Abstract
A clear perception of gene essentiality in bacterial pathogens is pivotal for identifying drug targets to combat emergence of new pathogens and antibiotic-resistant bacteria, for synthetic biology, and for understanding the origins of life. We have constructed a comprehensive set of deletion mutants and systematically identified a clearly defined set of essential genes for Streptococcus sanguinis. Our results were confirmed by growing S. sanguinis in minimal medium and by double-knockout of paralogous or isozyme genes. Careful examination revealed that these essential genes were associated with only three basic categories of biological functions: maintenance of the cell envelope, energy production, and processing of genetic information. Our finding was subsequently validated in two other pathogenic streptococcal species, Streptococcus pneumoniae and Streptococcus mutans and in two other gram-positive pathogens, Bacillus subtilis and Staphylococcus aureus. Our analysis has thus led to a simplified model that permits reliable prediction of gene essentiality.
The search for essential genes has long been a challenge. An essential gene is defined as one whose loss is lethal under a certain environmental condition. The identification of essential genes in bacteria promises to (i) identify critical genes and pathways for controlling pathogenic bacteria by identifying potential targets for antimicrobial drug development1; (ii) reveal the minimal gene set for living organisms and to shed light on the origin of life2,3; and (iii) reveal bacterial relationships during evolution4,5. Several genome-wide mutant libraries of model microbes have been constructed3,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22. Although these libraries are invaluable for research in systems biology they show inconsistent essential gene results, even for closely related strains11,15,23. This lack of consensus has prevented reliable prediction of essential genes or pathways in species that have not yet been examined.
To obtain a reliable account of essential genes, we turned to an opportunistic pathogen, Streptococcus sanguinis strain SK36, after completing its genome sequence24. The streptococci encompass a large group of important human pathogens. Many Streptococcus species are responsible for infectious diseases, such as pneumonia, bacteremia, strep throat, rheumatic fever, scarlet fever, meningitis, infective endocarditis, and dental caries. S. sanguinis has long been recognized as a principal causative agent of infective endocarditis25, and its virulence factors have been the subject of a number of investigations26,27,28. It also initiates biofilm formation on tooth surfaces. The complete genome sequence provides an opportunity to greatly advance our understanding of this organism by enabling the construction of a comprehensive set of genome-wide mutants. S. sanguinis is in many ways an ideal candidate for such a study. The SK36 chromosome is 2.39 Mb and contains 2270 putative protein-coding genes, far fewer than that of most microbes used in previous genome-wide gene replacement mutagenesis studies; e.g., Acinetobacter baylyi with ∼3310 genes21, Bacillus subtilis with ∼4100 genes13, E. coli with ∼4300 genes11, and S. cerevisiae with ∼6600 genes7. More importantly, S. sanguinis is highly competent. In our laboratory, up to 20% of S. sanguinis cells can be transformed by a simple method using ≤50 ng of DNA. Mutants of non-essential genes are therefore readily obtained, facilitating identification of essential genes. We report here identification of essential genes in S. sanguinis and a simplified picture of gene essentiality in streptococci and other bacteria that has emerged from our findings.
Results
Generation of S. sanguinis mutants
We set out to identify the essential genes of S. sanguinis by systematic gene replacement. Taking advantage of the completed S. sanguinis genome, we designed a PCR method (Figure S1A) to precisely replace target genes with the aphA-3 kanamycin resistance (Kmr) gene30. The cassette used initially lacked a promoter to avoid potential dysregulation of downstream genes, which could complicate subsequent use of the mutant library in functional studies. To facilitate translation, we provided ribosome binding sites (RBS) for the aphA-3 and adjacent downstream S. sanguinis genes. To ensure efficient homologous recombination, we created long (∼1 kb) flanking sequences upstream and downstream of each targeted gene. Flanking sequences were limited to 1 kb based on two considerations: (i) the total length of the amplicon (∼3 kb; 1 kb for the aphA-3 gene plus 2 kb of flanking sequences) allowed ready PCR amplification; and (ii) the feasibility of sequencing flanking regions by Sanger's method from both ends (∼1 kb). To test our approach, we randomly selected 10 and then 96 genes for mutagenesis by double cross-over homologous recombination. We successfully created 10 and 93 mutants, respectively, suggesting that the approach was efficient. We then designed and synthesized primers for replacement of 2266 of the 2270 ORFs in S. sanguinis, excluding only those four ORFs contained entirely within larger ORFs. In total, we synthesized over 10,000 primers in the construction of the 2266 gene replacement constructs (Table S1). These constructs were created by high-throughput PCR and purification in a 96-well format. Three PCR reactions were required for each replacement and one or more PCR reactions for mutant confirmation (Figure S1A and Figure 1). Over 9,000 PCR fragments were amplified for mutant creation and confirmation. To preclude false identification of genes as essential due to low transformation frequencies, we performed experiments to optimize the transformation efficiency of S. sanguinis SK3626. Using our optimized method, up to 2x106 mutant colonies could be obtained from 107 bacterial cells for non-essential gene transformations.
Figure 1. Flowchart of mutant construction.
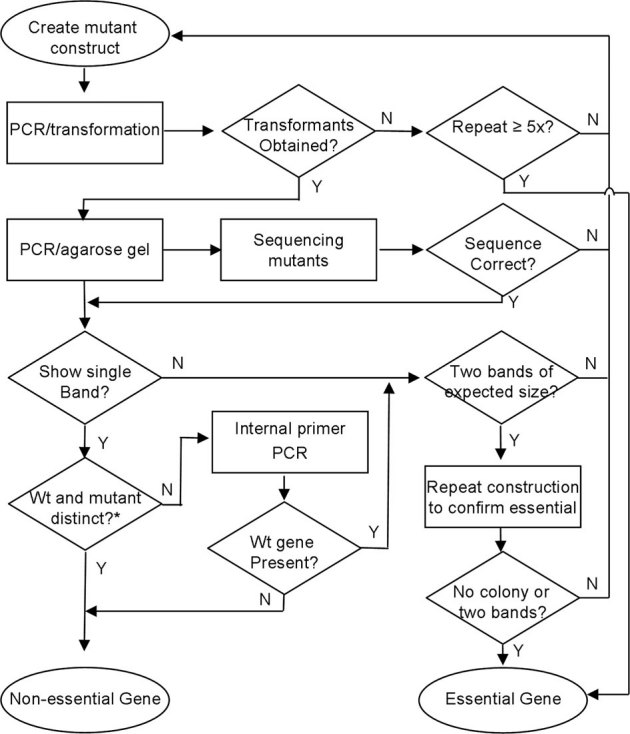
Procedures for creation and identification of essential genes. Wt, wild type. *Wild-type and mutant band sizes expected to differ by ≥100 bp.
