A general molecular framework for spindle checkpoint inactivation is lacking. The Mad2 inhibitor, p31comet, has roles independent of the ubiquitin-proteasome pathway. This key finding allows the delineation of two partially redundant pathways for mitotic exit.
Abstract
The spindle checkpoint senses unattached or improperly attached kinetochores during mitosis, inhibits the anaphase-promoting complex or cyclosome (APC/C), and delays anaphase onset to prevent aneuploidy. The mitotic checkpoint complex (MCC) consisting of BubR1, Bub3, Mad2, and Cdc20 is a critical APC/C-inhibitory checkpoint complex in human cells. At the metaphase–anaphase transition, the spindle checkpoint turns off, and MCC disassembles to allow anaphase onset. The molecular mechanisms of checkpoint inactivation are poorly understood. A major unresolved issue is the role of Cdc20 autoubiquitination in this process. Although Cdc20 autoubiquitination can promote Mad2 dissociation from Cdc20, a nonubiquitinatable Cdc20 mutant still dissociates from Mad2 during checkpoint inactivation. Here, we show that depletion of p31comet delays Mad2 dissociation from Cdc20 mutants that cannot undergo autoubiquitination. Thus both p31comet and ubiquitination of Cdc20 are critical mechanisms of checkpoint inactivation. They act redundantly to promote Mad2 dissociation from Cdc20.
INTRODUCTION
The spindle checkpoint is a cellular surveillance system that prevents premature sister-chromatid separation in mitosis and ensures the fidelity of chromosome segregation (Bharadwaj and Yu, 2004; Musacchio and Salmon, 2007). It senses the existence of unattached kinetochores and kinetochores not under tension in a mitotic cell and inhibits the function of Cdc20, the mitotic activator of the anaphase-promoting complex or cyclosome (APC/C), thus stabilizing securin and cyclin B and delaying anaphase onset (Peters, 2006; Yu, 2007). On checkpoint activation, unattached kinetochores recruit checkpoint proteins in a hierarchical manner and promote their enzymatic and conformational activation, thus producing diffusible APC/C-inhibitory signals (Bharadwaj and Yu, 2004; Musacchio and Salmon, 2007).
The mitotic checkpoint complex (MCC) consisting of BubR1 (Mad3 in yeast), Bub3, Mad2, and Cdc20 is a critical checkpoint inhibitor of APC/C (Sudakin et al., 2001; Tang et al., 2001; Yu, 2002). It inhibits APC/C by competitively blocking substrate binding and altering the mode of Cdc20 binding to APC/C (Burton and Solomon, 2007; King et al., 2007; Herzog et al., 2009; Izawa and Pines, 2011). All four MCC components are enriched at unattached kinetochores, which promote MCC assembly in mitosis. MCC formation involves the conformational activation of Mad2, an unusual two-state protein that has two natively folded states, referred to as O-Mad2 (or N1-Mad2) and C-Mad2 (or N2-Mad2) (De Antoni et al., 2005; Luo et al., 2000, 2002, 2004; Luo and Yu, 2008; Mapelli et al., 2007; Mapelli and Musacchio, 2007; Yang et al., 2008). Cytosolic Mad2 adopts the O-Mad2 conformation. Unattached kinetochores recruit a Mad1–C-Mad2 core complex, which in turn recruits cytosolic O-Mad2 through O–C Mad2 dimerization. The loosely bound O-Mad2 undergoes conformational activation to become intermediate Mad2 (I-Mad2) or unliganded C-Mad2, which then binds to Cdc20. Binding of Mad2 to Cdc20 primes Cdc20 for BubR1–Bub3 binding, forming the MCC (Kulukian et al., 2009).
