protein interface as xenobiotic target site: kinetic analysis of the nicotinic acetylcholine receptor. J. Biol. Chem. 276, 42191–42195 (2001). [DOI] [PubMed] [Google Scholar]
- Cheng M. H., Xu Y., & Tang P. Anionic lipid and cholesterol interactions with alpha4beta2 nAChR: insights from MD simulations. J. Phys. Chem. B 113, 6964–6970 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. P., Brooks B. R., & Klauda J. B. Binding and release of cholesterol in the Osh4 protein of yeast. Proteins 75, 468–477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest J. P., De L. H., Dohlman J. G., Brouillette C. G., & Anantharamaiah G. M. Amphipathic helix motif: classes and properties. Proteins 8, 103–117 (1990). [DOI] [PubMed] [Google Scholar]
- Strandberg E. & Killian J. A. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 544, 69–73 (2003). [DOI] [PubMed] [Google Scholar]
- Bocquet N. et al.. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445, 116–119 (2007). [DOI] [PubMed] [Google Scholar]
- Hilf R. J. & Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008). [DOI] [PubMed] [Google Scholar]
- Pearson A., Budin M., & Brocks J. J. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U. S. A 100, 15352–15357 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summons R. E., Bradley A. S., Jahnke L. L., & Waldbauer J. R. Steroids, triterpenoids and molecular oxygen. Philos. Trans. R. Soc. Lond B Biol. Sci. 361, 951–968 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrantes F. J. Cholesterol effects on nicotinic acetylcholine receptor. J Neurochem. 103 Suppl 1 , 72–80 (2007). [DOI] [PubMed] [Google Scholar]
- Rezác J. & Salahub D. R. Multilevel Fragment-Based Approach (MFBA): A Novel Hybrid Computational Method for the Study of Large Molecules. J. Chem. Theory Comput. 6, 91–99 (2010). [DOI] [PubMed] [Google Scholar]
- Yahi N., Aulas A., & Fantini J. How cholesterol constrains glycolipid conformation for optimal recognition of Alzheimer's beta amyloid peptide (Abeta1-40). PLoS. One. 5, e9079 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R. & Christensen M. H. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 49, 3315–3321 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information