In the initial round of replacements, we found that many of the genes for which mutants could not be obtained were annotated as acquired via horizontal gene transfer or as encoding hypothetical proteins24. We were curious as to the explanation for this finding. The expression of these genes was therefore examined by microarray analysis. Many had undetectable expression (Table S2). This led us to suspect that many of the unrecoverable mutants might have resulted from insufficient expression of the promoterless aphA-3 gene. To account for this possibility, we created a second mutagenic construct that retained the native aphA-3 promoter30, and used it to re-mutagenize the 142 genes that had generated no recoverable mutants (Figures S1). We obtained over 60 additional mutants (Table S3), confirming our hypothesis. Lastly, the Kmr cassette-gene junctions of every mutant were sequenced for confirmation.
Identification of essential genes in S. sanguinis
Two types of essential genes were identified in S. sanguinis. The first type was genes whose attempted mutagenesis yielded no transformants. New amplicons for these genes were re-amplified and re-transformed in a second cycle. A non-essential gene, SSA_016928, was used as a positive transformation control to assess whether the failed replacements were due to essential target genes or due to low transformation efficiency of the competent cells. The genes that were not successfully mutagenized after five independent attempts were classified as essential (Figure 1). There were 60 essential genes of this type in S. sanguinis (Figure S1B).
For the second type of essential gene, mutant colonies produced double-bands in PCR amplifications using F1 and R3 flanking primers (Figure S1A). The size of one DNA band typically corresponded to the size expected for the replacement mutant while the other matched the wild-type gene. We interpreted this as indicating that these genes were also essential, such that selection for Km resistance resulted in duplication of the target gene21. To precisely identify double-band mutants, after sequence confirmation, we examined all PCR amplicons by 1% agarose gel electrophoresis for 4 hrs. Under this agarose gel electrophoresis condition, any amplicons with ≥ 100 bp difference were clearly identified. When bands resulting from amplification of the Kmr cassette and the wild-type gene were anticipated to differ by < 100 bp, an internal T1 primer was designed to determine whether a wild-type gene could be detected by PCR. Of 498 mutants examined with internal T1 primers, 57 produced an amplicon with the same size as in the wild type strain, indicating a “double-band” mutant. (Tables S1 and S3). We identified a total of 158 double-band essential genes (Figure S1B). Our final result was the identification of 218 essential genes including those that we could not mutagenize and those that gave rise to double-bands (Figure S1B, Table S3).
Essential genes associated with specific pathways
The distribution of essential genes in specific categories based on KEGG pathway maps31 was examined. The categories of translation, carbohydrate metabolism, nucleotide metabolism, and replication and repair had the greatest number of essential genes, with over 30 each (Figure S2). The categories with the highest percentage of essential genes were translation (76%), transcription (67%), protein folding, sorting and degradation (57%), and glycan biosynthesis (50%).
We next assigned genes to biochemical pathways based on their annotations in KEGG (see Figure 2 for an overview and Table S4 for a detailed analysis). The essential genes were clustered in many pathways: (i) glycolysis, (ii) pentose phosphate pathway (PPP), (iii) peptidoglycan biosynthesis, (iv) terpenoid backbone biosynthesis, (v) glycerophospholipid metabolism, (vi) glycerolipid metabolism, (vii) fatty acid biosynthesis, (viii) nucleotide biosynthesis, (ix) metabolism of cofactors and vitamins including folate biosynthesis, (x) energy metabolism (production of ATP, NADH and NADPH), (xi) DNA replication, (xii) transcription, (xiii) protein biosynthesis, (xiv) GTP-binding proteins and (xv) cell division.
Figure 2. Concise pathways of S. sanguinis SK36 essential genes.
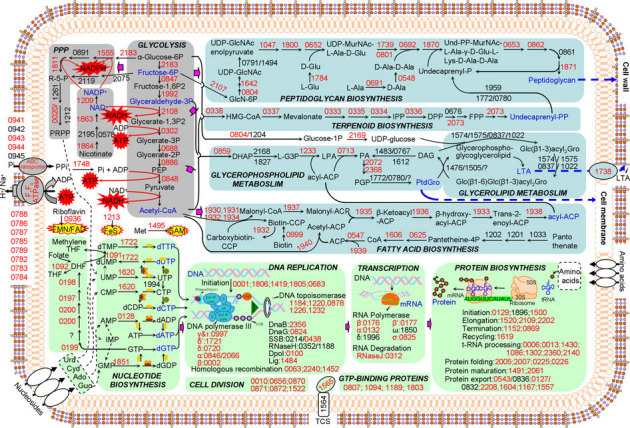
The three functions associated with essential pathways are indicated by color: pale blue, cell envelope; gray, energy production; light green, processing of genetic information. Numbers are SSA#; red number, essential gene; black number, nonessential gene; solid arrow, an enzymatic reaction; dashed arrow, multistep pathway; blue dashed arrow, products involved in cell wall and membrane formation; block arrow, product from one pathway serving as input to another pathway; oval with bold arrow, transporter; slash, separating paralogs. Ado, adenosine; Cyd, cytidine; DAG, 1,2-diacylglycerol; DHAP, dihydroxyacetone phosphate; DHF, dihydrofolate; L-G3P, sn-glycerol 3-phosphate; Guo, guanosine; LPA, lysophosphatidic acid; LTA, lipoteichoic acid; PA, phosphatidic acid; PGP, phosphatidylglycerophosphate; PPP, pentose phosphate pathway; TCS, two-component system; THF, tetrahydrofolate; Urd, uridine. In nucleotide biosynthesis, symbols are used to represent essential genes that participate in multistep reactions: SSA_1865 (green circle), SSA_0848 (yellow oval), SSA_2263, SSA_0771 (orange circles) and SSA_0768 and SSA_0770 (brown oval). Essential genes associated with ribosome and aminoacyl-tRNA biosynthesis are not indicated here, nor are SSA_0575, SSA_0800 and SSA_1903, which could not be categorized into these pathways.
