Abstract
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) after base-specific cleavage of PCR amplified and in vitro-transcribed 16S rRNA gene (rDNA) was used for the identification of mycobacteria. Full-length 16S rDNA reference sequences of 12 type strains of Mycobacterium spp. frequently isolated from clinical specimens were determined by PCR, cloning, and sequencing. For MALDI-TOF MS-based comparative sequence analysis, mycobacterial 16S rDNA signature sequences (∼500 bp) of the 12 type strains and 24 clinical isolates were PCR amplified using RNA promoter-tagged forward primers. T7 RNA polymerase-mediated transcription of forward strands in the presence of 5-methyl ribo-CTP maximized mass differences of fragments generated by base-specific cleavage. In vitro transcripts were subsequently treated with RNase T1, resulting in G-specific cleavage. Sample analysis by MALDI-TOF MS showed a specific mass signal pattern for each of the 12 type strains, allowing unambiguous identification. All 24 clinical isolates were identified unequivocally by comparing their detected mass signal pattern to the reference sequence-derived in silico pattern of the type strains and to the in silico mass patterns of published 16S rDNA sequences. A 16S rDNA microheterogeneity of the Mycobacterium xenopi type strain (DSM 43995) was detected by MALDI-TOF MS and later confirmed by Sanger dideoxy sequencing. In conclusion, analysis of 16S rDNA amplicons by MS after base-specific cleavage of RNA transcripts allowed fast and reliable identification of the Mycobacterium tuberculosis complex and ubiquitous mycobacteria (mycobacteria other than tuberculosis). The technology delivers an open platform for high-throughput microbial identification on the basis of any specific genotypic marker region.
Mycobacteria are important pathogens that cause disease in humans and animals. The most important mycobacterial disease, tuberculosis, remains a global public health problem, with an estimated 8.4 million new cases in 1999 and 3 million deaths each year (41). Mycobacteria other than tuberculosis have become increasingly important as opportunistic pathogens, causing severe disease in patients with or without immune dysfunction (1, 2, 9, 11, 25, 27, 28). Since the introduction of broth culture systems for the isolation of mycobacteria, time for identification has decreased substantially. However, species identification by conventional phenotypic traits is still time-consuming and may frequently result in erroneous identification (33). Chromatographic analysis of cell wall lipids such as high-performance liquid chromatography and gas-liquid chromatography are technically demanding and expensive and therefore available in very few specialized clinical laboratories only (6, 21, 22).
Laboratories have begun to rely on amplification-based methods for genotypic characterization of mycobacteria (3, 18, 20, 23, 26, 31). The 16S rRNA gene (rDNA) is the most widely accepted gene used for bacterial identification of both cultured and as-yet-uncultured bacteria, with high impact on the discovery of new species of the Mycobacterium genus. 16S rDNA sequences constitute one of the largest gene-specific data sets, with constantly increasing numbers of entries in publicly available databases.
New genotypic techniques for fast identification of microorganisms have been developed during the past few years. High-density oligonucleotide arrays (DNA chip) were able to identify mycobacteria and detected rifampin resistance in a single assay (14, 36). Using fluorescence in situ hybridization with peptide nucleic acid probes the M. tuberculosis complex and mycobacteria other than tuberculosis were differentiated by direct visualization, which is especially useful in cases of mixed mycobacterial infections (10). Nucleic acid probes for hybridization-based identification are capable of identifying a narrow range of mycobacterial species (29). They are widely used, but problems with sensitivity and specificity have been described (7, 12).
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has been widely applied for the phenotypic characterization of bacterial whole cells, simple cell lysates, or bacterial products such as lipopolysaccharides and proteins (4, 5, 32, 37, 40). The potential to analyze bacterial DNA and RNA has been shown during the last few years. Hurst et al. (17) described a protocol for fast and accurate differentiation of PCR products from methanotrophic bacteria by MALDI-TOF MS according to their length. Rapid analysis of PCR products and restriction fragment length polymorphism patterns of microbial samples by MALDI-TOF MS was reported by Taranenko and coworkers (35) using UV MALDI for size determination of double-stranded amplicons and restriction fragments. However, these approaches are limited by length heterogeneities of specific marker genes, which diminish their discriminatory power. Kirpekar et al. (19) monitored posttranscriptional modifications of bacterial RNA after G- and pyrimidine-specific digestion, using MALDI-TOF MS for direct analysis of the digest.
Rapid bacterial identification via base-specific cleavage of amplified 16S rDNA and MS was described by von Wintzingerode et al. (39) for the first time. Amplification of 16S rDNA signature sequences in the presence of dUTP instead of dTTP was followed by strand separation and uracil-DNA-glycosylase (UDG)-mediated cleavage at each T-specific site. Fragment pattern detection was performed by MALDI-TOF MS and resulted in unambiguous identification of a panel of cultured Bordetella spp. and as-yet-uncultured bacteria of anaerobic, organochlorine-reducing microbial consortia.
Here we present an alternative homogeneous in vitro transcription-RNase cleavage system followed by MALDI-TOF MS as a method for rapid 16S rDNA-based identification of mycobacteria to the species level. PCR amplification of mycobacterial 16S rDNA was combined with in vitro RNA transcription and subsequent G-specific cleavage of the RNA transcript. The resulting fragment pattern was analyzed by MALDI-TOF MS. The obtained mass signal pattern allows unambiguous and reliable identification of each species. Our concept combines the discriminatory power of 16S rDNA sequence information with the accuracy and data acquisition speed of MALDI-TOF MS.
MATERIALS AND METHODS
Bacterial strains.
