Abstract
Pre-mRNA splicing deposits multi-protein complexes, termed exon junction complexes (EJCs), on mRNAs near exon-exon junctions. The core of EJC consists of four proteins, eIF4AIII, MLN51, Y14 and Magoh. Y14 is a nuclear protein that can shuttle between the nucleus and the cytoplasm, and binds specifically to Magoh. Here we delineate a Y14 nuclear localization signal that also confers its nuclear export, which we name YNS. We further identified a 12-amino-acid peptide near Y14's carboxyl terminus that is required for its association with spliced mRNAs, as well as for Magoh binding. Furthermore, the Y14 mutants, which are deficient in binding to Magoh, could still be localized to the nucleus, suggesting the existence of both the nuclear import pathway and function for Y14 unaccompanied by Magoh.
Pre-mRNA splicing is an essential step for gene expression in metazoans as it is the process that removes introns from mRNA precursors and thereby produces mRNAs. The spliceosome that carries out the splicing of each intron recruits to the mRNA-specific proteins, including Y14, that remain bound to the mRNA after splicing and nuclear export, communicating to the cytoplasm the splicing history of the mRNA1. A specific group of proteins binds near the exon-exon junctions of mRNAs in the nucleus2,3,4. This complex is called an exon junction complex (EJC)2. Although many proteins including transiently associated proteins were identified as components of this complex, the core of EJC consists of four proteins, eIF4AIII, MLN51, Y14 and Magoh5,6,7. EJC provides a link between pre-mRNA splicing and its downstream events. For example, EJC components tethered in the coding region of mRNA enhance translation8, and EJC plays a role in nonsense-mediated mRNA decay (NMD). NMD is a quality control process that selectively degrades mRNAs harboring pre-mature termination codons9,10. Indeed, one of the NMD factors, Upf3, was shown to be an EJC component2,4,6,11,12,13,14, and two of the EJC components, RNPS1 and, to a lesser extent, Y14, were demonstrated to cause NMD when they are tethered downstream of a stop codon15,16.
Among the EJC components, EJC core components are thought to remain bound to mRNAs in the cytoplasm to form ‘a cytoplasmic EJC' after export from the nucleus1,2,4,17,18. In Drosophila, it has been shown that eIF4AIII, Barentsz (Drosophila MLN51), Tsunagi (Drosophila Y14) and Mago nashi are required for posterior localization of oskar mRNA in oocytes19,20. It has also been demonstrated that splicing of the first intron in oskar pre-mRNA is required for its posterior localization in oocytes, which likely indicates that deposition of EJC in the first exon is critical for oskar mRNA localization21. New physiological roles of EJC have been continuously reported. The eIF4AIII protein, one of the core components of EJC, has an important role in neuronal protein expression, likely as cEJC22. Additional roles of EJC in controlling splicing were also demonstrated in Drosophila and human23,24,25.
Y14 was first isolated as a specific interactor of Importin 51. This protein binds preferentially to mRNAs that are produced by splicing, but not to pre-mRNA, intron or intronless mRNA1. Y14 is predominantly nuclear but it rapidly shuttles between the nucleus and the cytoplasm1. Therefore, it was expected that Y14 contains both functional nuclear localization signal (NLS) and functional nuclear export signal (NES). Y14 was also demonstrated to form a heterodimer specifically with Magoh17,18. Crystal structure studies of Y14-Magoh heterodimer revealed an atypical function of the RNA-binding domain of Y14 in mediating interaction with its protein binding partner Magoh26,27,28. It was also shown that Y14-Magoh complex is imported into the nucleus by Importin 1329,30.
In this paper, we report functional domain analyses of the Y14 protein. We show that Y14 has overlapping but non-identical NLS and NES in the amino terminal (N-terminal) region. Since Y14 NLS also confers NES activity, this region is named Y14 nuclear shuttling sequence (YNS). Deletion analysis of the carboxyl terminus (C-terminus) of Y14 reveals that amino acids (aa) 148 to 159 are required not only for binding to its protein partner Magoh but also for association with spliced mRNAs. Y14 mutants deficient in Magoh binding still localize to the nucleus, suggesting that Y14 could be imported into the nucleus independent of its association with Magoh via YNS-mediated pathway.