These results were in keeping with expectation, given our current understanding of bacterial metabolism. Among the metabolic pathways containing essential genes, glycolysis and pyruvate oxidation from α-D-glucose 6-phosphate to acetyl-CoA play a pivotal role because they generate energy and provide input molecules for other essential metabolic pathways. Terpenoid backbone biosynthesis feeds into the pathway for synthesis of peptidoglycan, which is the main component of the cell wall. Glycerophospholipid metabolism in conjunction with glycerolipid produces lipoteichoic acid (LTA), a component of the cell wall in gram-positive bacteria. Phospholipids biosynthesized from glycerophospholipid metabolism and fatty acid biosynthesis compose the cell membrane. The nucleotides synthesized from PRPP derived from the pentose phosphate pathway (PPP) or from nucleosides imported from the growth medium supply materials for DNA replication and transcription. Several vitamins and cofactors including folate, biotin, pantothenate, nicotinate, riboflavin, SAM and FeS clusters are involved in the pathways above. ATP and NADH/NADPH produced from glycolysis, pyruvate oxidation and PPP provide energy.
We then analyzed essential genes by network functions using systems biology. All essential genes and their related functions were linked together in pathways and studied as a whole. This picture was simplified dramatically when it became apparent that all of these pathways could be linked to three basic biological functions: maintenance of the cell envelope, energy production and processing of genetic information. Remarkably, we found only three essential genes (none of which have been assigned exact functions) that could not be linked to these three functions (SSA_0575, SSA_0800, and SSA_1903; Table S4). SSA_0575 is annotated as a haloacid dehalogenase-like hydrolase, SSA_0800 as a glutamine amidotransferase and SSA_1903 as a conserved hypothetical protein.
Identification of additional essential genes in minimal medium
From our analysis, we found that the three functions of maintenance of the cell envelope, energy production and processing of genetic information, and no others, are essential to S. sanguinis. Based on the genome sequence, we predicted that S. sanguinis possesses de novo synthesis pathways for all amino acids and nucleotides from glycolysis and PPP (Figure 3A). We identified 96 genes responsible for biosyntheses of 19 amino acids starting from glycolysis, leaving only the undetermined gene responsible for L-asparagine biosynthesis from L-aspartate (Figure 3A). Two pathways each were present for biosyntheses of L-glutamate, L-serine and L-glycine and L-threonine. Yet, in our initial screen, we found few essential genes related to amino acid synthesis or nucleotide synthesis from PRPP to UMP/GMP/AMP, suggesting amino acids and nucleotide precursors were provided by the rich brain heart infusion (BHI) medium. To examine this possibility, we compared the growth of strains with mutations in genes responsible for amino acid and nucleotide de novo synthesis in a chemically defined medium (CDM) with that in BHI, as described previously27 (Figure 3B; Table S5). CDM contains only 3 amino acids (L-glutamate, L-cysteine and L-leucine) and lacks nucleosides and nucleobases. All of these mutants grew to levels similar to SK36 in BHI medium (Figure 3B). However, the growth of the mutants was significantly lower than that of SK36 in CDM medium, with the following exceptions: (i) the mutants whose deleted genes were involved in the synthesis of the amino acids that were supplied in CDM medium (Glu, Cys and Leu); (ii) the mutants possessing alternative pathways (Ser, Gly and Thr); and (iii) the mutants whose replaced genes had paralogs or isozymes. Although dramatically reduced, some of the mutants still grew to low levels, perhaps benefitting from carry-over of nutrients from the inocula. Similar results were obtained with 23 mutants associated with nucleotide biosynthesis from PRPP and/or L-glutamine (Figure 3B). The combined results suggest that S. sanguinis does indeed obtain required amino acids and nucleotide precursors from the BHI medium. Provision of nutrients by BHI appears to explain another result that initially appeared discrepant with our model. We predicted that SSA_1201, SSA_1202 and SSA_1033 would be essential because they were required for biosynthesis of CoA from pantothenate, but mutants of these three genes were obtained (Figure 2). We found, however, that these mutants exhibited minimal or undetectable growth in CDM, suggesting pantetheine-4P is provided by BHI (Table S5).
Figure 3. Growth of S. sanguinis mutants related to amino acid and nucleotide biosyntheses in BHI and CDM media.
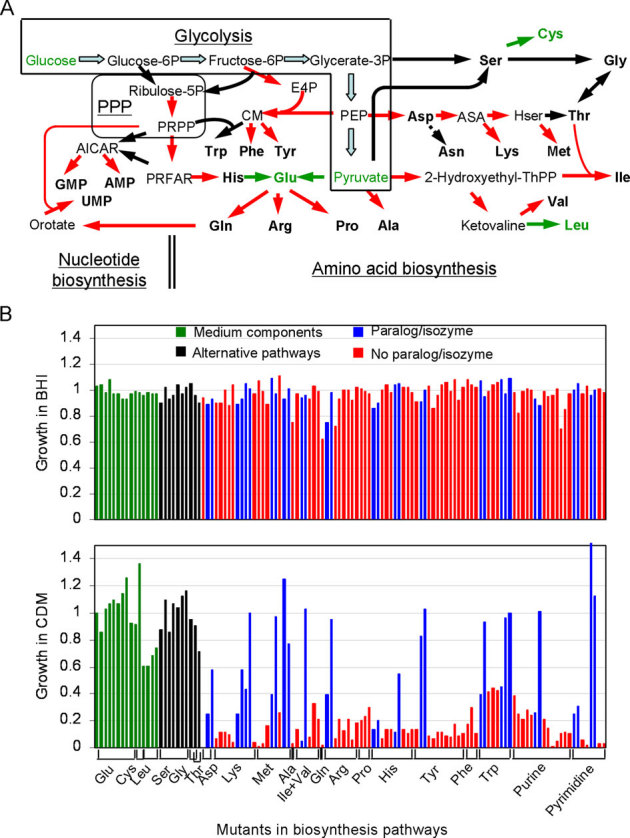
(A) amino acid and nucleotide biosynthesis pathways in which mutants grew (black arrow) or did not grow (red arrow) in CDM medium. Components of the CDM medium are indicated in green. PPP, pentose phosphate pathway, dashed arrow, undetermined gene responsible for asparagine biosynthesis. Note the two pathways for serine, glycine, threonine, glutamate and AICAR biosyntheses. ASA, L-aspartate 4-semialdehyde; CM, chorismate; E4P, D-Erythrose 4-phosphate; Hser, homoserine or o-phospho-L-homoserine; PRFAR, phosphoribulosyl-formimino-AICAR-phosphate. (B) growth of mutants in BHI and CDM media. Relative growth was represented by mutant OD450 / SK36 OD450 after 2d anaerobic incubation.