Twelve type strains purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and the Institute for Standardization and Documentation in Medical Laboratories (Instand e.V., Düsseldorf, Germany) as well as 24 mycobacterial isolates from our clinical microbiology department were used in the present study (Table 1). Mycobacteria were grown in liquid medium (MGIT liquid medium; Becton Dickinson Europe, Le Pont de Claix, France) with enrichment supplement (MGIT system oleic acid-albumin-dextrose-citric acid) and antimicrobial supplement (MGIT system PANTA [polymyxin B, nalidixic acid, trimethoprim, and azlocillin]) at 37 and 30°C (for M. marinum), respectively. When an MGIT vial was detected as positive, mycobacteria were concentrated in 0.5 ml of broth by centrifugation (3,300 × g, 20 min) followed by DNA extraction using a commercially available respiratory specimen preparation kit (AMPLICOR; Roche Molecular Systems, Inc., Branchburg, N.J.). In brief, 100 μl of resuspended mycobacterial pellet was transferred into a 1.5-ml polypropylene tube, washed with washing solution (500 μl) provided with the kit, and centrifuged (14,000 × g) for 10 min. The supernatant was discarded, and the bacterial pellet was suspended in a lysis reagent (100 μl). After incubation in a heating block at 60°C for 45 min the lysate was neutralized with the neutralizing reagent (100 μl) provided with the kit and stored at 4°C.
TABLE 1.
Mycobacterial strains used in this study and typing results of repeated MALDI-TOF MS analysis of clinical isolates
| Species | Source | EMBL accession no. |
|---|---|---|
| Reference strains (n = 12) | ||
| Mycobacterium abscessus | DSM 44196 | AJ536038 |
| Mycobacterium avium subsp. avium | DSM 44156 | AJ536037 |
| Mycobacterium celatum | DSM 44243 | AJ536040 |
| Mycobacterium fortuitum subsp. fortuitum | DSM 46621 | AJ536039 |
| Mycobacterium gordonae | DSM 44160 | AJ536042 |
| Mycobacterium intracellulare | DSM 43223 | AJ536036 |
| Mycobacterium kansasii | DSM 44162 | AJ536035 |
| Mycobacterium marinum | DSM 44344 | AJ536032 |
| Mycobacterium scrofulaceum | DSM 43992 | AJ536034 |
| Mycobacterium smegmatis | DSM 43758 | AJ536041 |
| Mycobacterium tuberculosis | H37RV | AJ536031 |
| Mycobacterium xenopi | DSM 43995 | AJ536033 |
| Clinical isolatesa (n = 24) | ||
| Mycobacterium aurumb | MT1323 | |
| Mycobacterium avium | MT1367 | |
| Mycobacterium avium | MT1951 | |
| Mycobacterium chelonae | MT1297 | |
| Mycobacterium fortuitum | MT2014 | |
| Mycobacterium gordonae | MT804 | |
| Mycobacterium gordonae | MT2843 | |
| Mycobacterium interjectumb | MT1223 | |
| Mycobacterium intracellulare | MT1881 | |
| Mycobacterium intracellulare | MT2544 | |
| Mycobacterium kansasii | MT1619 | |
| Mycobacterium marinum | MT3065 | |
| Mycobacterium paraffinicumb | MT1423 | |
| Mycobacterium smegmatis | MT3344 | |
| Mycobacterium tuberculosis | MT1551 | |
| Mycobacterium tuberculosis | MT1598 | |
| Mycobacterium tuberculosis | MT1692 | |
| Mycobacterium tuberculosis | MT1905 | |
| Mycobacterium tuberculosis | MT2068 | |
| Mycobacterium tuberculosis | MT2125 | |
| Mycobacterium tuberculosis | MT2271 | |
| Mycobacterium tuberculosis | MT2924 | |
| Mycobacterium xenopi | MT309 | |
| Mycobacterium xenopi | MT2236 |
Species determined by partial 16S rDNA sequencing and phenotypic methods (M. chelonae, M. kansasii, M. marinum, and M. tuberculosis). All clinical isolates were correctly identified by repeated MALDI-TOF MS (in triplicate) unless otherwise indicated.
Correct identification after addition of calculated-mass signal pattern from published 16S rDNA sequences into the database.
Identification by PCR and sequencing.
Full-length 16S rRNA genes of the 12 type strains were analyzed as described in detail elsewhere (38). Briefly, 16S rDNA was amplified using eubacterial primers TPU1 (AGA GTT TGA TCM TGG CTC AG; corresponding to Escherichia coli positions 8 to 27) and RTU8 (AAG GAG GTG ATC CAK CCR CA; corresponding to E. coli positions 1541 to 1522), according to a standard protocol. PCR products were inserted into pCR2.1 (TA Cloning Kit; Invitrogen, de Schelp, The Netherlands) and transformed into E. coli according to the manufacturer's instructions. Recombinant plasmid DNA was purified using the GFX plasmid preparation kit (Amersham Pharmacia, Freiburg, Germany) and used directly for cycle sequencing (Thermosequenase fluorescent labeled primer cycle sequencing kit; Amersham Pharmacia). Sequencing reaction mixtures were analyzed on a LICOR 4000L automated DNA sequencer (MWG-Biotech, Ebersberg, Germany). Alignments were generated with ARB software (http://www.arb-home.de/) (38).
