Abstract
The agreement of the microwell plate AMPLICOR HIV-1 MONITOR version 1.0 (MWP 1.0), the microwell plate AMPLICOR HIV-1 MONITOR version 1.5 (MWP 1.5), and the COBAS AMPLICOR HIV-1 MONITOR version 1.5 (COBAS 1.5) tests was evaluated using clinical specimens and well-characterized control material. Two hundred patient plasma specimens and a panel of known human immunodeficiency virus type 1 (HIV-1) subtypes were tested. All data were log10 transformed prior to analysis. The 95% limits of agreement for the three tests at the average of 3.66 log10 copies/ml were ± 0.28 log10, ± 0.34 log10, and ± 0.34 log10 copies/ml for MWP 1.0-MWP 1.5, MWP 1.0-COBAS 1.5, and MWP 1.5-COBAS 1.5, respectively. Ten specimens (6.1%) had differences exceeding the limits of agreement for the MWP 1.0 and MWP 1.5 tests. Correlation coefficients among the three tests were high (r ≥ 0.96). The viral-load values obtained with the MWP 1.0 test were only 2.1% higher on average than those measured with the MWP 1.5 test and 1.6% higher than those seen with the COBAS 1.5 test. The MWP 1.5 test values were 0.8% higher than the COBAS 1.5 test values. Overall, there was less agreement among the different tests for viral-load values near the lower limit of quantification. The MWP 1.0 test underquantified subtypes A, E, F, G, and H by 1.0 to 2.0 log10 copies/ml; this problem was not observed with the MWP 1.5 test. The close agreement among the results obtained with the different test versions and formats suggests that it is not necessary to reestablish a baseline viral load when changing AMPLICOR HIV-1 MONITOR tests, unless the patient is known to be infected with a non-B subtype.
The quantification of human immunodeficiency virus type 1 (HIV-1) RNA from plasma has become the standard of care in managing persons with HIV-1 infection. Clinical-practice guidelines for the use of HIV-1 viral-load testing have been established (9, 11). Viral-load testing is used to make decisions regarding the initiation of highly active antiretroviral therapy, to monitor response to therapy, and to determine the prognosis. Three HIV-1 viral-load assays have been approved by the Food and Drug Administration (FDA): the AMPLICOR HIV-1 MONITOR test (Roche Diagnostics, Indianapolis, Ind.), the Quantiplex HIV-1 assay (bDNA; Bayer Corp., Tarrytown, N.J.), and the NucliSens HIV-1 QT assay (bioMerieux, Inc., Durham, N.C.). Viral-load values do not always agree among the three tests; therefore, it is recommended that a single assay be used when monitoring patients on highly active antiretroviral therapy (3, 6, 10). Factors that affect the reproducibility of viral-load assays include intra-assay variation and biological variation. The intra-assay variation of the AMPLICOR HIV-1 MONITOR test has been reported to range from 0.1 to 0.2 log10 copies/ml (2, 5). A comprehensive evaluation of the assay variation for the AMPLICOR HIV-1 MONITOR version 1.0 test has shown that the total variation (standard deviation) was ∼0.26 log10 copies/ml. This included intra-assay, interassay, and biological variation, with biological variation comprising over half of the total variation (2). When the 95% confidence limits are considered, these data (2) support other published reports (5, 9) that the variation in plasma HIV-1 RNA levels for a given individual is approximately ±0.5 log10 copies/ml.
The microwell plate AMPLICOR MONITOR version 1.0 (MWP 1.0) test has been shown to underquantify non-B subtypes of HIV-1 RNA. Studies have shown that subtype A virus is underquantified by 2 log10 copies/ml, while subtypes E and F are underquantified by 1 log10 copies/ml (4, 10; N. Michael, M. Robb, D. Birx, J. R. Mascola, J. Wang, K. Dreyer, K. McDonough, C. Christopherson, S. D. Lu, S. Kwok, and S. Herman, 4th Conf. Retrovir. Opportunistic Infect., poster 279, 1997). As a result of modifications in the primer sequences, the microwell plate AMPLICOR MONITOR version 1.5 (MWP 1.5) and COBAS AMPLICOR MONITOR HIV-1 version 1.5 (COBAS 1.5) tests can accurately quantify HIV-1 RNA subtypes A to H (4, 7, 8). The MWP 1.5 and COBAS 1.5 tests have recently been approved by the FDA and have replaced the MWP 1.0 assay in clinical use. In spite of the availability of the version 1.5 tests, very little is known about how viral-load values compare between the version 1.0 and 1.5 tests. In this study, we evaluated the agreement among the MWP 1.0, MWP 1.5, and COBAS 1.5 tests using clinical specimens and well-standardized control material. The goal was to establish whether there is a need to redetermine a patient's baseline viral load when changing from version 1.0 to version 1.5 of the AMPLICOR MONITOR test.