Examination of paralogs and isozymes with double-knockout mutants
In our analysis, we also found that some genes predicted to be critical in one of these three functions were not identified as essential. We considered that their functions may be performed by genes encoding paralogs or isozymes with similar functions. To examine this possibility, we identified gene paralogs based on protein sequence homology (Table S3). We also examined the genome annotation and literature for potential isozymes. When we re-examined the linked essential genes in the network, we found paralogs or isozymes in every case in which a series of linked essential genes was interrupted by a “non-essential” gene in the same pathway (Figure 2; for example, paralogs SSA_0791/SSA_1494 involved in peptidoglycan biosynthesis, isozymes SSA_0578/SSA_2195 involved in NAD+/NADP+ biosynthesis). This suggested that these non-essential genes might substitute for one another in performing essential functions. To test this hypothesis, we selected four pairs of paralogous or isozyme genes for double-gene knockouts to examine essentiality: SSA_0791/SSA_1494, SSA_1827/SSA_2168, SSA_0578/SSA_2195, and SSA_0352/SSA_1188. As a control, double mutants were created by combining the replacement of each gene of interest with a replacement of the SSA_0169 gene, which is a hypothetical gene with no known function28. As we anticipated, double mutants could not be constructed for the gene pairs SSA_0791/SSA_1494, SSA_0578/SSA_2195, or SSA_0352/SSA_1188, whereas SSA_0169 double mutants were readily obtained (Table 1). Unexpectedly, the double mutant SSA_1827/SSA_2168 was viable. This may result from the existence of an alternative pathway (composed of SSA_0049/SSA_0050, SSA_0287 and SSA_1826) in glycerolipid metabolism.
Table 1. Double-gene knockouts in S. sanguinis.
| Double mutant | Donor | Product Name | Acceptor | CFU | Gel | Essential |
|---|---|---|---|---|---|---|
| Ssx_0791::1494 | SSA_0791 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | Ssx_1494 | 0 | yes | |
| Ssx_0578::2195 | SSA_0578 | nicotinic acid mononucleotide adenylyltransferase | Ssx_2195 | 0 | yes | |
| Ssx_0352::1188 | SSA_0352 | ribonuclease HIII | Ssx_1188 | 0 | yes | |
| Ssx_2168::1827 | SSA_2168 | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase | Ssx_1827 | >5000 | 1-band | No |
| Ssx_0169::1494 | SSA_0169 | hypothetical protein | Ssx_1494 | >5000 | 1-band | No |
| Ssx_0169::2195 | SSA_0169 | Donor Product Name | Ssx_2195 | >5000 | 1-band | No |
| Ssx_0169::1188 | SSA_0169 | hypothetical protein | Ssx_1188 | >5000 | 1-band | No |
| Ssx_0169::1827 | SSA_0169 | hypothetical protein | Ssx_1827 | >5000 | 1-band | No |
All mutants with double crossover replacement are named Ssx. The number indicates the replaced gene.
Identification of essential genes in other streptococcal species
To validate the hypothesis that essential genes are associated with these three functions, we examined representative genes in two more pathogenic streptococcal species. We first selected 8 genes in S. pneumoniae strain TIGR4 that were not previously identified as essential in either of two S. pneumoniae genome-wide essential gene screens10,17. These 8 genes are predicted to be involved in maintenance of the cell envelope or in energy production (SP_0382, SP_0383, SP_0384 and SP_0262, involved in terpenoid backbone biosynthesis; SP_1511, SP_1513 and SP_1514 encoding F1Fo-ATPase subunits; and SP_0261 involved in glycerophospholipid metabolism). We also selected one gene (SP_0489) as a control that encodes a protein unrelated to these three essential functions whose ortholog was non-essential in S. sanguinis. The S. pneumoniae genes were mutagenized and then characterized in the same manner as for S. sanguinis. As shown in Table 2, none of the eight genes could be mutagenized, although the control produced numerous transformants. The process was repeated for another pathogen, S. mutans strain UA159. The orthologs of the above 8 genes in S. mutans were selected and examined for gene essentiality, yielding identical results (Table 2). Again, all tested genes were identified as essential in S. mutans. These results suggested the accuracy of our essential gene predictions for other streptococci.
Table 2. Confirmation of essential genes in S. pneumoniae and S. mutans.
| SP_ID* | Product Name | CFU† | Gel‡ | Essential | SMU_ID* | CFU | Gel‡ | Essential |
|---|---|---|---|---|---|---|---|---|
| SP_0382 | diphosphomevalonate decarboxylase | 0 | yes | SMU.937 | 0 | yes | ||
| SP_0383 | phosphomevalonate kinase | 0 | yes | SMU.938 | 0 | yes | ||
| SP_0384 | Isopentenyl pyrophosphate isomerase | 0 | yes | SMU.939 | 0 | yes | ||
| SP_1514 | F0F1 ATP synthase subunit C | 2 | 2-band | yes | SMU.1534 | 1 | 2-band | yes |
| SP_1513 | F0F1 ATP synthase subunit A | 2 | 2-band | yes | SMU.1533 | 0 | yes | |
| SP_1511 | F0F1 ATP synthase subunit delta | 6 | 2-band | yes | SMU.1531 | 1 | 2-band | yes |
| SP_0262 | phosphatidate cytidylyltransferase | 0 | yes | SMU.1785 | 0 | yes | ||
| SP_0261 | undecaprenyl pyrophosphate synthase | 0 | yes | SMU.1786 | 1 | 2-band | yes | |
| SP_0489 | PAP2 family protein | 242 | 1-band | no | SMU.1702c | 874 | 1-band | no |
*Gene identifiers. IDs in the same row indicate orthologous genes
†Number of transformants obtained from attempt to mutagenize the gene indicated
‡Appearance of PCR products resulting from amplification of the target gene locus from selected transformants
We used comparative genomics32 to examine the conservation among streptococcal species of the genes we identified as essential. We downloaded 49 complete streptococcal genomes publicly available from the NCBI database and compared them with the S. sanguinis essential genes. The vast majority of S. sanguinis essential genes (202 of 218) had orthologs in all of the other 48 streptococcal genomes (Table S6A). Thus, in agreement with expectation, most of the genes we identified as essential were highly conserved within streptococci.
Comparison with essential genes in other gram-positive bacteria
We then compared essential genes of S. sanguinis with those in other species that have been examined experimentally. All essential genes of the 13 bacterial species presently included in the Database of Essential Gene (DEG)33 were collected and searched against the S. sanguinis protein database by BLASTP to find homologs (Table S7). The sequence comparison suggested that there were many differences. In the other species, about half (33 to 66%) of the essential genes with homologs in S. sanguinis matched S. sanguinis essential genes, with the rest matching non-essential S. sanguinis genes.