Identification of mycobacteria from clinical sources was performed by PCR amplification of partial 16S rDNA and direct sequencing focusing on hypervariable regions A and B corresponding to E. coli 16S rDNA positions 129 to 267 and 430 to 500 according to a protocol of Springer et al. (33). Resulting sequences were compared to all 16S rRNA gene entries in EMBL and GenBank databases using BLASTN and FASTA of the Husar program package (version 4.0; Heidelberg Unix Sequence Analysis Resources, DKFZ, Heidelberg, Germany). Clinical isolates were identified to the species level on the basis of sequence identity in both hypervariable regions with a database entry and a total sequence identity of >99%, respectively.
Identification by PCR, base-specific cleavage, and MALDI-TOF.
Experiments for each mycobacterial strain were run in triplicate to validate the reproducibility of the PCR, base-specific cleavage, and MALDI-TOF methods.
PCR.
Fifty-microliter amplification mixtures contained 1× PCR buffer [Tris-HCl, KCl, (NH4)2SO4, MgCl2 [pH 8.7]; final MgCl2 concentration of 2.5 mM], a 200 μM concentration of each deoxynucleoside triphosphate, 1 U of HotStarTaq (QIAGEN GmbH, Hilden, Germany), 10 pmol of primer Myko109-T7 (5′-GTAATACGACTCACTATAGGG ACG GGT GAG TAA CAC GT-3′; corresponding to E. coli 16S rDNA positions 105 to 121) and 10 pmol of primer R259-SP6 (5′-ATTTAGGTGACACTATAGAA TTT CAC GAA CAA CGC GAC AA-3′; corresponding to E. coli 16S rDNA positions 609 to 590) and 5 μl of DNA. The temperature profile consisted of 40 cycles of denaturation (1 min, 95°C), annealing (1 min, 58°C), and extension (1 min 30 s, 72°C) after an initial step of HotStarTaq activation (15 min, 95°C) (Thermocyler Goldblock; Biometra, Göttingen, Germany).
RNA transcription.
Forward-strand RNA transcription was performed by incubation of 2.4 μl of PCR product, 10 U of T7 RNA polymerase (Epicentre), a 0.5 mM concentration of each nucleoside triphosphate, and 1× transcription buffer (6 mM MgCl2, 10 mM dithiothreitol, 10 mM NaCl, 10 mM spermidine, 40 mM Tris-Cl [pH 7.9] at 20°C) at 37°C for 2 h. Ribo-CTP was replaced by its chemically modified analog 5-methyl ribo-CTP (ribo-CTP; Trilink, San Diego, Calif.).
RNase T1 cleavage.
Complete G-specific cleavage was achieved by adding 20 U of RNase T1 and 1 U of shrimp alkaline phosphatase and incubating this mixture at 30°C for 30 min.
Sample conditioning.
Each sample was diluted by adding 21 μl of H2O. Conditioning of the phosphate backbone was achieved with 6 mg of SpectroCLEAN (SEQUENOM, San Diego, Calif.).
MALDI-TOF MS analysis.
Samples were analyzed from SpectroCHIP arrays. Aliquots of 15 nl were dispensed robotically onto a silicon chip (SpectroCHIP; SEQUENOM). Mass spectra were recorded using a Biflex III mass spectrometer (Bruker Daltonik, Bremen, Germany). Exclusively positive ions were analyzed, and ∼50 single-shot spectra were accumulated per sample. All samples were analyzed in linear TOF mode using delayed ion extraction and a total acceleration voltage of 20 kV. Analysis of all mass spectra and automated data interpretation were performed with in-house software developed by SEQUENOM, Inc.
Nucleotide sequence accession numbers.
Full-length 16S rRNA gene sequences of the 12 type strains were deposited in the EMBL nucleotide sequence database under accession numbers AJ536031 to AJ536042 (Table 1).
RESULTS
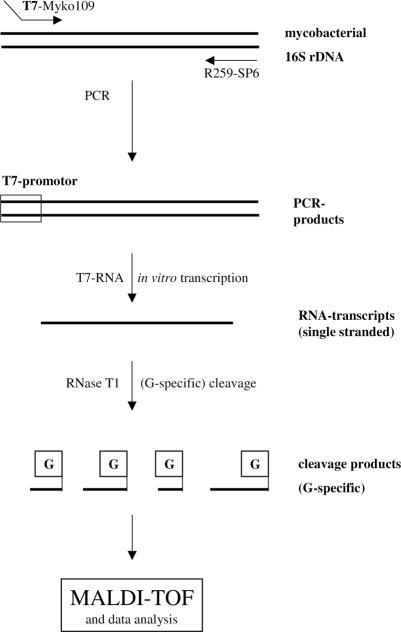
An ∼500-bp region of the 16S rRNA gene corresponding to E. coli 16S rDNA positions 105 to 609 was PCR amplified from all type strains and clinical isolates. RNA transcription and base-specific cleavage resulted in unique MALDI-TOF mass spectra for all tested type strains. The basic principle of the method is depicted in Fig. 1.
FIG. 1.
Schematic representation of the base-specific cleavage of amplified and reverse-transcribed mycobacterial 16S rDNA.