When all kinetochores achieve proper microtubule attachment and are under tension, the spindle checkpoint turns off to allow APC/C activation and chromosome segregation. Several mechanisms have been implicated in checkpoint inactivation in mammalian cells, including depletion of checkpoint proteins from attached kinetochores (Howell et al., 2001), capping of the Mad1–Mad2 core complex by the Mad2 inhibitor p31comet (Mapelli et al., 2006; Yang et al., 2007), p31comet- and UbcH10-dependent Cdc20 autoubiquitination (Xia et al., 2004; Reddy et al., 2007; Stegmeier et al., 2007), and proteasomal degradation (Visconti et al., 2010; Zeng et al., 2010; Ma and Poon, 2011). The first two mechanisms regulate the kinetochore-mediated production of APC/C-inhibitory checkpoint complexes, including MCC, whereas the two latter mechanisms promote MCC disassembly. How these mechanisms are coordinated during checkpoint inactivation and mitotic exit is not understood. In particular, the role of Cdc20 autoubiquitination in checkpoint inactivation has remained controversial because preventing Cdc20 ubiquitination by mutating all lysines in Cdc20 to arginines does not prevent MCC disassembly (Nilsson et al., 2008).
In this study, we further investigate the roles and coordination of p31comet, proteasomal degradation, and Cdc20 ubiquitination in MCC disassembly and checkpoint inactivation. We show that both the proteasome and p31comet are required for timely MCC disassembly in HeLa cells. Depletion of p31comet by RNA interference (RNAi) further delays MCC disassembly in cells treated with the proteasome inhibitor MG132, suggesting that p31comet has a proteasome-independent role in this process. Consistently, p31comet depletion delays Mad2–Cdc20 dissociation and mitotic exit in cells expressing Cdc20 mutants deficient for autoubiquitination. Taken together, our results reconcile earlier conflicting data and suggest that p31comet and Cdc20 ubiquitination act in somewhat redundant pathways to promote MCC disassembly and checkpoint inactivation.
RESULTS
p31comet is required for progression of unperturbed mitosis
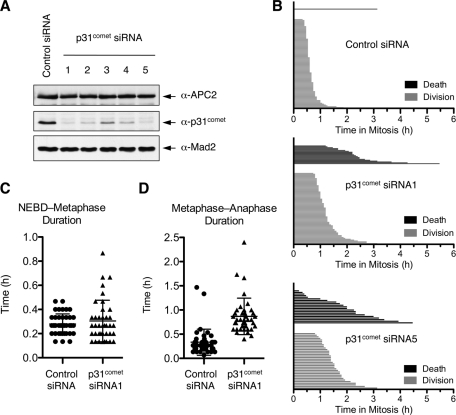
We previously showed that depletion of p31comet by RNAi in HeLa cells caused a delay in mitotic exit following the removal of nocodazole (Xia et al., 2004). To examine whether p31comet was required for normal mitotic progression in the absence of spindle poisons, we performed time-lapse, live-cell imaging of HeLa cells treated with either a control small interfering RNA (siRNA) or siRNAs against p31comet. p31comet siRNA1 and siRNA5 efficiently depleted p31comet (Figure 1A). Both were used in live-cell imaging experiments. Mitotic duration of each individual control- or p31comet-RNAi cell is shown in a waterfall plot (Figure 1B). Representative images of cells at various time points of the time-lapse movies are shown in Supplemental Figure S1.
FIGURE 1:
p31comet is required for proper mitotic progression. (A) Lysates of HeLa Tet-on cells transfected with control or p31comet siRNAs were blotted with the indicated antibodies. APC2 and Mad2 were used as loading controls. (B) Waterfall plots showing the mitotic duration and fate of each individual HeLa cell transfected with control or p31comet siRNAs. Mitotic duration is defined as the time between nuclear envelope breakdown (NEBD) and anaphase onset. (C) Duration from NEBD to the formation of a metaphase plate (metaphase) of cells transfected with the control or p31comet siRNAs. Mean and SD are indicated. (D) Duration from metaphase to the onset of anaphase of cells transfected with the control or p31comet siRNAs. Mean and SD are indicated.