MATERIALS AND METHODS
Specimens and control material.
Plasma specimens used for the study were randomly selected from those sent to the Molecular Diagnostics Laboratory at Emory University Hospital for HIV-1 viral-load testing. Plasma remaining after clinical testing using the MWP 1.0 test was frozen at −70°C until it was tested in the version 1.5 tests (both MWP and COBAS). The use of clinical specimens in this study was approved by the Emory University Institutional Review Board (IRB no. 247-2001).
A total of 200 plasma specimens were used for the study; 100 of the specimens were tested by the ultrasensitive test, and 100 were tested by the standard test. Reproducibility studies were done of pooled plasma specimens representing low- and midrange values of the dynamic ranges of both the standard and ultrasensitive assays. Six replicates of the low- and midrange pools were tested in the MWP 1.5 assay. In addition, 12 replicates of the AcroMetrix (Benicia, Calif.) control (100 copies/ml) were tested in a single run in the ultrasensitive MWP 1.0 and MWP 1.5 tests. This was done to examine the reproducibility near the limit of detection of the ultrasensitive assay, where the greatest variability was expected. The subtype evaluation was done using a panel of samples representing subtypes A to H (Boston Biomedica Inc., West Bridgewater, Mass.). The viral loads of the panel members were determined by the supplier with the MWP 1.5 test and were as follows: subtype A, 3.6 log10 copies/ml; subtype C, 4.3 log10 copies/ml; subtypes B, D, E, and H, 5.3 log10 copies/ml; and subtypes F and G, 5.7 log10 copies/ml. The panel members were diluted in normal human plasma to a nominal concentration of 4.3 log10 copies/ml for all subtypes except subtype A, which was used undiluted. The subtype panel was tested in triplicate in both the MWP 1.0 and 1.5 tests.
Viral-load tests.
Viral-load testing was done using the MWP 1.0, MWP 1.5, and COBAS 1.5 tests. Both the ultrasensitive and standard tests were used in this study. Testing was performed according to the manufacturer's protocols, using 200 and 500 μl of specimen in the ultrasensitive and standard tests, respectively. The standard test has a linear range of 2.6 to 5.88 log10 copies/ml (400 to 750,000 copies/ml) for both the version 1.0 and 1.5 tests. The lower limit of quantification for both the version 1.0 and 1.5 ultrasensitive tests is 1.7 log10 copies/ml (50 copies/ml). The upper limit of the linear range is 4.88 log10 copies/ml (75,000 copies/ml) for the MWP 1.0 test and 5.0 log10 copies/ml (100,000 copies/ml) for the MWP 1.5 and COBAS 1.5 tests, respectively. One lot of reagents was used for each of the three tests.
Statistics.
All data were log10 transformed prior to analysis. Only specimens that were within the dynamic range of the MWP 1.0 assay were used for statistical analysis; 165 specimens (85 ultrasensitive and 80 standard) were used in the analysis. The data from the standard and ultrasensitive tests were combined when calculating the mean viral load for each version of the assay. Specimens that were undetectable in the MWP 1.5 and COBAS 1.5 ultrasensitive and standard assays were recorded as 1.30 (n = 7) and 2.3 (n = 2) log10 copies/ml, respectively. Specimens that were above the linear range of the standard version 1.5 tests were recorded as 6.0 log10 copies/ml (n = 2). There were no specimens above the linear range of the version 1.5 ultrasensitive tests. Mean differences in log10 viral load between pairs of tests were compared to see if they differed from zero by using a one-sample, two-sided paired t test. Agreement between the results from the different test versions was assessed by plotting the difference between the log10 copies per milliliter against the mean log10 copies per milliliter for each specimen (1). Limits of agreement (95%) between tests were estimated as the mean difference ± 1.96 standard deviations of the differences. The relationships among the three tests were also analyzed by fitting a linear regression through the origin for each pair of tests.