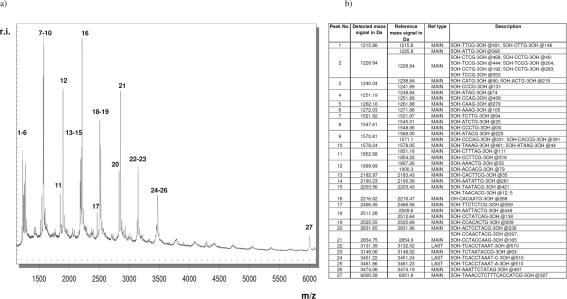
A representative mass spectrum of M. tuberculosis H37Rv is shown in Fig. 2a. The main cleavage products corresponding to peaks 1 to 27 are listed in Fig. 2b. Their nucleic acid composition and exact location within the uncleaved PCR amplicon are assigned. Reference mass signals have been calculated from the reference sequence by in silico cleavage at all positions of guanine and correlated to mass signals detected by MALDI-TOF MS. Calculated fragments with a mass difference smaller than 4 Da could not be separated in the linear, axial MALDI-TOF MS employed in this study. Corresponding detected cleavage products were assessed as one fragment only (peaks 2, 3, 4, 8, 9, 11, 12, and 18).
FIG. 2.
(a) M. tuberculosis mass spectrum displaying all signals generated by G-specific cleavage of the 16S rDNA amplicon after RNA transcription. To simplify identification, all mass signals corresponding to main cleavage products are marked with numbers. (b) List of detected mass signals corresponding to the spectrum depicted in panel a. For all signals the sequence and location within the uncleaved PCR amplicon is provided in the description column (position follows “@”). MAIN, main cleavage product; LAST, product at the 3′ end of the amplicon, often elongated by one nucleotide as a result of 3′-terminal transferase activity of T7 RNA polymerase.
Mass signals 22, 24, and 25 (Fig. 2b) are classified LAST, because they represent cleavage products at the 3′ end of the transcript (all at position 510) differing by the addition of one 5-methyl-CTP (3′ fragment + 319.2 Da) or one ATP (3′ fragment + 329.2 Da), respectively. Nontemplated addition of a nucleotide to the 3′ end of the RNA transcript most probably reflects terminal transferase activity of T7-RNA polymerase, a feature well known for Taq DNA polymerases. The nontemplated addition of nucleotides to the terminal fragments has been included in the software-automated identification of fragments for all mycobacterial species to avoid misinterpretation.
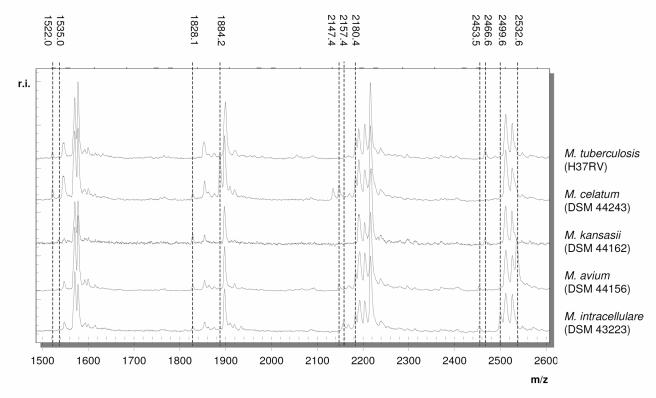
Characteristic mass spectra of five representative mycobacterial type strains in a mass range between 1,500 and 2,600 Da are shown in Fig. 3. M. tuberculosis, M. avium, M. intracellulare, M. kansasii, and M. celatum can be clearly differentiated by their unique mass spectra. M. tuberculosis is the only species lacking a fragment at 1,828 Da. M. celatum shows a signal at 1,884 Da not present within all other mass patterns. The spectrum of M. kansasii displays no signal at 2,180 Da. Mass spectra of M. avium and M. intracellulare differ from those of the other species by fragments at 2,532 and 2,157 Da, respectively.
FIG. 3.
Overlay of mass spectra of five representative mycobacterial strains in a mass range between 1,500 and 2,600 Da.
In silico discriminatory peak patterns of all mycobacterial species used in this study are compiled in Table 2. The ranking was performed according to the number of missing and additional peaks compared to the mass spectrum of M. tuberculosis. Only discriminatory peaks that are not present throughout all Mycobacteria spp. are included. M. tuberculosis can be clearly differentiated from other species on the basis of multiple additional or missing mass signals. M. celatum and M. kansasii are the species closest to M. tuberculosis, showing one missing and three additional peaks or two missing and two additional peaks, respectively. M. marinum and M. scrofulaceum differ by only two fragments (2,453.5 and 2,795.8 Da). All calculated mass patterns have been confirmed experimentally. A comparison of all mass spectra resulted in unambiguous identification of all mycobacterial species.
TABLE 2.