Depletion of p31comet caused two phenotypes (Figure 1B). In the case of p31comet siRNA1, 27% of the p31comet RNAi cells underwent a mitotic delay of 1.8 h on average and then died in mitosis. The rest of the p31comet RNAi cells experienced a shorter mitotic delay of ∼40 min on average and then completed cell division normally (Figure 1B). For the latter group of cells, the duration from nuclear envelope breakdown (NEBD) to metaphase was similar to that of the control cells (Figure 1C). The delay in mitotic progression of these cells occurred at the metaphase–anaphase transition, that is, from the formation of a metaphase plate to the onset of chromosome segregation (Figure 1D). Even for the first group of cells that eventually died in mitosis, ∼50% of those also exhibited discernible chromosome alignment at the metaphase plate. p31comet siRNA5 caused a similar phenotype (Figure 1B). These results indicate that p31comet is required for the metaphase–anaphase transition during normal mitosis.
We used siRNA1 in subsequent experiments because ectopic expression of Myc-p31comet resistant to siRNA1 rescued both the mitotic delay and cell death phenotypes caused by this siRNA (Supplemental Figure S2). We also note that a moderate mitotic delay does not necessarily trigger cell death. It is conceivable that p31comet has a rather direct role in suppressing mitotic cell death.
MCC disassembly requires proteasome activity
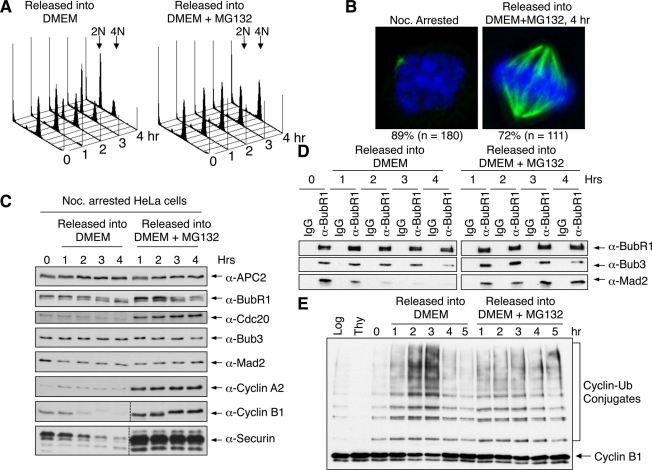
Recent studies implicated proteasomal degradation in checkpoint inactivation (Visconti et al., 2010; Zeng et al., 2010; Ma and Poon, 2011). To confirm the role of the proteasome in MCC disassembly, we released nocodazole-arrested HeLa cells into fresh medium with and without the proteasome inhibitor MG132 (Figure 2). On the basis of fluorescence-activated cell sorting (FACS) analysis and immunoblotting of cell cycle markers, cells efficiently underwent mitotic exit in the absence of MG132 (Figure 2, A and C). As expected, the amounts of Mad2 present in BubR1 immunoprecipitates dropped sharply within 1 h of release from nocodazole and reached basal levels at 2 h after release (Figure 2D), indicating that MCC disassembled when cells exited from mitosis. By contrast, ∼70–80% of cells remained arrested in mitosis, with bipolar spindles and chromosomes aligned at the metaphase plate, at 4 h after the release from the nocodazole arrest into MG132-containing medium (Figure 2B). Consistent with earlier findings (Visconti et al., 2010; Zeng et al., 2010; Ma and Poon, 2011), MCC persisted in cells arrested at metaphase by MG132 (Figure 2D).
FIGURE 2:
The proteasome activity is required for MCC disassembly. (A) FACS analysis of nocodazole-arrested HeLa cells released into fresh medium with or without MG132 for indicated times. (B) Nocodazole-arrested cells or cells released into MG132-containing medium for 4 h were stained with 4′,6-diamidino-2-phenylindole (blue) or anti-tubulin (green). The percentage of cells with the indicated phenotypes is indicated. (C) Lysates of cells in A were blotted with the indicated antibodies. (D) Lysates of cells in A were immunoprecipitated with IgG or anti-BubR1. The immunoprecipitates were blotted with the indicated antibodies. (E) APC/C was immunoprecipitated from nocodazole-arrested HeLa cells released into fresh medium with or without MG132 for indicated times and assayed for ligase activity.