To find the reason for this inconsistency, we analyzed more carefully the other two gram-positive species in the database, Staphylococcus aureus NCTC8325 and B. subtilis 168 (Figure 4 and Table S8), that were subjected to genome-wide screens13,23 in similar nutrient-rich media (BHI and LB). Some essential genes in S. sanguinis had homologs identified as non-essential in S. aureus and B. subtilis, and vice versa although many essential genes in S. sanguinis were shared with these two species. However, we found that most of the differences could be explained by gene paralogs, isozymes, or alternative pathways. As shown in Figure 4, B. subtilis synthesizes peptidoglycan from UDP-N-acetylmuramoyl-L-alanyl-D-glutamate via an alternative pathway using meso-2,6-diaminopimelate rather than L-lysine that results in the genes responsible for producing meso-2, 6-diaminopimelate from L-aspartate 4-semialdehyde via lysine biosynthesis pathway being essential. S. aureus cross-links peptidoglycan via the essential femXAB operon rather than the streptococcal murMN. The essential mevalonate pathway in the terpenoid backbone pathway in S. sanguinis and S. aureus is replaced by an essential MEP/DOXP pathway in B. subtilis. In S. sanguinis, LTA and wall teichoic acid (WTA, a component of cell wall in gram-positive bacteria) are likely synthesized by the same set of enzymes because both have identical components in S. sanguinis34 and repeating units with identical structures in S. pneumoniae35, whereas S. aureus and B. subtilis use a set of enzymes different from LTA synthesis to produce wall teichoic acid, and the genes encoding them are essential. S. sanguinis lacks a respiratory chain and undergoes fermentation to produce acids such as lactate and acetate, and the F1Fo-ATPase uses ATP hydrolysis to pump intracellular protons out. This is indispensible for creating a protonmotive force for a variety of transport processes and to maintain pH homeostasis, which likely results in the genes encoding F1Fo-ATPase being essential in S. sanguinis. In contrast, S. aureus and B. subtilis use the electron transport chain to generate protonmotive force, and the genes associated with its biosynthesis (i.e. menaquinone) are essential. B. subtilis and S. aureus likely maintain cellular pH homeostasis via a Na+/H+ transporter, the genes for which are essential in B. subtilis and paralogous in S. aureus. S. sanguinis lacks this transporter. In genetic information processing, the set of essential genes in S. sanguinis is smaller than those in other two species, but their essential pathways are still identical.
Figure 4. Essential gene comparisons among S. sanguinis, S. aureus and B. subtilis.
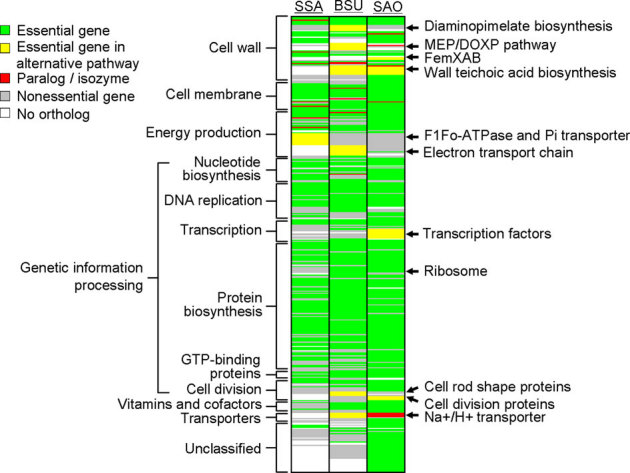
Essential gene comparisons were based on sequence homologs, and genes were grouped according to pathways or functions. SSA, S. sanguinis SK36 genes; BSU, B. subtilis 168 genes; SAO, S. aureus NCTC8325 genes. Colors represent gene categorizations, as indicated in the figure.
Discussion
Accurate prediction of essential genes is important for identification of drug targets to combat the emergence of pathogens and antibiotic-resistant bacteria20, especially for serious infectious agents for which there is no research model system available. In addition, the rational design of bacterial cells through synthetic biology, as is currently possible, requires an understanding of the minimal gene set for further advances3,36. However, the lack of consensus among experimental essential gene lists has prevented prediction of essential genes in other species. In our essential gene comparisons, we found significant differences of essential genes identified for S. sanguinis and those identified in previous studies with other species. We suggest several possible explanations for these differences. (i) We would expect some different genes due to genetic inheritance among species. For example, the essential genes for cell wall composition in gram-positive bacteria are different from those in gram-negative bacteria. (ii) Identified essential genes using single-gene knockouts will vary when the bacteria being compared differ in protein paralogs, isozymes or alternative pathways for essential functions. These cases can be tested by creating double-gene knockouts, as we have demonstrated (Table 1). (iii) Differences in screening conditions may have caused differences in essential gene identification, although the most favorable environmental conditions for each bacterium were used in most screens. As we found for S. sanguinis above, the use of a minimal medium obviously results in the identification of additional essential genes. (iv) False-negative results may have been obtained in some cases due to partial gene inactivation, which is known to occur frequently in random insertion mutagenesis37. (v) Some genes may have been falsely identified as essential because of genetic system limitations. Many bacterial species have low transformation frequencies. In these cases, even a moderate reduction in plating efficiency may result in lack of mutant recovery, resulting in categorization of the gene as “essential” in a large-scale, genome-wide mutagenesis. (vi) Finally, insufficient expression of the selection marker, as occurred with our promoterless Kmr gene, may result in false-positive identification of essential genes.
Many in silico methods have also been established to predict essential genes. These methods include ortholog identification23, genomic intrinsic feature analysis38, gene evolution rate39, phylogenetic conservation40, network analysis41 and machine learning based integrative approaches38,42. Of these, essential gene prediction via phyletic conservation is the most commonly used. Our results indicate that the vast majority of S. sanguinis essential genes are indeed conserved in closely related species (Table S6A). We also found that essential genes had greater sequence conservation among the streptococci than non-essential genes. The average identity for the essential gene protein sequences was 79.71% while for the non-essential gene protein sequences, it was 58.96% (Table S6B). It should be noted, however, that phyletic conservation alone was a poor predictor of gene essentiality. We found 787 non-essential S. sanguinis genes with orthologs in all 48 streptococcal genomes. Inclusion of bacteria from other genera in our analysis would have reduced the number of conserved genes identified, likely increasing the specificity of the phyletic method for essential gene prediction. However, the sensitivity of this method then decreases, as we found a number of essential S. sanguinis genes with orthologs that can be detected in different genera by identity of annotated function, but not by sequence similarity (data not shown).