RESULTS
There was good correlation between the viral-load values obtained with the MWP 1.0 and MWP 1.5 tests (r = 0.97). Similar results were observed with the MWP 1.0 and COBAS 1.5 tests (r = 0.96) and with the MWP 1.5 and COBAS 1.5 tests (r = 0.97) (data not shown).
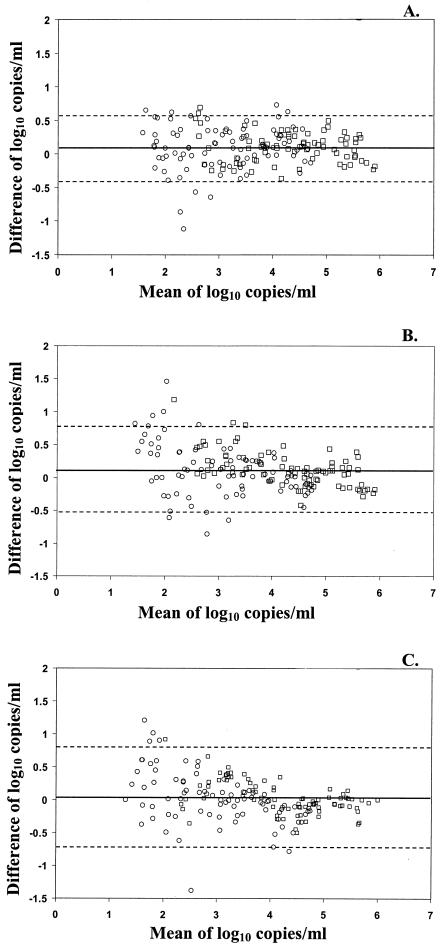
The limits of agreement for the different tests were determined by plotting the difference between the log10 copies/ml for each specimen (version 1.0 minus version 1.5) versus the mean log10 copies/ml. Comparing the MWP 1.0 and MWP 1.5 tests, the mean log10 difference was 0.09 log10 copies/ml, with a standard deviation of 0.28 log10 copies/ml (Fig. 1A). According to this analysis, 95% of the results for the two versions of the test would be expected to agree within 0.56 log10 copies/ml (±2 standard deviations). The observed results showed that the differences between viral loads in the MWP 1.0 and MWP 1.5 tests for 10 (6.1%) specimens were >0.56 log10 copies/ml. The mean difference in the viral loads for these 10 specimens was 0.70 log10 copies/ml, with a range of 0.57 log10 to 1.12 log10 copies/ml. Of these 10 specimens, 7 had viral-load values of <3.0 log10 copies/ml. Similar results were obtained when comparing the MWP 1.0 and COBAS 1.5 tests (Fig. 1B). The mean log10 difference was 0.12 log10 copies/ml, with a standard deviation of 0.34 log10 copies/ml, with the limits of agreement calculated to be 0.68 log10 copies/ml. Eleven (6.7%) specimens were found to have a difference in viral load of >0.68 log10 copies/ml. The mean difference in viral load for these 11 specimens was 0.93 log10 copies/ml, with a range of 0.73 log10 to 1.45 log10 copies/ml. Eight of the 11 specimens had viral-load values of <3.0 log10 copies/ml. The agreement between the version 1.0 and 1.5 assays was poorer near the lower limit of the linear range for both the ultrasensitive and standard tests.
FIG. 1.
Agreement of viral-load values between the MWP 1.0 and MWP 1.5 (A), the MWP 1.0 and COBAS 1.5 (B), and the MWP 1.5 and COBAS 1.5 (C) tests. ○, ultrasensitive test; □, standard test. The solid lines represent the mean differences; the dashed lines represent ±2 standard deviations.
Comparing the MWP 1.5 and COBAS 1.5 tests, the mean log10 difference was 0.03 log10 copies/ml with a standard deviation of 0.34 log10 copies/ml (Fig. 1C). According to this analysis, 95% of the results would be expected to agree within 0.68 log10 copies/ml. Seven of the 165 (4.2%) specimens were found to have a viral-load difference of >0.68 log10 copies/ml between the two platforms for the version 1.5 test. The mean difference in viral load was 1.01 log10 copies/ml, with a range of 0.78 log10 to 1.38 log10 copies/ml. Of these seven specimens, five had viral-load values <3.0 log10 copies/ml in the MWP 1.5 test.
The mean viral load (± standard deviation) for the 165 specimens that were within the linear range of the MWP 1.0 assay were as follows: MWP 1.0, 3.74 ± 1.13 log10 copies/ml; MWP 1.5, 3.64 ± 1.13 log10 copies/ml; and COBAS 1.5, 3.61 ± 1.26 log10 copies/ml. The mean differences and standard deviations of the differences in viral load for the three tests are shown in Table 1. The viral-load values obtained with the MWP 1.0 test are statistically higher than those seen with the MWP 1.5 and COBAS 1.5 tests (P < 0.001), but this is clinically unimportant, since the bias estimated from the mean differences (0.09 and 0.12) was small. Slope estimates (1.021 and 1.016) obtained by linear regression, forcing the analysis through the origin, indicated that the MWP 1.0 values were 2.1 and 1.6% higher on average than those measured with the MWP 1.5 and COBAS 1.5 tests, respectively. There was no significant difference between the viral-load values obtained with the MWP 1.5 and the COBAS 1.5 tests (P = 0.3).
TABLE 1.
Mean differences in viral loads determined for 165 clinical specimens by MWP 1.0, MWP 1.5, and COBAS 1.5 tests
| Comparison | Mean difference (log10 copies/ml) | SDa (log10 copies/ml) | P value |
|---|---|---|---|
| MWP 1.0 minus MWP 1.5 | 0.09 | 0.28 | <0.001 |
| MWP 1.0 minus COBAS 1.5 | 0.12 | 0.34 | <0.001 |
| MWP 1.5 minus COBAS 1.5 | 0.03 | 0.34 | 0.3 |
SD, standard deviation of the differences.
The subtype analysis (Table 2) revealed that the MWP 1.0 test underquantified subtypes A, E, F, G, and H by 1 log10 to 2 log10 copies/ml compared to the MWP 1.5 test. The input concentration for subtype A was lower than those for the other subtypes due to the concentration of that panel member supplied by the manufacturer. For the other subtypes, there was no difference between the viral load values in the MWP 1.0 and MWP 1.5 tests. The reproducibility of the MWP 1.5 assay was determined by testing six replicates of pooled plasma specimens representing the low- and midrange values of the dynamic ranges of both the standard and ultrasensitive tests. For the ultrasensitive test, the six replicates of the low-range pool had a mean viral load of 2.89 log10 copies/ml, with a standard deviation of 0.15 log10 copies/ml. The midrange sample had a mean of 4.06 log10 copies/ml, with a standard deviation of 0.09 log10 copies/ml. For the standard test, the means (standard deviations) of the low-range and midrange pools were 3.44 log10 (0.09 log10) and 4.92 log10 (0.08 log10) copies/ml, respectively.
TABLE 2.
Quantification of group M subtype A through H HIV-1 RNAs using MWP 1.0 and MWP 1.5 tests
| Subtype | log10 nominal copies/ml | MWP 1.0
|
MWP 1.5
|
||
|---|---|---|---|---|---|
| log10 copies/mla | SD | log10 copies/mla | SD | ||
| A | 3.60 | <2.6 | 3.85 | 0.07 | |
| B | 4.30 | 4.23 | 0.02 | 4.36 | 0.05 |
| C | 4.30 | 4.25 | 0.09 | 4.43 | 0.03 |
| D | 4.30 | 4.54 | 0.07 | 4.47 | 0.08 |
| E | 4.30 | 3.09 | 0.10 | 4.30 | 0.11 |
| F | 4.30 | 3.15 | 0.07 | 4.28 | 0.10 |
| G | 4.30 | <2.6b | 4.32 | 0.09 | |
| H | 4.30 | <2.6b | 4.34 | 0.14 | |
Mean of three replicates.
HIV-1 RNA detected but below the level of quantification.
To further evaluate the agreement and reproducibility at the lower end of the range of the ultrasensitive test, 12 replicates of a 2.0-log10 copy/ml control were assayed in both the MWP 1.0 and MWP 1.5 tests. The mean (standard deviation) viral load for the MWP 1.0 test was 2.39 log10 (0.13 log10) copies/ml, while the mean viral load in the MWP 1.5 test was 2.29 log10 (0.13 log10) copies/ml.
DISCUSSION
With their recent FDA approval, the MWP 1.5 and COBAS 1.5 tests have replaced the MWP 1.0 test currently used in clinical laboratories. In this study, we assessed the agreement between the MWP 1.0 and 1.5 tests to establish whether it is necessary to redetermine a patient's baseline viral load when changing versions or formats of the test. Our data show that there is a good correlation among the viral-load values of the different formats and versions of the test (r ≥ 0.96). The agreement among the different tests was also assessed, as this is the parameter most useful to clinicians in determining whether there is a need to rebaseline patients when changing tests. According to this analysis, 94% of the values between the MWP 1.0 and MWP 1.5 tests were within 0.56 log10 copies/ml. This value is near 0.5 log10 copies/ml (threefold), which is the total variation that can be expected in viral-load measurements (2, 5, 9). For the MWP 1.0 and COBAS 1.5 tests, 93% of the viral-load values were within 0.68 log10 copies/ml, and when the MWP 1.5 and COBAS 1.5 tests were compared, 96% of the viral-load values were within 0.68 log10 copies/ml. Differences among these sets of tests are in the fivefold range. The correlation and agreement between the two different formats (MWP and COBAS) of the version 1.5 test were similar to those seen for the two different versions (versions 1.0 and 1.5) of the MWP test. Since only one lot of reagents was used for each of the three tests, we are unable to comment on how lot-to-lot variation may influence agreement.
Overall, there is less agreement among the different versions of the test and assay formats for viral-load values near the lower limit of quantification for the tests. Of the samples that fell outside the 2-standard deviation range around the mean difference, 20 of 28 (71%) had viral loads of <3.0 log10 copies/ml. There was not a difference between the numbers of specimens with <1.7 log10 copies/ml in the two versions of the test, which implies similar sensitivities for the version 1.0 and 1.5 tests. However, this needs to be thoroughly investigated with a large number of specimens.
The MWP 1.0 test gave viral-load values that were higher than the values measured with either the MWP 1.5 or COBAS 1.5 test. These differences, though statistically significant, are small (1.6 to 2.1%) and do not appear to be clinically relevant. There was no difference between the viral load values for the MWP 1.5 and COBAS 1.5 tests.
The reproducibility of the MWP 1.5 test is similar to that previously established for the MWP 1.0 test, with standard deviation values ranging from 0.08 log10 to 0.15 log10 copies/ml. A standard deviation of 0.15 log10 copies/ml has been used by the Virology Quality Assessment Laboratory program to ensure that a laboratory can maintain the precision required to have 90% power to detect a fivefold (0.69 log10 copies/ml) difference in copy number between two samples in the same batch (12). Based on our data, it appears that the same standard will apply to the version 1.5 test, although establishing the reproducibility of the version 1.5 test in a multisite study is required.
Our data support the findings of previous studies (4, 7, 8) that have shown that the MWP 1.0 test underquantifies subtypes A, E, F, G, and H, which does not appear to be a problem for the version 1.5 test. In our study, there were several samples that had a higher viral load in the MWP 1.5 test than in the MWP 1.0 test; there was not an adequate volume of the samples remaining to determine the virus subtype.
In summary, the close agreement among the results obtained with the different test versions and formats suggests that it is not necessary to reestablish a baseline viral load when changing AMPLICOR HIV-1 MONITOR tests, unless the person is known to be infected with subtype A, E, F, G, or H. However, there is less agreement near the lower limit of quantification, so small changes in the viral load near the limit of quantification of the test should be interpreted with caution, as they may not represent biologically relevant changes in viral replication.
REFERENCES
- 1.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed]
- 2.Brambilla, D., P. S. Reichelderfer, J. W. Bremer, D. E. Shapiro, R. C. Hershow, D. A. Katzenstein, S. M. Hammer, B. Jackson, A. C. Collier, R. S. Sperling, M. G. Fowler, and R. W. Coombs, for participating Adult and Pediatric AIDS Clinical Trials Groups, the Women Infant Transmission Study Clinics, and laboratories participating in the Virology Quality Assurance Program. 1999. The contribution of assay variation and biological variation to the total variability of plasma HIV-1 RNA measurements. AIDS 13:2269-2279. [DOI] [PubMed] [Google Scholar]
- 3.Dyer, J. R., C. D. Pilcher, R. Shepard, J. Schock, J. J. Eron, and S. A. Fiscus. 1999. Comparison of NucliSens and Roche Monitor assays for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 37:447-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jagodzinski, L. L., D. L. Wiggins, J. L. McManis, S. Emery, J. Overbaugh, M. Robb, S. Bodrug, and N. L. Michael. 2000. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J. Clin. Microbiol. 38:1247-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lew, J., P. Reichelderfer, M. Fowler, J. Bremer, R. Carrol, S. Cassol, D. Chernoff, R. Coombs, M. Cronin, R. Dickover, S. Fiscus, S. Herman, B. Jackson, J. Kornegay, A. Kovacs, K. McIntosh, W. Meyer, N. Michael, L. Mofenson, J. Moye, T. Quinn, M. Robb, M. Vahey, B. Weiser, and T. Yeghiazarian, for the TUBE Meeting Workshop attendees. 1998. Determinations of levels of human immunodeficiency virus type 1 RNA in plasma: reassessment of parameters affecting assay outcome. J. Clin. Microbiol. 36:1471-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolte, F. S., J. Boysza, C. Thurmond, W. S. Clark, and J. L. Lennox. 1998. Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 36:716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parekh, B., S. Phillips, T. C. Granade, J. Baggs, D. J. Hu, and R. Respess. 1999. Impact of HIV type 1 subtype variation on viral RNA quantitation. AIDS Res. Hum. Retrovir. 15:133-142. [DOI] [PubMed] [Google Scholar]
- 8.Pasquier, C., K. Sandres, G. Salama, J. Puel, and J. Izopet. 1999. Using RT-PCR and bDNA assays to measure non-clade B HIV-1 subtype RNA. J. Virol. Methods 81:123-129. [DOI] [PubMed] [Google Scholar]
- 9.Saag, M. S., M. Holodniy, D. R. Kuritzkes, W. A. O'Brien, R. Coombs, M. E. Poscher, D. M. Jacobsen, G. M. Shaw, D. D. Richman, and P. A. Volberding. 1996. HIV viral load markers in clinical practice. Nat. Med. 2:625-629. [DOI] [PubMed] [Google Scholar]
- 10.Segondy, M., J. Izopet, I. Pellegrin, B. Montes, B. Dumon, C. Pasquier, M. Peeters, H. J. Fleury, J. Puel, and J. Reynes. 1998. Comparison of the Quantiplex HIV-1 RNA 2.0 assay with the Amplicor HIV-1 Monitor 1.0 assay for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma of patients receiving stavudine-didanosine combination therapy. J. Clin. Microbiol. 36:3392-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeni, P., S. Hammer, C. Carpenter, D. Cooper, M. Fischl, J. Gatell, B. Gazzard, M. Hirsch, D. Jacobsen, D. Katzenstein, J. Montaner, D. Richman, M. Saag, M. Schechter, R. Schooley, M. Thompson, S. Vella, and P. Volberding. 2002. Antiretroviral treatment for adult HIV infection in 2002. JAMA 288:222-235. [DOI] [PubMed] [Google Scholar]
- 12.Yen-Lieberman, B., D. Brambilla, B. Jackson, J. Bremer, R. Coombs, M. Cronin, S. Herman, D. Katzenstein, S. Leung, H. J. Lin, P. Palumbo, S. Rasheed, J. Todd, M. Vahey, and P. Reichelderfer. 1996. Evaluation of a quality assurance program for quantification of human immunodeficiency virus type 1 RNA in plasma and AIDS Clinical Trials Group virology laboratories. J. Clin. Microbiol. 34:2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]