We next tested whether APC/CCdc20 was active in MG132-arrested cells. APC/C isolated from nocodazole-arrested HeLa cells had basal level activity (Figure 2E). APC/C became more active at 2 h after release from nocodazole, concurrent with the completion of MCC disassembly (Figure 2, D and E). By contrast, APC/C in cells arrested by MG132 exhibited only basal level activity similar to that of nocodazole-arrested cells (Figure 2E). Therefore, despite apparent chromosome alignment, MCC still existed and APC/C remained inactive in MG132-arrested HeLa cells, indicating that the spindle checkpoint was not silenced in these cells. The levels of Cdc20, cyclin A2, cyclin B1, and securin were higher in MG132-treated cells (Figure 2C). Thus APC/C ubiquitinated its substrates and targeted them for degradation at a basal rate even when the checkpoint was on.
It was generally believed that cells arrested in mitosis by MG132 had silenced their spindle checkpoint and activated APC/CCdc20. They were arrested in metaphase because MG132 prevented the proteasome-dependent degradation of securin and cyclin B1. Our findings now suggest that MCC disassembly and APC/C activation require proteasome-mediated protein degradation and create an important caveat in the interpretation of results obtained using MG132-arrested metaphase cells. Although MG132 can clearly be expected to arrest cells in metaphase by blocking the degradation of securin and cyclin B1 independent of the spindle checkpoint, it also blocks MCC disassembly and some aspects of checkpoint inactivation.
p31comet promotes Mad2 dissociation from Cdc20 even when the proteasome is inhibited
Both proteasome and p31comet are required for timely checkpoint inactivation. We next wanted to examine the relationship between the two in this process. p31comet has been proposed to play two roles in checkpoint inactivation (see also later discussion of Figure 6). First, it caps the Mad1–Mad2 core complex and prevents Mad2 conformational activation, thus limiting the production of MCC. Second, it binds to Cdc20-bound Mad2, stimulating APC/C activity and promoting MCC disassembly. To separate the two roles of p31comet, we used the Aurora B inhibitor ZM447439 (referred to as ZM hereafter) to block the kinetochore-dependent pathway of MCC production in Taxol-arrested mitotic cells and investigated the fate of the existing MCC.
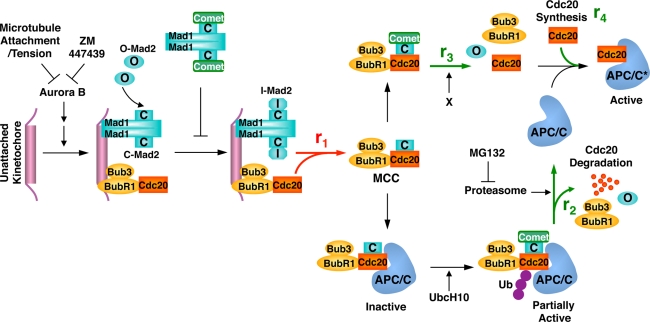
FIGURE 6:
Model illustrating the dynamics of MCC formation and disassembly and the roles of p31comet, Cdc20 ubiquitination, and the proteasome. See Discussion for details.
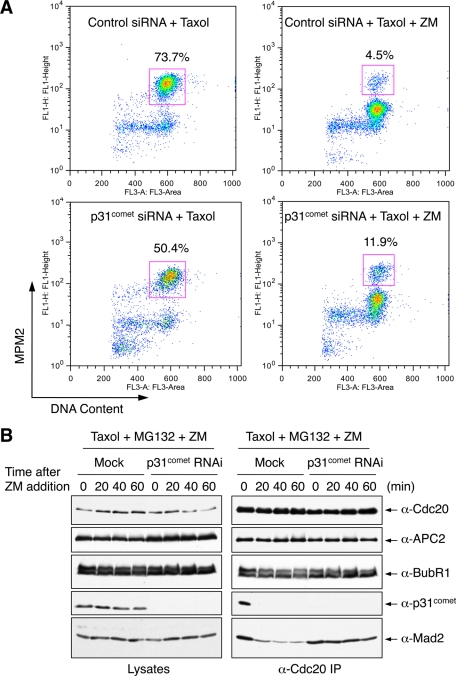
We first tested whether p31comet was required for checkpoint inactivation in HeLa cells arrested in mitosis by Taxol and then treated with ZM. p31comet depletion did not block mitotic exit induced by Aurora B inhibition (Figure 3A). At 90 min after ZM addition, the majority of both control and p31comet RNAi cells exited from mitosis. Fewer p31comet siRNA cells arrested in mitosis in the presence of Taxol, due to higher incidences of cell death.