Using our findings, we believe that most essential genes in other bacteria can now be predicted based on their genome annotations. We have established a model of essential pathways (Figure 5), in which the essential genes are linked by crucial chemical compounds. Although essential genes may differ among species due to different final products or alternative pathways, we propose they will contribute to the three basic functions of maintenance of the cell envelope, energy production, and processing of genetic information. We used this model for predicting essentiality of paralogs and isozymes by double-gene knockouts in S. sanguinis and for predicting essential genes in S. mutans, S. pneumoniae, B. subtilis and S. aureus. Our predictions were largely confirmed.
Figure 5. Deduced essential pathways based on S. sanguinis essential genes.
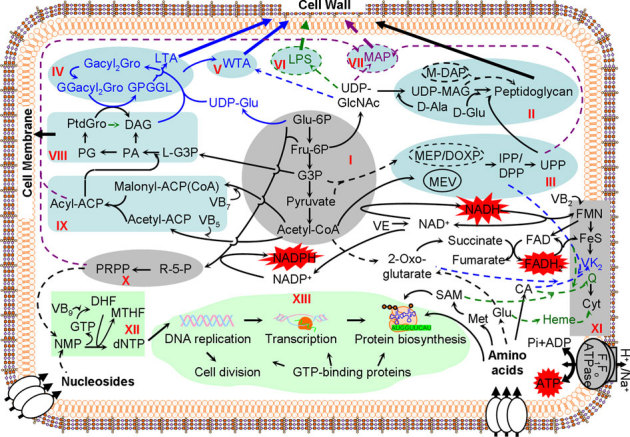
Colors indicate the three functions associated with essential pathways, as in Figure 2. I, glycolysis and pyruvate oxidation; II, peptidoglycan biosynthesis; III, terpenoid backbone biosynthesis; IV, glycerolipid metabolism (LTA biosynthesis); V, WTA biosynthesis; VI, lipopolysaccharide biosynthesis; VII, MAP biosynthesis; VIII, phosphoglycerolipid metabolism; IX, fatty acid biosynthesis; X, electron transport chain; XI, pentose phosphate pathway; XII, nucleotide biosynthesis; XIII, genetic information processing including DNA replication, transcription, protein biosynthesis, GTP-binding proteins and cell division. Black arrow, all bacteria; black dashed arrow or oval, alternative pathways; blue arrow, gram-positive bacteria only; green arrow, gram-negative bacteria only; violet arrow, mycobacteria only; bold arrow, synthesis of cell envelope. CA, chorismate; Cyt, cytochrome; DAG, 1,2-diacylglycerol; DG-DAG, diglucosyl-diacylglycerol; G3P,D-glyceraldehyde 3-phosphate; GPGGL, glycerophosphoglycoglycerolipid; L-G3P, glycerol-3-phosphate; LPS, lipopolysaccharide; Malonyl-ACP(CoA), malonyl-ACP or malonyl-CoA; M-DAP, meso-2,6-diaminopimelate; MEV, mevalonate; MG-DAG, monoglucosyl-diacylglycerol; PA, phosphatidate; PG, phosphatidylglycerophosphate; R-5-P, D-Rribulose 5-phosphate; PPP, pentose phosphate pathway; PtdGro, phosphatidylglycerol; UDP-MAG, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate; UPP, undecaprenyl diphosphate.
This model can also be used to explain many of the apparent inconsistencies observed previously among different organisms that were not predictable based on gene sequence conservation alone. Bacteria can be categorized into gram-negatives, gram-positives and Mycobacteria due to the differences in their cell envelope compositions. The greatest difference is that gram-negative bacteria contain an outer membrane with lipopolysaccharides while LTA and WTA are found in gram-positive bacteria and mycolyl-arabinogalactan-peptidoglycan complex in Mycobacteria. This leads to different sets of essential genes responsible for the respective envelope components. In the gram-negative bacteria E. coli18 and Pseudomonas aeruginosa19, the genes responsible for lipopolysaccharide biosynthesis have been demonstrated essential. Many genes responsible for biosynthesis of mycolyl-arabinogalactan-peptidoglycan complex were identified as being essential in Mycobacterium tuberculosis14. In B. subtilis and S. aureus13,23, the genes responsible for the biosynthesis of WTA and LTA were essential. Our results indicated most S. sanguinis orthologs of genes responsible for LTA biosynthesis in S. aureus were nonessential. Although this can be explained by the existence of paralogs (Figure 2), we cannot exclude the possibility of an alternative biosynthetic pathway for S. sanguinis LTA synthesis, as glycerophosphate residues in the LTA of S. sanguinis DSM 20567 may be substituted with D-alanine ester, α-D-glucosyl and α-isomalto-oligosaccharide residues43.
Most bacteria possess a respiratory chain that is important for energy production and maintenance of redox balance. It has been demonstrated that the genes involved in synthesis of electron transport chain components, such as CoQ, menaquinone or heme, are essential in E. coli18, P. aeruginosa19, B. subtilis and S. aureus13,23. Although an important function of electron transport is generation of ATP via the F1Fo-ATPase, there was only one instance of an F1Fo-ATPase gene being identified as essential in previous studies performed with the latter two species9,13,23. One possibility is that these genes were missed. Most replacements of F1Fo-ATPase genes in S. sanguinis, S. pneumoniae, and S. mutans generated “double-band” mutants (Table 2; Table S3). This would lead to identifying F1Fo-ATPase genes as nonessential in studies in which the presence of double-bands was not investigated. Nevertheless, the ability of these species to grow anaerobically indicates that ATP generation by F1Fo-ATPase is not essential, suggesting that electron transport is required for other purposes, such as secondary active transport. In S. sanguinis, all eight subunits of the F1Fo-ATPase were identified as essential. Since streptococci lack a respiratory chain, the essential function of the F1Fo-ATPase in these species is also not generation of ATP and is, instead, likely to be export of protons using energy from ATP hydrolysis. A previous analysis of essential genes in S. pneumoniae identified five of the eight F1Fo-ATPase subunit genes as essential10,17. Here, we demonstrated the other three genes encoding F1Fo-ATPase components were also essential in S. pneumoniae, as were their orthologs in S. mutans (Table 2).
In conclusion, by choosing an ideal test organism and by employing exhaustive measures to avoid false-positive and false-negative identifications, we have reliably identified the essential genes of S. sanguinis. The validity of our findings is suggested by the virtually perfect association between our list of essential genes and the list of genes expected to be required for the three functions of cell envelope maintenance, energy production, and processing of genetic information. Although S. sanguinis is important in its own right, the relative ease with which these results can be used to identify essential genes in other prominent streptococcal pathogens including S. pyogenes and S. pneumoniae lends increased importance to this work. Moreover, our study suggests that with minimal additional effort, our results can be used to predict essential genes for most bacteria for which an annotated genome sequence is available.