Ranking of base-specific mycobacterial 16S rDNA cleavage mass patterns according to the number of identical and nonidentical mass peaks relative to the mass spectrum of M. tuberculosisa
| Organism | Peak(s) with calculated mass(es) (Da) of: |
|||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1,215.8 | 1,261.9 | 1,522.0 | 1,535.0 | 1,555.0, 1,558.0 | 1,591.0 | 1,828.1 | 1,864.2 | 1,877.3 | 1,884.2, 1,887.3 | 2,147.4 | 2,157.4 | 2,170.4 | 2,180.4, 2,183.4 | 2,203.4, 2,206.5 | 2,229.5 | 2,453.5 | 2,466.6 | 2,499.6 | 2,532.6, 2,535.7 | 2,795.8 | 2,808.8 | 2,818.8 | 2,841.9 | 2,867.9 | 3,490.3 | 4,079.6 | 4,408.8 | 4,421.8 | 4,764.1 | 5,373.4 | 6,001.8 | |
| M. tuberculosis | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | |||||||||||||||||||||||||
| M. celatum | ▪ | ▪ | ▪ | ░⃞ | ░⃞ | ░⃞ | ▪ | ▪ | ▪ | |||||||||||||||||||||||
| M. kansasii | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ▪ | ▪ | |||||||||||||||||||||||||
| M. marinum | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | ▪ | ||||||||||||||||||||||||
| M. scrofulaceum | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | ▪ | ||||||||||||||||||||||||
| M. paraffinicum | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | ▪ | ||||||||||||||||||||||||
| M. avium | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | |||||||||||||||||||||||
| M. intracellulare | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | |||||||||||||||||||||||
| M. gordonae | ▪ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ||||||||||||||||||||||||
| M. xenopi | ▪ | ▪ | ░⃞ | ▪ | ▪ | ░⃞ | ░⃞ | ░⃞ | ||||||||||||||||||||||||
| M. interjectum | ▪ | ▪ | ░⃞ | ░⃞ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | |||||||||||||||||||||||
| M. smegmatis | ▪ | ▪ | ░⃞ | ░⃞ | ▪ | ░⃞ | ░⃞ | |||||||||||||||||||||||||
| M. fortuitum | ▪ | ░⃞ | ░⃞ | ░⃞ | ░⃞ | ▪ | ░⃞ | |||||||||||||||||||||||||
| M. abscessus | ▪ | ░⃞ | ░⃞ | ░⃞ | ▪ | ▪ | ░⃞ | ░⃞ | ░⃞ | ░⃞ | ||||||||||||||||||||||
| M. aurum | ▪ | ▪ | ░⃞ | ░⃞ | ░⃞ | ░⃞ | ░⃞ | ░⃞ | ||||||||||||||||||||||||
Only discriminating mass peaks are shown. Mass peaks identical to those of M. tuberculosis are shown as black boxes. Additional mass peaks are shown as gray boxes.
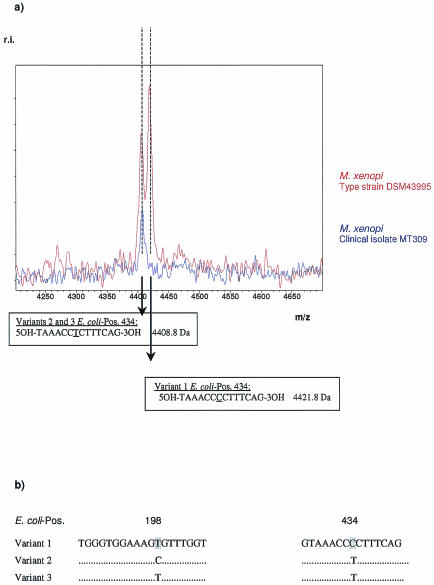
In the case of the M. xenopi type strain, DSM 43995, comparison of experimental and calculated mass patterns revealed an additional mass peak at 4,408.8 Da in MALDI-TOF analysis (Fig. 4a). Cloning of the respective M. xenopi 16S rDNA amplicon and repeated sequencing of several plasmids resulted in the detection of three sequence variants differing in 1 to 2 bp at E. coli position 198 (T/C) and 434 (T/C) (Fig. 4b). The sequence variation at E. coli position 198 is not detected in a G-specific cleavage reaction. The resulting dimeric fragments (5OH-TG-3p and 5OH-CG-3p) overlap with cleavage products of the same composition originating from different positions in the amplicon. Base-specific cleavage of an ∼500-bp amplicon statistically results in all possible combinations of dimers represented multiple times. In addition, the mass range below 1,000 Da can be affected by background noise signals caused by matrix molecules, a feature specific to the use of 3-hydroxypicolinic acid matrices in MALDI-TOF MS.
FIG. 4.
Identification of M. xenopi variants. (a) Experimental mass pattern. (b) Sanger dideoxy sequencing of cloned 16S rDNA.
Sequence variation at E. coli position 434 (T/C) affects a 14-bp G-specific cleavage product. The nucleotide mass difference between a T (corresponding to U in cleaved RNA) and a C diminishes the mass of the expected fragment by 13 Da. The detection of mass signals at both 4,408.8 and 4,421.8 Da leads to the conclusion that the analyzed amplicon of the type strain consists of a mixture of both sequence variants.
After establishing a database including the 12 mycobacterial type strains 24 clinical isolates have been analyzed automatically with MALDI-TOF MS. G-specific cleavage of RNA-transcribed 16S rDNA amplification products and MS led to unambiguous identification of 21 isolates. Results are presented in Table 1. All isolates representing species from the type strain database were identified correctly in repeated experiments. Three clinical isolates representing M. aurum (MT1323), M. paraffinicum (MT1423), and M. interjectum (MT1223) could not be identified after MALDI-TOF analysis of their RNA cleavage products. The database lacked the corresponding in silico mass pattern of all three species. An extension of the database with the species-specific mass signal pattern calculated from published 16S rDNA sequences of M. aurum (MT1323), M. paraffinicum (MT1423), and M. interjectum (MT1223) led to correct identification in all corresponding experiments.
DISCUSSION
The advantage of using molecular analysis based upon 16S rDNA sequence determination over using phenotypic methods for the identification of mycobacterial isolates has been extensively evaluated. In contrast to phenotypic (e.g., biochemical) features, the 16S rRNA sequence of a species is a stable property specific for most mycobacteria at the species level. Unfortunately members of the Mycobacterium tuberculosis complex (M. tuberculosis, M. bovis, M. africanum, and M. microti) share identical rDNA sequences. Instead, polymorphisms in the gyrB sequences facilitate the differentiation of all four species (24). Springer et al. found two 16S rDNA sequence regions, termed A and B, corresponding to E. coli 16S positions 129 to 267 and 430 to 500, suitable to identify most mycobacteria (33). RIDOM (http://www.ridom.de/) (15) and the MicroSeq 500 16S rDNA bacterial sequencing kit (PE Applied Biosystems) (8) employ the same regions of 16S rDNA in a stretch of 440 to 500 bp for mycobacterial identification.
In this study, we present comparative sequence analysis by MALDI-TOF MS as a novel tool for the analysis of a similar 500-bp region covering variable sequence regions A and B for 16S rDNA-based genotyping of mycobacteria. With the introduction of capillary systems, standard 16S rDNA sequencing has developed further during the past decade, and same-day turnaround times can be achieved. Although DNA extraction, PCR amplification, RNA transcription, and G-specific cleavage require similar time frames as processing steps in conventional sequencing, the speed and accuracy of MALDI-TOF MS are very appealing. Currently available mass spectrometers allow analysis of all cleavage products in less than 2 s. Sample processing can be automated (96- and 384-microtiterplate format), and analysis by MALDI-TOF MS can be completed within 4 h post-PCR processing using the described protocol.
According to Hartmer et al. (16) the small mass difference of 1 Da between uracil and cytosine is a serious limitation for the analysis of RNA fragments by MALDI-TOF MS. Therefore, CTP was exchanged for its 5-methyl base-modified ribonucleotide triphosphate analogue to increase the mass difference to 13 Da without loss in transcription yield. By using 5-methyl-CTP instead of CTP we were able to achieve good fragment separation with MALDI-TOF MS, limited only by fragments differing by less than 4 Da. RNA cleavage was performed with RNase T1, resulting in complete G-specific cleavage of the RNA transcripts (16). Addition of shrimp alkaline phosphatase leads to removal of 3′-phosphate groups at each fragment, further avoiding the development of 2′, 3′-cyclic phosphate groups as potential side products. Recently, von Wintzingerode et al. (39) described the use of MALDI-TOF MS for analysis of UDG-mediated base-specific cleavage of 16S rDNA PCR products for bacterial identification. After strand separation, UDG-mediated phosphate backbone cleavage resulted in a mixture of hydrolysis products (fragments applied for identification) and side products (nonspecific cleavage products without discriminatory power), increasing the complexity of the mass spectrum. Our approach generated no side products, and all resulting cleavage products were used for identification of the underlying sequence. The use of post-PCR in vitro RNA transcription mediated an additional amplification step resulting in a higher analyte concentration for MS analysis. Furthermore, the process generates a single-stranded nucleic acid, thus eliminating the need for strand separation. Moreover, the stability of RNA during the desorption-ionization process is higher than that of DNA due to the balancing effect of the 2′-hydroxy group on the polarization of the N-glycosidic bond of protonated bases (34).
MALDI-TOF analysis of base-specific 16S rRNA fragments resulted in unique mass spectra for each of the mycobacterial type strains. According to Fig. 3 and Table 1 some single mass peaks seem to be species specific (e.g., 2,157.4 Da for M. intracellulare, 2,229.5 Da for M. gordonae, and 3,490.3 Da for M. smegmatis). However, the number of unique mass signals is expected to decrease within a growing database. In consequence, analysis based on single species-specific peaks could lead to erroneous assignments of sample spectra to the corresponding reference spectra. Hence, the process for identification of species was based on a comparison of whole mass peak patterns.
Based on a database of in silico patterns derived from in-house sequences and database 16S rDNA sequences, 24 mycobacterial isolates were identified correctly, and repeated experiments showed a high degree of reproducibility. This illustrates the potential of enlarging the database with more mycobacterial species-specific cleavage patterns of published 16S rDNA sequences. Even species differing in only three bases of their reference sequence, like M. paraffinicum and M. scrofulaceum, are unambiguously identified by G-specific cleavage and the corresponding mass signal pattern.
MALDI-TOF MS of cleavage products derived from M. xenopi type strain DSM 43995 resulted in discrepancies between expected mass signal patterns and sequencing data, illustrating the capabilities of our method, but also the limitations when only a single cleavage reaction is employed. A single nucleotide sequence variation embedded in a 14-mer cleavage product resulted in the unambiguous detection of an additional mass signal, whereas a second sequence variation as part of a 2-bp fragment did not induce a discernible change in the detected mass signal pattern. Due to overlapping cleavage products of the same composition and due to the small mass, the latter could not be detected. An opportunity to overcome this issue is alteration of the fragment compositions by a change of the cleavage base and by performing additional cleavage reactions (e.g., base-specific cleavage at C or T; unpublished data). This facilitates detection, identification, and localization of sequence changes and microheterogeneities (30).
Identification of the detected sequence variations in RIDOM (http://www.ridom.de/) showed 100% similarity with three sequevars of M. xenopi (variant 1 = DSM 43995, variant 2 = S88, variant 3 = S91). Variants S91 and S88 differ from type strain DSM 43995 by 1 to 2 bp and exhibit a 4- or 5-bp insertion in the 16S-23S rDNA internal transcribed spacer sequence (31). Interestingly, patient isolates MT309 and MT2236 revealed a missing signal at 4,421.8 Da and an additional signal at 4,408.8 Da, corresponding to the reference sequences of variants 2 and 3 of type strain M. xenopi DSM 43995 (Fig. 4). Results of partial 16S rDNA sequencing of both isolates verified the sequence variation (T instead of C) at E. coli position 434 and detected a C at E. coli position 198, resulting in 100% sequence identity with M. xenopi strain S88 as derived from the RIDOM database.
The method presented here uses 16S rDNA for identification of cultured mycobacterial strains. As described recently (39), 16S rDNA-based identification can be extended to yet-uncultured bacteria, and the application can be utilized for phylogenetic classification and typing of unknown bacteria. In contrast, protein- or whole-cell-based identification by MS requires cultivation and is prone to growth-dependent variations, resulting in problems of low reproducibility (4, 5, 35). Compared to conventional 16S rDNA sequencing for identification of bacteria, the absence of any time- and cost-consuming electrophoresis step is an important advantage. The experimental procedure for PCR amplification, RNA transcription, and subsequent base-specific cleavage is straightforward, and a homogeneous format avoids any purification steps.
So far we have restricted our approach to the analysis of ∼500-bp amplicons. Comparative sequence analysis of larger target regions (e.g., 1,500 bp) covering the whole 16S rDNA appears to be promising. Further experiments will show whether a larger number of cleavage products will result in higher discriminatory power or will obscure important peaks by increasing cross talk and background noise due to a higher number of overlapping mass signals.
Resequencing of nucleic acids by base-specific cleavage after virtually all four bases has been described elsewhere (30). A single-stranded copy of the target sequence is cleaved to completion in four separate reactions at positions corresponding to each of the four bases. After MALDI-TOF MS each resulting peak can be unambiguously identified using a reference sequence. Changes in sequence have discernible effects on a maximum of four cleavage patterns. With reference sequences available for many bacterial pathogens, comparative sequence analysis with validation by MALDI-TOF seems to be an ideal tool for phylogenetic and diagnostic purposes as well as microheterogeneity detection and localization.
The platform is not restricted to 16S rDNA identification, but can be extended to other genotypic markers, e.g., gyrB sequence polymorphism analysis for M. tuberculosis complex differentiation, multidrug-resistant regions or multilocus-sequence typing, which further broadens its application.
The approach presented here is a promising prototype for high-throughput applications in clinical diagnostics as well as pharmaceutical and environmental microbiology, combining the accuracy of MALDI-TOF MS with the phylogenetic information of genotypic marker regions.
Acknowledgments
This work has been supported in part by a grant of the Deutsche Forschungsgemeinschaft (DFG FOR 299/1) to U. B. Göbel.
REFERENCES
- 1.American Thoracic Society. 1997. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am. J. Respir. Crit. Care Med. 156:1-25. [DOI] [PubMed] [Google Scholar]
- 2.Ashford, D. A., E. Whitney, P. Raghunathan, and O. Cosivi. 2001. Epidemiology of selected mycobacteria that infect human and other animals. Rev. Sci. Tech. 20:235-337. [DOI] [PubMed] [Google Scholar]
- 3.Bahrmand, A. R., T. G. Bakayeva, and V. V. Bakayev. 1998. Use of restriction enzyme analysis of amplified DNA coding for the hsp65 gene and polymerase chain reaction with universal primer for rapid differentiation of Mycobacterium species in the clinical laboratory. Scand. J. Infect. Dis. 30:477-480. [DOI] [PubMed] [Google Scholar]
- 4.Bernardo, K., N. Pakulat, M. Macht, O. Krut, H. Seifert, S. Fleer, F. Hünger, and M. Krönke. 2002. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:747-753. [DOI] [PubMed] [Google Scholar]
- 5.Bright, J. J., M. A. Claydon, M. Soufian, and D. B. Gordon. 2002. Rapid typing of bacteria using matrix-assisted laser desorption ionisation time-of-flight mass spectrometry and pattern recognition software. J. Microbiol. Methods 48:127-138. [DOI] [PubMed] [Google Scholar]
- 6.Butler, W., K. Jost, and J. Kilburn. 1991. Identification of mycobacteria by high-performance liquid chromatography. J. Clin. Microbiol. 29:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler, W. R., S. P. O′Connor, M. A. Yakrus, and W. M. Gross. 1994. Cross-reactivity of genetic probe for detection of Mycobacterium tuberculosis with the newly described Mycobacterium celatum. J. Clin. Microbiol. 32:536-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloud, J. L., H. Neal, R. Rosenberry, C. Y. Turenne, M. JAMA, D. R. Hillyard, and K. C. Carroll. 2002. Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J. Clin. Microbiol. 40:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbett, E. L., L. Blumberg, G. J. Churchyard, N. Moloi, K. Mallory, T. Clayton, B. G. Williams, R. E. Chaisson, R. J. Hayes, and K. M. De Cock. 1999. Nontuberculous mycobacteria. Defining disease in a prospective cohort of South African miners. Am. J. Respir. Crit. Care Med. 160:15-21. [DOI] [PubMed] [Google Scholar]
- 10.Drobniewski, F. A., P. G. More, and G. S. Harris. 2000. Differentiation of Mycobacterium tuberculosis complex and nontuberculous mycobacterial liquid cultures by using peptide nucleic acid-fluorescence in situ hybridization probes. J. Clin. Microbiol. 38:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkinham III, J. O. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford, E. G., S. J. Snead, J. Todd, and N. G. Warren. 1993. Strains of Mycobacterium terrae complex which react with DNA probes for Mycobacterium tuberculosis complex. J. Clin. Microbiol. 31:2805-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredericks, D. N., and D. A. Relman. 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. J. Clin. Microbiol. Rev. 9:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis fo generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 15.Harmsen, D., J. Rothgänger, C. Singer, J. Albert, and M. Frosch. 1999. Intuitive hypertext-based molecular identification of micro-organisms. Lancet 353:291. [DOI] [PubMed] [Google Scholar]
- 16.Hartmer, R., N. Storm, S. Böcker, C. P. Rodi, F. Hillenkamp, C. Jurinke, and D. van den Boom. 2003. RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high throughput comparative sequence analysis. Nucleic Acids Res. 31:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurst, G. B., K. Weaver, M. J. Doktycz, M. V. Buchanan, A. M. Costello, and M. E. Lidstrom. 1998. MALDI-TOF analysis of polymerase chain reaction products from methanotrophic bacteria. Anal. Chem. 70:2693-2698. [DOI] [PubMed] [Google Scholar]
- 18.Kim, B.-J., K.-H. Lee, B.-N. Park, S.-J. Kim, G.-H. Bai, S.-J. Kim, and Y.-H. Kook. 2001. Differentiation of mycobacterial species by PCR-restriction analysis of DNA (342 base pairs) of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 39:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirpekar, F., D. Douthwaite, and P. Roepstorff. 2000. Mapping posttranscriptional modifications in 5S ribosomal RNA by MALDI mass spectrometry. RNA 6:296-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louro, A. P. S., K. B. Waites, E. Georgescu, and W. H. Benjamin JR. 2001. Direct identification of Mycobacterium avium complex and Mycobacterium gordonae from MB/BacT bottles using AccuProbe. J. Clin. Microbiol. 39:570-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luquin, M., V. Ausina, F. López Calahorra, F. Belda, M. Garcia Barceló, C. Celma, and G. Prats. 1991. Evaluation of practical chromatographic procedures for identification of clinical isolates of mycobacteria. J. Clin. Microbiol. 29:120-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metchock, B. G., F. S. Nolte, and R. J. Wallace JR. 1999. Mycobacterium, p. 399-437. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 23.Miller, N., S. Infante, and T. Cleary. 2000. Evaluation of the LiPa MYCOBACTERIA assay for the identification of mycobacterial species from BACTEC 12B bottles. J. Clin. Microbiol. 38:1915-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niemann, S., D. Harmsen, S. Ruesch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O′Brien, D. P., B. J. Currie, and V. L. Krause. 2000. Nontuberculous mycobacterial disease in northern Australia: a case series and review of the literature. Clin. Infect. Dis. 31:958-967. [DOI] [PubMed] [Google Scholar]
- 26.Oh, E.-J., Y.-J. Park, C. L. Chang, B. K. Kim, and S. M. Kim. 2001. Improved detection and differentiation of mycobacteria with combination of Mycobacterium Growth Indicator Tube and Roche COBAS AMPLICOR System in conjunction with duplex PCR. J. Microbiol. Methods 46:29-36. [DOI] [PubMed] [Google Scholar]
- 27.Oliver, A., L. Maiz, R. Canton, H. Escobar, F. Banquero, and E. Gomez-Mampaso. 2001. Nontuberculous mycobacteria in patients with cystic fibrosis. Clin. Infect. Dis. 32:1298-1303. [DOI] [PubMed] [Google Scholar]
- 28.Perlman, D. C., R. D'Amico, and N. Salomon. 2001. Mycobacterial infections of the head and neck. Curr. Infect. Dis. Rep. 3:233-241. [DOI] [PubMed] [Google Scholar]
- 29.Richter, R., S. Niemann, S. Rüsch-Gerdes, and S. Hoffner. 1999. Identification of Mycobacterium kansasii by using a DNA probe (AccuProbe) and molecular techniques. J. Clin. Microbiol. 37:964-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodi, C. P., B. Darnhofer-Patel, P. Stanssens, M. Zabeau, and D. van den Boom. 2002. A strategy for the rapid discovery of disease markers using the MassARRAY system. BioTechniques 32:62-69. [PubMed] [Google Scholar]
- 31.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smole, S. C., L. A. King, P. E. Leopold, and R. D. Arbeit. 2002. Sample preparation of gram-positive bacteria for identification by matrix assisted laser desorption/ionization time-of-flight. J. Microbiol. Methods 48:107-115. [DOI] [PubMed] [Google Scholar]
- 33.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, W., L. Zhu, and L. M. Smith. 1997. Controlling DNA fragmentation in MALDI-MS by chemical modification. Anal. Chem. 69:302-312. [DOI] [PubMed] [Google Scholar]
- 35.Taranenko, N. I., R. Hurt, J. Z. Zhou, N. R. Isola, H. Huang, S. H. Lee, and C. H. Chen. 2002. Laser desorption mass spectrometry for microbial DNA analysis. J. Microbiol. Methods 48:101-106. [DOI] [PubMed] [Google Scholar]
- 36.Troesch, A., H. Nguyen, C. G. Miyada, S. Desvarenne, T. R. Gingeras, P. M. Kaplan, P. Cros, and C. Mabilat. 1999. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J. Clin. Microbiol. 37:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Baar, B. L. M. 2000. Characterisation of bacteria by matrix assisted laser desorption/ionisation and electrospray mass spectrometry. FEMS Microbiol. Rev. 24:193-219. [DOI] [PubMed] [Google Scholar]
- 38.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Wintzingerode, F., S. Böcker, C. Schlötelburg, N. H. L. Chiu, N. Storm, C. Jurinke, C. R. Cantor, U. B. Göbel, and D. van den Boom. 2002. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: a tool for rapid bacterial identification. Proc. Natl. Acad. Sci. USA 99:7039-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker, J., A. J. Fox, V. Edwards-Jones, and D. B. Gordon. 2002. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J. Microbiol. Methods 48:117-126. [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization. 2001. World Health Organization report W.H.O./CDS/TB/2001.287. World Health Organization, Geneva, Switzerland.