FIGURE 3:
p31comet depletion delays MCC disassembly even with the proteasome inhibited. (A) FACS analysis of HeLa Tet-on cells transfected with control or p31comet siRNA and then treated with Taxol followed by Aurora B inhibition with ZM447439. The percentage of mitotic cells (cells that have 4N DNA content and are MPM2 positive) is shown for each sample. (B) HeLa Tet-on cells were either mock transfected or transfected with p31comet siRNA1 and arrested in mitosis by Taxol. MG132 and ZM447439 were then added to the cells. Samples were taken at the indicated times following ZM447439 addition. Cell lysates and anti-Cdc20 immunoprecipitates were blotted with the indicated antibodies.
Next we examined the relative contributions of proteasome and p31comet during checkpoint inactivation triggered by Aurora B inhibition. Addition of MG132 alone did not prevent Mad2 dissociation from Cdc20 (Figure 3B). Thus proteasomal degradation was not the sole mechanism for MCC disassembly. During this short time window of ZM addition, BubR1 did not completely dissociate from Cdc20, suggesting that Mad2 dissociation from Cdc20 could occur prior to, and independently of, BubR1 dissociation. Depletion of p31comet delayed Mad2 dissociation from Cdc20 in the presence of MG132 (Figure 3B). Thus p31comet has a function in MCC disassembly that is independent of proteasome activity.
p31comet promotes Mad2 dissociation from lysine-less Cdc20
Cdc20 autoubiquitination stimulated by p31comet and UbcH10 had been proposed to mediate MCC disassembly (Reddy et al., 2007; Stegmeier et al., 2007). On the other hand, a Cdc20 mutant with all lysines mutated to arginine (referred to as Cdc20 K-less) was capable of dissociating from Mad2 and BubR1 during checkpoint inactivation induced by Aurora B inhibition (Nilsson et al., 2008). Furthermore, cells expressing Cdc20 K-less established, but failed to maintain, their spindle checkpoint and underwent mitotic exit in the presence of spindle poisons. These results called into question the requirement of Cdc20 ubiquitination in MCC disassembly and suggested that Cdc20 ubiquitination and possibly degradation were required for checkpoint maintenance.
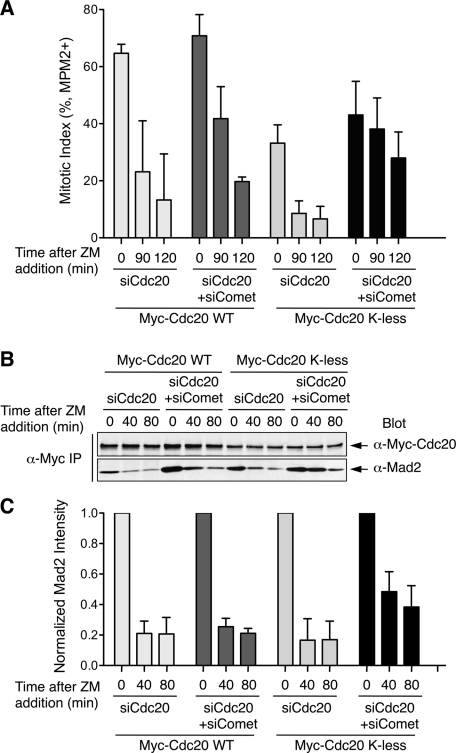
Because p31comet promoted Mad2 dissociation in the absence of proteasome activity, we tested whether p31comet was involved in the observed Mad2 dissociation from Cdc20 K-less. We constructed HeLa cell lines that stably expressed siRNA-resistant Myc-Cdc20 wild type (WT) or K-less transgenes. Consistent with earlier findings (Nilsson et al., 2008; Huang et al., 2009), depletion of Cdc20 from HeLa cells by RNAi caused mitotic arrest (Supplemental Figure S3). Ectopic expression of either Myc-Cdc20 WT or K-less rescued this mitotic block, indicating that both Cdc20 WT and K-less supported mitotic progression. Consistent with the previous report (Nilsson et al., 2008), cells expressing Myc-Cdc20 K-less at levels lower than those of Myc-Cdc20 WT failed to maintain their spindle checkpoint and exited from mitosis after a transient mitotic delay of ∼2 h in the presence of Taxol (data not shown).
We then transfected these cells with Cdc20 siRNA alone or both Cdc20 and p31comet siRNAs, synchronized these cells at the G1/S boundary, released them into Taxol, and treated them with ZM to initiate checkpoint inactivation. As expected, p31comet depletion from Myc-Cdc20 WT–expressing cells delayed mitotic exit by ∼30 min (Figure 4A), as observed in parental HeLa cells. Fewer Myc-Cdc20 K-less–expressing cells accumulated in mitosis at the time of ZM addition, consistent with a defect in checkpoint maintenance (Figure 4A). Addition of ZM greatly reduced the mitotic index of Cdc20 WT– or K-less–expressing cells, indicating that the mitotic delay seen in these cells was checkpoint dependent. Depletion of p31comet from Cdc20 K-less–expressing cells delayed mitotic exit following Aurora B inhibition (Figure 4A). On the basis of quantitative Western blotting, depletion of p31comet also delayed Mad2 dissociation from Myc-Cdc20 K-less (Figure 4, B and C). These results indicate that p31comet can promote mitotic exit and Mad2 dissociation from Cdc20, independently of Cdc20 ubiquitination. Of interest, compared with Myc-Cdc20 WT cells, depletion of p31comet from Cdc20 K-less–expressing cells had apparently a greater effect on the kinetics of mitotic exit and MCC disassembly (Figure 4). It is possible that, in the absence of Cdc20 autoubiquitination, the p31comet-dependent MCC disassembly pathway becomes particularly active.
FIGURE 4:
p31comet depletion delays Mad2 dissociation from lysine-less Cdc20. (A) Mitotic indices of HeLa Tet-on cells stably expressing Myc-Cdc20 WT or K-less that were transfected with siCdc20 or siCdc20/si-p31comet and then treated with Taxol followed by ZM addition for the indicated durations. Mitotic index is determined by FACS. Mean and SD from four independent experiments are shown. (B) HeLa Tet-on cells stably expressing Myc-Cdc20 WT or K-less were transfected with siCdc20 or siCdc20/si-p31comet and then treated with Taxol followed by ZM addition for the indicated durations. Anti-Myc immunoprecipitate from these cell lysates was blotted with anti-Myc and anti-Mad2. The blots were analyzed with an Odyssey Infrared Imaging System. (C) Quantification of Mad2 intensity in the anti-Myc immunoprecipitate in B. The Mad2 intensity is normalized to that of time 0 for each sample. Mean and SD of four independent experiments are shown.