It should be noted that although we examined all protein coding regions for their essentiality, it is well known that some non-coding regions are also essential if they contain DNA sequences for important biological functions. Such sequences include the chromosomal origin of replication, promoters, tRNAs, rRNAs and perhaps small RNAs.
Finally, although the focus of the current study was the identification of essential genes, the study has also produced an ordered, comprehensive library of non-essential gene mutants of S. sanguinis SK36. The design of the mutagenic constructs to (i) ensure near-complete deletion of each gene; (ii) retain expression signals for adjacent genes; and (iii) introduce a promoter only in cases where it was required, in combination with the care with which each mutant was characterized ensures that the library will be of great value. We are currently employing the mutants for a number of investigations of gene function involving conventional and systems biology approaches.
Methods
Bacterial strains and growth
S. sanguinis strain SK36 was grown at 37°C in BHI broth (BD) as described previously27. S. pneumoniae TIGR4 and S. mutans UA159 strains purchased from ATCC were grown in Todd Hewitt (TH; BD) broth plus 0.5% yeast extract and in BHI broth under microaerobic conditions (6% O2, 7.2% CO2, 7.2% H2 and 79.6% N2).
Primer design and PCR
We developed a recombinant PCR method for in vitro creation of linear constructs for the replacement of every protein-coding gene in the S. sanguinis SK36 genome (Figure S1A). Based on the complete S. sanguinis SK36 genome sequence24, three sets of primers (F1/R1, F2/R2 and F3/R3) were designed to amplify the S. sanguinis sequence upstream from each targeted gene, the aphA-3 gene, encoding Kmr45 and the S. sanguinis sequence downstream from each targeted gene, respectively.
For most of the mutagenized genes, the R1 and F3 primers were designed to delete the coding region from 6 bp after the start codon to 30 bp before the stop codon. Stop codons were inserted in all three frames to prevent fusion of the N-terminus of the targeted open reading frame with the Kmr protein. The last 30 bp were retained to preserve potential ribosomal binding sites used by adjacent downstream genes. The upstream retained region was extended from 6 bp to 100 bp when two neighboring genes were located head-to-head in opposite orientation to prevent deletion of potential promoters for flanking genes. Primers R1 and F3 contained 25-nt sequences that are complementary with the antibiotic selection cassette at their 5' ends. The P1, P2, and various T1 primers were designed for sequencing to confirm mutants. The sequence of every primer is documented in Table S1.
Three PCR amplicons were created using F1/R1, F2/R2 and F3/R3. All PCR reactions were performed at 94°C for 1 min, and 30 cycles of 94°C for 30 sec, 54°C for 30 sec and 68°C for 1.5 min. After DNA purification by PureLink 96 PCR purification kits (Invitrogen), the three PCR amplicons were combined in equal amounts in one tube and amplified again using the F1 and R3 primers to obtain the final linear recombinant PCR amplicon. Conditions were 94°C for 2 min, 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 68°C for 3.5 min, and finally 68°C for 4 min. High-fidelity Platinum® Taq DNA polymerase (Invitrogen) was used in all reactions.
Promoterless and promoter-containing cassette construction
We initially created a promoterless Kmr cassette46 to eliminate possible polar transcriptional effects on neighboring genes. To address instances of poor expression of the ahpA-3 gene, we created a second construct in which the native promoter of aphA-3 gene30 was included in the Kmr cassette. Apart from the design of new R1_Promoter and F2 primers to include the promoter, all other construction steps were the same as for the promoterless constructs.
Gene replacement and mutant storage
The above linear PCR amplicons (∼50 ng) were directly transformed into S. sanguinis as described previously26. Allelic exchange mutants generated by double cross-over homologous recombination were selected by two-day microaerobic incubation on BHI agar plates containing 500 μg/ml kanamycin. For each replacement mutant, one colony was randomly picked and cultured in BHI with Km. To determine whether the mutant contained the expected gene replacement, colony PCR was performed using F1 and R3 primers. The PCR amplicon was examined by 20-cm long agarose gel electrophoresis. The amplicon was further confirmed by Applied Biosystems Big Dye terminator DNA sequencing. The sequencing confirmation was performed using the P1 primer, which binds to the Kmr cassette (Figure S1A). The mutants which gave rise to a DNA amplicon of the expected size by long gel electrophoresis and with the expected junction sequence determined from the P1 primer or the T1 primer were collected as the final confirmed mutants. These were retained and cryopreserved in 30% glycerol at −80 °C.
The methods for construction of gene replacement amplicons with the promoter-containing Kmr cassette for S. mutans UA159 and S. pneumoniae TIGR4 were the same as for S. sanguinis. Transformation of S. mutans UA159 was conducted similarly to that of S. sanguinis except S. mutans CSP was used. Transformation of S. pneumoniae TIGR4 was performed as described by Bricker and Camilli47.
Microarray analysis
S. sanguinis cultures at late log phase were used for microarray analysis. RNA from each of three independent samples was isolated by RNeasy mini kit (Qiagen, Valencia, CA). Spotted microarray slides were obtained from the Pathogen Functional Genomics Resource Center at JCVI. The microarray was performed according to the manufacturer's protocol. Each sample was divided into two parts and labeled separately by Cy5 and Cy3 dyes for microarray. The microarray data were analyzed using the programs Spotfinder and Midas to obtain the expression ratio of each gene labeled with each dye. All ratios were within the range of 0.6 to 1.5, indicating consistency in the labeling and analysis. Additionally, the ratio of dye intensity to background in a microarray slide was obtained after Spotfinder analysis. Absolute expression of each gene was represented by the average ratio of dye intensity to background for each slide. The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) with record GSE25340.
Growth comparison of mutants in CDM and BHI media
Selected mutants were cultured overnight at 37°C in 96-well blocks with 1 ml BHI broth under anaerobic conditions (10% CO2, 10% H2 and 80% N2 with a palladium catalyst). The overnight cultures were inoculated into 1 ml either CDM27 or BHI medium in 96-well plates by dipping with multichannel tips. The inocula were incubated for 2 d under the same conditions as above. Cultures were then mixed by pipetting several times with a multichannel pipette, and 200 µl cultures were transferred into 96-well plates for measuring OD450. The relative growth of each mutant was calculated as mutant OD450/SK36 OD450.
Gene function analysis
The pathway distributions of essential genes were analyzed via KEGG. KEGG annotations were downloaded to a local computer from /pub/kegg/genes/organisms/ssa/. The assigned KO numbers and path numbers of S. sanguinis genes were extracted. Multiple KO numbers or path numbers from KEGG were often assigned for a single gene if it was involved in different pathways. To view the essential pathways, these genes were assigned to the pathway with the greatest percentage of essential genes. To study essential genes as a whole, as many essential genes as possible were linked together via pathways. Based on a product in one pathway being the substrate of another pathway, the essential pathways were linked together and then integrated into their possible functions.