Results
Amino terminus of Y14 confers both NLS and NES activities
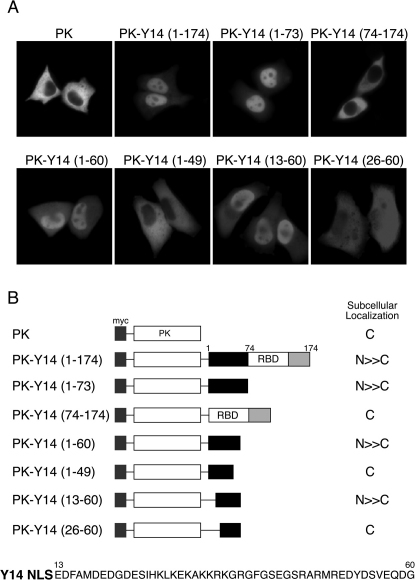
As previously shown, Y14 is mostly localized in the nucleus, while a very small amount of Y14 can be detected in the cytoplasm1. We have also demonstrated that Y14 can shuttle between the nucleus and the cytoplasm1 and it remains associated with mRNAs in the cytoplasm until they are translated31. From these findings, we concluded that Y14 has both a functional nuclear localization signal (NLS) and a functional nuclear export signal (NES). However, neither of these signal sequences of Y14 was identified. First, in order to identify Y14 NLS, a series of expression vectors encoding myc-tagged pyruvate kinase (PK)32 were prepared. PK has been used as a reporter protein for NLS identification, since it is exclusively localized in the cytoplasm (Figure 1A)32,33. A myc-tagged epitope was fused to the N-terminus of PK, which allowed us to detect the localization of a chimera protein. Similar to the localization of endogenous Y141, full-length Y14 fused to PK is localized mostly in the nucleus with a weak signal in the cytoplasm (Figure 1A). We found that the fusion of N-terminal 73 aa of Y14 to PK (PK-Y14(1-73)) produced mostly nuclear protein, while PK fused to the rest of Y14 protein (PK-Y14(74-174)) showed exclusively cytoplasmic staining (Figure 1A), suggesting that Y14 has one NLS in its N-terminal region. Further deletions of Y14(1-73) from both N- and C-termini revealed that the smallest fragment conferring nuclear localization mostly the same as that with full-length Y14 corresponds to aa 13-60. Figure 1B summarizes the results described above and displays the minimal sequence of Y14 NLS. Y14 NLS contains a stretch of basic residues in the middle, resembling a classical basic NLS. However, this basic stretch alone is not sufficient to function as an NLS, as PK-Y14(26-60) does not localize to the nucleus (Figure 1A).
Figure 1. Delineation of the nuclear localization signal (NLS) of Y14.
(A) HeLa cells were transfected with expression vectors encoding myc-tagged pyruvate kinase (PK) fusion proteins. The numbers above each panel correspond to the aa within Y14 that were fused to PK. The subcellular localization of each protein was determined by indirect immunofluorescence using anti-myc monoclonal antibody, 9E10. (B) Schematic drawing of the PK fusion proteins and summary of their intracellular localization as displayed in (A). Also shown is the sequence of the minimally defined Y14 NLS.