Comparative genomics
Comparative genomic analyses were performed to identify conserved proteins in S. sanguinis as previously described24. We downloaded 48 other completed streptococcal genomes and their annotations from public databases. S. sanguinis proteins were compared to other streptococcal protein databases by BLASTP. Significant matches (E < 1e-5) were analyzed to find homologs in other streptococcal genomes. The S. sanguinis database was also BLASTP searched against itself to identify paralogs. Protein sequence conservation was calculated by percent amino acid identity of orthologs. Essential genes in Staphylococcus aureus NCTC 8325, Bacillus subtilis 168 and S. pneumoniae were obtained from the Database of Essential Genes (DEG)33. The S. sanguinis orthologs were identified using BLASTP.
Author Contributions
PX conceived the main idea and the conceptual design of the experiments. PX, XG, LC, XW, YD, JX, JP, VS, MT and KE constructed mutants; PX designed primers and performed bioinformatics analysis; XG established the high throughput transformation system and performed microarray and gene functional analysis; XW performed double-knockout and mutant growth comparison; PX, XG, LC, JX, TK and DB analyzed the data. PX, XG, TK, DB and GB wrote the manuscript.
Supplementary Material
Supplementary Figures
Supplementary Tables
Acknowledgments
This work was supported by the National Institutes of Health grant R01DE018138 (PX) and, in part, by Virginia Commonwealth University Presidential Research Incentive Program (PRIP) 144602-3 (PX). We thank the DNA Core Facility at Virginia Commonwealth University for ABI Big Dye sequencing. We thank the Pathogen Functional Genomics Resource Center at JCVI for providing S. sanguinis spotted microarray slides.
References
- Gerdes S. Y. et al.. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol 184, 4555–72 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1, 127–36 (2003). [DOI] [PubMed] [Google Scholar]
- Glass J. I. et al.. Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A 103, 425–30 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I. K. et al.. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res 12, 962–8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B. Y., Scott N. M. and Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol 23, 2072–80 (2006). [DOI] [PubMed] [Google Scholar]
- Ji Y., et al.. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293, 2266–9 (2001). [DOI] [PubMed] [Google Scholar]
- Giaever G. et al.. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–91 (2002). [DOI] [PubMed] [Google Scholar]
- Akerley B. J. et al.. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci U S A 99, 966–71 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth R. A. et al.. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43, 1387–400 (2002). [DOI] [PubMed] [Google Scholar]
- Thanassi J. A. et al.. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res 30, 3152–62 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes S. Y. et al.. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185, 5673–84 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M. A. et al.. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100, 14339–44 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. et al.. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A 100, 4678–83 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H. and Rubin E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48, 77–84 (2003). [DOI] [PubMed] [Google Scholar]
- Knuth K. et al.. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol Microbiol 51, 1729–44 (2004). [DOI] [PubMed] [Google Scholar]
- Salama N. R., Shepherd B. and Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol 186, 7926–35 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. H. et al.. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol Cells 19, 365–74 (2005). [PubMed] [Google Scholar]
- Baba T. et al.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati N. T. et al.. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103, 2833–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A. et al.. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A 104, 1009–14 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Berardinis V. et al.. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol 4, 174 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. T. et al.. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol Microbiol 69, 67–76 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R. et al.. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics 10, 291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P. et al.. Genome of the opportunistic pathogen Streptococcus sanguinis. J Bacteriol 189, 3166–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Filippo S. et al.. Current patterns of infective endocarditis in congenital heart disease. Heart 92, 1490–95 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S. et al.. Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect Immun 73, 6064–74 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. et al.. Identification of Streptococcus sanguinis genes required for biofilm formation and examination of their role in endocarditis virulence. Infect Immun 76, 2551–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L. S. et al.. Development of genetic tools for in vivo virulence analysis of Streptococcus sanguinis. Microbiology 155, 2573–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. E. and London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol 175, 3247–52 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Klier A. and Courvalin P. DNA sequences specifying the transcription of the streptococcal kanamycin resistance gene in Escherichia coli and Bacillus subtilis. Mol Gen Genet 198, 348–52 (1985). [DOI] [PubMed] [Google Scholar]
- Kanehisa M. et al.. The KEGG resource for deciphering the genome. Nucleic Acids Res 32 Database issue D277–D280 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P. et al.. The genome of Cryptosporidium hominis. Nature 431, 1107–12 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang R. and Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res 37(Database issue) D455–D458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T. H., Emdur L. I. and Platt D. Lipoteichoic acids from Streptococcus sanguis. J Bacteriol 118, 471–9 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H. S. et al.. A new model of pneumococcal lipoteichoic acid structure resolves biochemical, biosynthetic, and serologic inconsistencies of the current model. J Bacteriol 190, 2379–87 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C. and Church G. M. Towards synthesis of a minimal cell. Mol Syst Biol 2, 45 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton B. P. and Raleigh E. A. Transposon-Mediated Linker Insertion Scanning Mutagenesis of the Escherichia coli McrA Endonuclease. J Bacteriol 186, 5699–707 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seringhaus M. et al.. Predicting essential genes in fungal genomes. Genome Res 16, 1126–35 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P. J. and Knop M. Evolution of mutational robustness in the yeast genome: a link to essential genes and meiotic recombination hotspots. PLoS Genet 5, e1000533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S. and Heber S. In silico prediction of yeast deletion phenotypes. Genet Mol Res 5, 224–32 (2006). [PubMed] [Google Scholar]
- Pandey G. et al.. An integrative multi-network and multi-classifier approach to predict genetic interactions. PLoS Comput Biol 6, e1000928 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J. et al.. Investigating the predictability of essential genes across distantly related organisms using an integrative approach. Nucleic Acids Res 39, 795–807 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanowski B., Leopold K. and Fischer W. Isomalto-oligosaccharide-containing lipoteichoic acid of Streptococcus sanguis. Microheterogeneity and distribution of chain substituents. Eur J Biochem 214, 757–61 (1993). [DOI] [PubMed] [Google Scholar]
- Kilian M. and Holmgren K. Ecology and nature of immunoglobulin A1 protease-producing streptococci in the human oral cavity and pharynx. Infect Immun 31, 868–73 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P. and Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene 23, 331–41 (1983). [DOI] [PubMed] [Google Scholar]
- Lukomski S. et al.. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun 68, 535–42 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker A. L. and Camilli A. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett 172, 131–5 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables