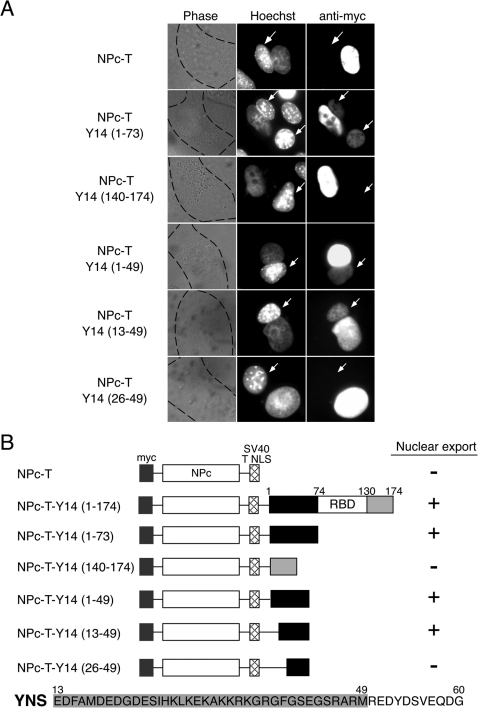
Next, we tried to identify the nuclear export signal (NES) of Y14. To achieve this, we carried out the heterokaryon assays based on a reporter construct containing nucleoplasmin core (NPc) domain fused with Simian Virus 40 (SV40) Large T antigen NLS34, designated as NPc-T. The NPc forms highly stable oligomers that remain in the nucleus unless coupled to a strong NES. This construct has been used to test whether proteins contain NESs or not32,34,35. A myc-peptide epitope was fused to the N-terminus of all constructs to allow detection of all the chimeric proteins. To monitor the shuttling of the chimera proteins, cells were fixed and stained with anti-myc antibodies after 2 hours of incubation. As the first series of experiments, we fused the NPc reporter protein with aa 1 to 73, 74 to 139 or 140 to 174 of Y14 and designated those proteins as NPc-T-Y14(1-73), NPc-T-Y14(74-140) and NPc-T-Y14(140-174), respectively. We have already demonstrated that the full-length Y14 fused to the Npc reporter protein can shuttle between the nucleus and the cytoplasm1. When NPc-T-Y14(1-73) was examined in the heterokaryon assay, it rapidly shuttled in heterokaryons, while the reporter protein alone (NPc-T) and NPc-T-Y14(140-174) did not show shuttling activity (Figure 2A). We could not test the shuttling activity of NPc-T-Y14(74-140) because we could not see the complete nuclear localization of chimera protein (data not shown). These results indicate that the N-terminus of Y14, which has NLS activity, also contains NES.
Figure 2. Delineation of the nuclear export signal (NES) of Y14.
(A) Expression vectors encoding various nucleoplasmin core (NPc) fusion proteins were transfected into HeLa cells. After expression of the transfected DNAs, the cells were fused with NIH3T3 cells to form heterokaryons and incubated in media containing 100μg/ml cycloheximide for a period of 2 hrs. The cells were then fixed and stained for immunofluorescence microscopy with a monoclonal antibody 9E10 (anti-myc panel) to detect the fusion proteins, and with Hoechst 33258 (Hoechst panel) that discriminates the human and mouse nuclei within the heterokaryon (the mouse nuclei are shown by arrows). The phase panel shows the phase contrast image of the heterokaryons with the cytoplasmic edge highlighted with black dashed lines. (B) Schematic drawing of the NPc fusion proteins and summary of the heterokaryon results as depicted in (A). The sequences of the minimally defined YNS domain are shown below, and the sequences sufficient for NES activity are highlighted in gray shading.
To determine the minimum NES sequence, further deletion constructs were made for the heterokaryon assays. The results shown in Figure 2A and summarized in Figure 2B show that aa 13-49 represent the smallest fragment that is sufficient to maintain NES activity of Y14. Further deletion from the N-terminus of this fragment severely inactivates the NES activity (Figure 2, NPc-T-Y14(26-49)). These results indicate that the NLS and NES activities of Y14 overlap, although NES is shorter than NLS. We refer to aa 13-60 of Y14 as Y14 nuclear shuttling sequence (YNS).
Carboxyl terminus of Y14 is required for its association with both spliced mRNA and Magoh
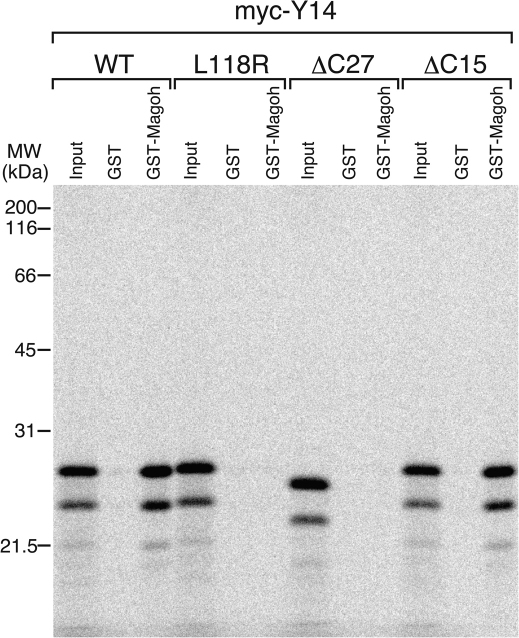
As described above, the N-terminus of Y14 confers NLS and NES activities, while, as reported previously, its C-terminus contains a single RNA-binding domain (RBD) that mediates interactions with its binding partners. In particular, RBD is a main domain for interaction with Mago nashi protein26,27,28 and its C-terminus binds Aly/REF17 and hUpf312. Y14's C-terminal domain also contains many basic residues and two arginine-serine dipeptide repeats that are characteristic of SR splicing factors17. It was demonstrated that two serine residues at the C-terminus of Y14 are phosphorylated and phosphorylated Y14 has reduced binding activity to Upf3, an essential NMD factor36. In order to determine whether the C-terminal region of Y14 has other function(s), we tested whether it is required for the interaction with Magoh, the human ortholog of Mago nashi protein. We carried out in vitro binding experiments using [35S] methionine-labeled, in vitro translated Y14 protein and purified recombinant GST-Magoh. As shown in Figure 3, wild-type Y14 binds efficiently to GST-Magoh, but not to GST, as previously reported17. As a negative control, we employed L118R mutant of Y14, which was shown to be defective in interacting with Magoh in vitro28. Indeed, this mutant had severely reduced binding activity to Magoh (Figure 3, lane 6). Surprisingly, ΔC27 mutant of Y14, which lacks C-terminal 27 aa, showed no specific binding to Magoh. In contrast, another deletion mutant, ΔC15, retained the ability to bind Y14. These results demonstrated that, although it is not likely to be a main contact domain of Y14 to Magoh, the 12 aa near Y14's C-terminus (aa 148-159) are required for binding to its partner, Magoh.
Figure 3. C-terminal of Y14 is also required for binding to Magoh in addition to the RNA-binding domain.
Wild-type and mutant myc-tagged Y14 proteins, produced and labeled with [35S]methionine in vitro, were incubated with 5 μg each of GST or GST-Magoh. After washing, bound proteins were eluted, separated on 15% SDS-PAGE gel and visualized by fluorography. The lanes marked Input contain 10% of the total protein used in each binding reaction. The position of molecular mass markers is shown on the left.
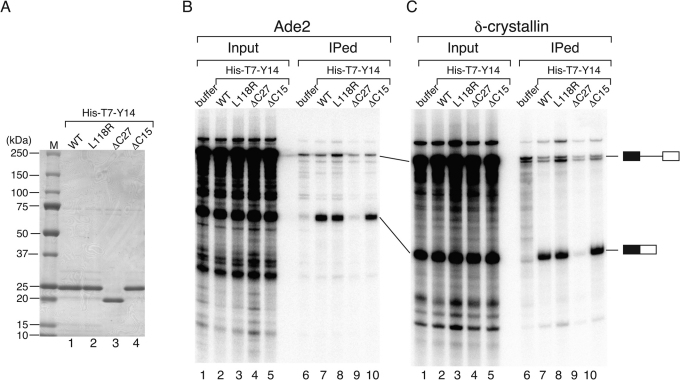
Next, we examined if the 12 aa of Y14 identified by in vitro binding to Magoh are necessary for its association to spliced mRNAs. In vitro splicing reaction with Ade2 pre-mRNA was carried out with either wild-type or mutant Y14 proteins. Recombinant proteins for wild-type and three different mutants of Y14 were overexpressed in E. coli and purified using Ni-NTA column (Figure 4A). The association of exogenously added recombinant proteins was tested by immunoprecipitation with anti-T7 antibody, since recombinant proteins contain T7-tag at the N-terminus. As expected, we could detect association of exogenously added wild-type Y14 with spliced mRNA compared with that upon adding buffer alone (Figure 4B, lanes 6 and 7). L118R, while deficient in binding to Magoh, could bind to spliced mRNA at least as efficiently as the wild-type protein (Figure 4B, lane 8). The ΔC27 mutant showed greatly reduced precipitation of spliced mRNA, whereas the ΔC15 mutant could precipitate spliced mRNA as efficiently as the wild type (Figure 4B, lanes 9 and 10). Similar results were obtained with δ-crystallin pre-mRNAs (Figure 4C, lanes 6-10). These results demonstrated that the 12 aa near C-terminus of Y14 are essential for its association to both spliced mRNAs and Magoh. In addition, these results strongly suggest that binding of Magoh is not required for Y14 to bind spliced mRNAs.
Figure 4. Y14 binding to spliced mRNA is Magoh-independent and requires its C-terminus.
(A) Preparation of various recombinant Y14 proteins in E. coli. Wild-type and three mutant proteins were expressed in E. coli cells and purified using Ni-NTA column. Five hundred ng of each purified protein was analyzed by SDS-PAGE. (B) Immunoprecipitation of RNAs from in vitro splicing reaction with Ade2 pre-mRNA in the presence of recombinant proteins was performed. 2.5 μg of anti-T7 tag antibody was used for immunoprecipitation. Pre-mRNAs and mRNA products are indicated with schematic drawings on the right. (C) RNA immunoprecipitation experiment with chicken δ-crystallin pre-mRNA was carried out as in (B).
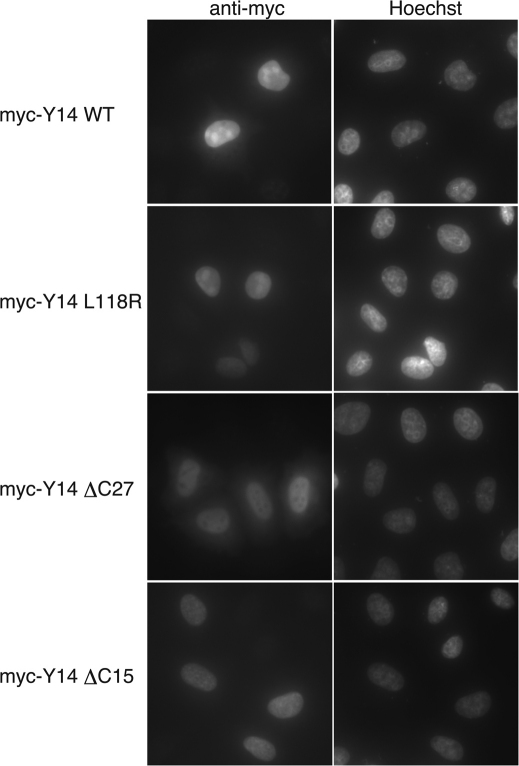
We have also determined the subcellular localization of both wild-type and mutant Y14 proteins used in Figure 4 with a myc-epitope tag. As reported previously1, wild-type myc Y14 protein was mostly nuclear (Figure 5). L118R and ΔC27 mutant proteins, which could not interact with Magoh (Figure 4), still localized predominantly in the nucleus (Figure 5). The ΔC15 protein was also capable of localizing in the nucleus (Figure 5). These results strongly suggest that nuclear localization of Y14 is independent of its interaction with Magoh.
Figure 5. Subcellular localization of myc-tagged wild-type and mutant Y14 in HeLa cells.
Both wild-type and mutant Y14 are mainly localized in the nucleus. Myc-Y14 cDNA encoding either wild type or several mutants was transfected into HeLa cells, which were subsequently immunostained using anti-myc-tag antibody (9E10) as described in the Materials and Methods.
Discussion
In this study, we analyzed the function of several domains of Y14. As previously shown, although Y14 is predominantly nuclear, it shuttles continuously between the nucleus and the cytoplasm1. We delineated that Y14's N-terminus functions both as an NLS and as an NES. The amino acid sequences of both signals overlap but are not identical. This is similar to the case of hnRNP K and HuR32,37. The hnRNP K protein has two functional NLSs. The second NLS also has NES activity and is therefore called K nuclear shuttling sequence (KNS). YNS does not show any significant sequence similarity to known NLSs and NESs, suggesting that YNS nuclear import is mediated by a novel nuclear transport receptor. Indeed, Importin 13 was identified as a nuclear import receptor for Y14-Magoh complex29. Recently, the crystal structure of Y14/Mago/Importin 13 complex was reported30. Although YNS was thought to interact with Importin 13, this region is not required for Importin 13 interaction in the context of the ternary nuclear import complex formation30. One possibility is that YNS is only required for the nuclear localization of Magoh-free Y14. Indeed, two Magoh-binding-deficient Y14 mutants could be localized in the nucleus of HeLa cells, as well as the wild type (Figure 5), providing evidence for the possibility that Y14 utilizes YNS-mediated nuclear import pathway when Magoh is absent. It remains to be determined whether Y14 has function(s) independent of Magoh in cells.
The results that we obtained with C-terminus truncated mutant of Y14 indicate that the 12 aa of Y14 near the C-terminus are required for efficient association with both Magoh and spliced mRNAs. Since ΔC15 mutant bound both spliced mRNAs and Magoh, the critical region for both associations lies between amino acids 148 and 159 of Y14. The analyses of the crystal structures of both Y14-Magoh heterodimer and the EJC core demonstrate that tryptophan residue at position 147 of Y14 contacts Magoh directly26,27,28. Although the main contact region of Y14 with Magoh is the central domain that encompasses the RNA-binding domain, the interaction of this tryptophan with Magoh also makes a large contribution to their heterodimer formation. Since this tryptophan residue is conserved among many species1, it is likely that the contribution of this contact is also conserved.
Neither L118R nor ΔC27 mutant could bind to Magoh (Figure 3). However, L118R could associate with spliced mRNAs, while ΔC27 mutant could not (Figure 4). The result that L118R could associate with spliced mRNAs strongly suggests that Magoh binding to Y14 is not required for Y14 incorporation into the EJC core. It is likely that L118R, while losing the ability to interact with Magoh, could still interact with other EJC components and splicing factors to participate in splicing. On the other hand, ΔC27 is defective in interacting with Magoh and likely with other EJC factors as well, thus failed to be recruited into the spliceosome. It was demonstrated that Magoh cannot associate with spliced mRNA without Y1418. Y14 could serve as an adaptor for Magoh to enter EJC. Previously, the C-terminus of Y14 was demonstrated to be an interaction domain for Aly/REF, hUpf3 and PRMT512,17,36,38. This domain was also shown to be able to associate with RNA in vitro28. Crystal structures of both Y14-Magoh heterodimer and the EJC core revealed that the amino acid region downstream of tryptophan does not contact Magoh and other EJC components26,27,28,39,40. Neither does this domain interact with RNA directly in the crystal structure model of the EJC core39,40. It is still unclear how these 12 aa of Y14 enable its association with spliced mRNAs. Further studies of the crystal structure of the EJC core with full-length proteins are required to define the interaction of each core protein in EJC.
Although we demonstrated that YNS confers both nuclear import and nuclear export activities of Y14, the nuclear transport receptor(s) for YNS has yet to be identified. We have not been able to detect binding of either RanBP5/Importin 5 or Importin 13 to YNS (data not shown). YNS may utilize a specific Importin for nuclear import. Although less likely, it is possible that YNS can interact with a nuclear protein to enter the nucleus by a ‘piggy-back' mechanism. It is also interesting to identify a nuclear export receptor for YNS, if any, since Y14 and Magoh were shown to have important roles in certain cytoplasmic events, such as cytoplasmic mRNA localization21,22. The nuclear export receptor for YNS may regulate these steps by transport. Identification of specific interactors to YNS is required to address these issues, and this approach may lead to novel function(s) of Y14 being uncovered in cells.
Methods
Plasmid construction, protein expression and purification
To create myc-PK/pCDNA3, HindIII-EcoRI-digested fragment of myc-PK/pCDNAI32 was subcloned to pCDNA3 (Invitrogen). For myc-PK-Y14 fusion constructs, plasmid myc-Y141 was used as a template in a series of PCRs. The primers were designed to include an EcoRI site at the 5' end and an XhoI site at the 3' end to amplify several fragments of the Y14 coding sequences and subcloned into similarly digested myc-PK/pCDNA3.
Plasmid myc-NPc-T has been described elsewhere32,34,35. For myc-NPc-T-Y14 constructs, full-length or portions of Y14 cDNA were PCR amplified and cloned between XhoI and XbaI sites.
To make the plasmids for Y14 L118R mutant, QuikChangeTM Site-Directed Mutagenesis Kit (STRATAGENE) was used with myc-Y14 plasmid1 in accordance with the manufacturer's recommendations. For preparation of the plasmids carrying C-terminus deletion mutants of Y14, the truncated Y14 cDNAs were PCR-amplified and cloned between BamHI and XhoI sites of myc-pCDNA3 and pET28a (Novagen). Protein expression and purification were carried out as described previously17.
Cell culture, transfection and indirect immunofluorescence
HeLa cell culture was performed as described previously41. For transfection, Lipofectamine2000 transfection reagent (Invitrogen) was used according to the manufacturer's recommendations. Immunofluorescence with anti-myc antibody (9E10) was carried out as described previously32.
Heterokaryon assay
The heterokaryon analysis was performed as described previously32,34,35. The co-cultures of HeLa cells and mouse NIH3T3 cells were incubated for 3 hr in the presence of 75 μg/ml cycloheximide and then fused exactly as described previously42. The heterokaryons were returned to media containing 100 μg/ml cycloheximide and incubated for 2 hr before fixation for indirect immunofluorescence analysis.
In vitro protein binding
In vitro protein binding experiments were performed as described previously17. Briefly, 5 µg of GST fusion proteins was immobilized on glutathione beads. In vitro translated proteins were prepared using a TNT-coupled rabbit reticulocyte lysate system (Promega). Approximately 2 × 105 c.p.m. of in vitro translated products was added to the beads in the binding buffer (50 mM Tris–HCl pH 7.4, 200 mM NaCl, 2 mM EDTA, 0.1% NP-40) and mixed at 4°C for 1 hr. The beads were then washed five times with binding buffer, and bound proteins were eluted with sample buffer and analyzed by SDS–PAGE.
In vitro splicing and immunoprecipitation
Template DNA preparation and in vitro transcription were carried out as described previously1,43. The in vitro splicing reactions with HeLa cell nuclear extracts in the presence of 20 μg of exogenously added recombinant proteins were performed in 50 μl reaction mixture as described previously, but without pre-incubating nuclear extract with recombinant proteins43. Immunoprecipitation was performed as described previously44. Briefly, 2.5 μg of anti-T7 antibody (Novagen) was used for each immunoprecipitation.
Author Contributions
N.K., G.D. and M.O. designed experiments and N.K., M.D., M.Y., C.H. and I.D. performed them. N.K. and M.O. analyzed the data and N.K., G.D., M.H. and M.O. wrote the manuscript.
Acknowledgments
We are grateful to Drs. Kensuke Ninomiya, Lili Wan and Hiroshi Sasaki for critical reading and comments on this manuscript. We also thank members of both Ohno laboratory and Hagiwara laboratory for discussions. This work was supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and grants from the National Institute of Health and the Human Frontier Science Program Organization. G.D. is an Investigator of the Howard Hughes Medical Institute. C.H. was supported by the 21st Century COE Program of the MEXT of Japan. N.K. was supported by Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology (SCF) commissioned by the MEXT of Japan.
References
- Kataoka N., et al.. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6, 673-682 (2000). [DOI] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E. & Moore M. J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20, 4987-4997 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde E., Maquat L. E. & Moore M. J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19, 6860-6869 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. N., et al.. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 20, 2062-2068 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T., Tange T. O., Sonenberg N. & Moore M. J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11, 346-351 (2004). [DOI] [PubMed] [Google Scholar]
- Tange T. O., Shibuya T., Jurica M. S. & Moore M. J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11, 1869-1883 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L., et al.. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 12, 861-869 (2005). [DOI] [PubMed] [Google Scholar]
- Nott A., Le Hir H. & Moore M. J. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 18, 210-222 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F. & Maquat L. E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 17, 309-315 (2005). [DOI] [PubMed] [Google Scholar]
- Maquat L. E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5, 89-99 (2004). [DOI] [PubMed] [Google Scholar]
- Kim V. N., Kataoka N. & Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293, 1832-1836 (2001). [DOI] [PubMed] [Google Scholar]
- Gehring N. H., Neu-Yilik G., Schell T., Hentze M. W. & Kulozik A. E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11, 939-949 (2003). [DOI] [PubMed] [Google Scholar]
- Gehring N. H., et al.. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20, 65-75 (2005). [DOI] [PubMed] [Google Scholar]
- Gehring N. H., Lamprinaki S., Hentze M. W. & Kulozik A. E. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 7, e1000120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M. D. & Steitz J. A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121-1131 (2000). [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M. D. & Steitz J. A. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293, 1836-1839 (2001). [DOI] [PubMed] [Google Scholar]
- Kataoka N., Diem M. D., Kim V. N., Yong J. & Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 20, 6424-6433 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Braun I. C., Forler D. & Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2, 1119-1124 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S. E., Dillon S. T. & Boswell R. E. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15, 2886-2899 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O. & Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11, 1666-1674 (2001). [DOI] [PubMed] [Google Scholar]
- Hachet O. & Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428, 959-963 (2004). [DOI] [PubMed] [Google Scholar]
- Giorgi C., et al.. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 130, 179-191 (2007). [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D., et al.. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143, 251-262 (2010). [DOI] [PubMed] [Google Scholar]
- Roignant J. Y. & Treisman J. E. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143, 238-250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb T. L., Lam B. J. & Hertel K. J. Retention of spliceosomal components along ligated exons ensures efficient removal of multiple introns. RNA 16, 1786-1796 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H. & Xu R. M. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev. 17, 971-976 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. K., Diem M. D., Dreyfuss G. & Van Duyne G. D. Structure of the Y14-Magoh core of the exon junction complex. Curr. Biol. 13, 933-941 (2003). [DOI] [PubMed] [Google Scholar]
- Fribourg S., Gatfield D., Izaurralde E. & Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol 10, 433-439 (2003). [DOI] [PubMed] [Google Scholar]
- Mingot J. M., Kostka S., Kraft R., Hartmann E. & Gorlich D. Importin 13: a novel mediator of nuclear import and export. EMBO J. 20, 3685-3694 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F., Cook A. G., Grunwald M., Ebert J. & Conti E. Nuclear import mechanism of the EJC component Mago-Y14 revealed by structural studies of importin 13. Mol. Cell 37, 211-222 (2010). [DOI] [PubMed] [Google Scholar]
- Dostie J. & Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12, 1060-1067 (2002). [DOI] [PubMed] [Google Scholar]
- Michael W. M., Eder P. S. & Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16, 3587-3598 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H. & Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129, 551-560 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W. M., Choi M. & Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83, 415-422 (1995). [DOI] [PubMed] [Google Scholar]
- Nakielny S. & Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 134, 1365-1373 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Ia W., et al.. Phosphorylation of Y14 modulates its interaction with proteins involved in mRNA metabolism and influences its methylation. J. Biol. Chem. 280, 34507-34512 (2005). [DOI] [PubMed] [Google Scholar]
- Fan X. C. & Steitz J. A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 95, 15293-15298 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T. W., Peng P. J. & Tarn W. Y. The exon junction complex component Y14 modulates the activity of the methylosome in biogenesis of spliceosomal snRNPs. J. Biol. Chem. 286, 8722-8728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F., Ebert J., Lorentzen E. & Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713-725 (2006). [DOI] [PubMed] [Google Scholar]
- Andersen C. B., et al.. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968-1972 (2006). [DOI] [PubMed] [Google Scholar]
- Siomi M. C., et al.. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 138, 1181-1192 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S. & Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355, 730-732 (1992). [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka N., Charroux B. & Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95, 615-624 (1998). [DOI] [PubMed] [Google Scholar]
- Hanamura A., Caceres J. F., Mayeda A., Franza B. R. Jr & Krainer A. R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4, 430-444 (1998). [PMC free article] [PubMed] [Google Scholar]