p31comet promotes Mad2 dissociation from a Cdc20 mutant deficient for autoubiquitination
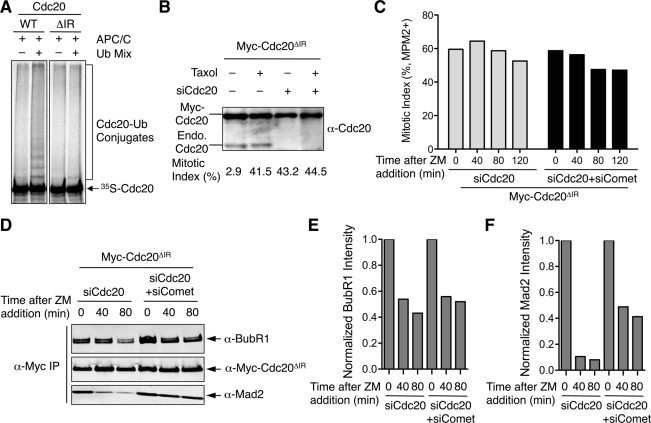
Cells expressing Cdc20 K-less cannot maintain mitotic arrest in the presence of Taxol. Furthermore, p31comet depletion delays both mitotic exit and Mad2 dissociation from Cdc20 K-less in these cells. It is formally possible that the delay in Mad2 dissociation from Cdc20 is a consequence of the delay in mitotic exit, not a direct effect of p31comet inactivation. To circumvent these two caveats, we constructed another Cdc20 mutant that was deficient for autoubiquitination. The C-terminal IR motif of Cdc20 is critical for Cdc20 to bind and activate APC/C. Indeed, the Cdc20 mutant with its C-terminal IR motif deleted (Cdc20ΔIR) did not undergo efficient autoubiquitination in vitro (Figure 5A). We next created HeLa Tet-on cell lines that stably expressed Myc-Cdc20ΔIR at levels severalfold higher than that of endogenous Cdc20 (Figure 5B). As expected, depletion of Cdc20 caused mitotic arrest of Cdc20ΔIR-expressing cells (Figure 5B), as Cdc20ΔIR could not bind to APC/C and was thus inactive. More important, inhibition of Aurora B by ZM did not trigger mitotic exit in Myc-Cdc20ΔIR–expressing cells depleted of Cdc20 alone or of both p31comet and Cdc20, indicating that this mitotic arrest was independent of Aurora B (Figure 5C). We next performed Myc immunoprecipitation from these cell lysates and blotted the immunoprecipitate with anti-Mad2 and anti-BubR1. Again, BubR1 did not completely dissociate from Cdc20ΔIR during this short time window of ZM addition (Figure 5, C and D). Depletion of p31comet did not appreciably alter the partial BubR1 dissociation from Cdc20ΔIR. By contrast, the majority of Mad2 dissociated from Cdc20ΔIR within 40 min of ZM addition. Depletion of p31comet significantly delayed Mad2 dissociation from Cdc20ΔIR. Therefore, these results suggest that p31comet can promote Mad2 dissociation from Cdc20 independent of Cdc20 autoubiquitination. In these experiments with Cdc20ΔIR-expressing cells, the cells are arrested in mitosis in a checkpoint-independent manner through Cdc20 RNAi, thus eliminating the two aforementioned caveats associated with Cdc20 K-less–expressing cells.
FIGURE 5:
p31comet depletion delays Mad2 dissociation from Cdc20ΔIR. (A) In vitro translated, 35S-labeled Myc-Cdc20 WT or ΔIR was incubated with Xenopus APC/C bound to anti-Cdc27 beads in the presence or absence of the ubiquitination reaction mixture. The reaction mixtures were resolved by SDS–PAGE and analyzed with a phosphoimager. (B) Myc-Cdc20ΔIR-expressing cells were mock transfected or transfected with siCdc20 and treated with or without Taxol. Cell lysates were blotted with anti-Cdc20. The bands of Myc-Cdc20ΔIR and the endogenous Cdc20 are indicated. The mitotic indices of each sample as determined by FACS are shown below each lane. (C) Myc-Cdc20ΔIR–expressing cells depleted of Cdc20 alone or depleted of both Cdc20 and p31comet were treated with Taxol and then ZM for the indicated durations. Mitotic index of these cells was determined by FACS and plotted. (D) Myc immunoprecipitate from lysates of cells described in C was blotted with the indicated antibodies. The blots were analyzed with an Odyssey Infrared Imaging System. (E) Quantification of BubR1 intensity in the anti-Myc immunoprecipitate in D. The BubR1 intensity is normalized to that of time 0 for each sample. (F) Quantification of Mad2 intensity in the anti-Myc immunoprecipitate in D. The Mad2 intensity is normalized to that of time 0 for each sample.
DISCUSSION
Redundant functions of p31comet and Cdc20 autoubiquitination in checkpoint inactivation
Recently, extensive efforts have led to the discovery of multiple mechanisms for checkpoint inactivation and MCC disassembly and have greatly advanced our molecular understanding of these processes. The coordination and cross-talk among the multiple checkpoint inactivation mechanisms are poorly understood, however. In particular, a key unresolved controversy in the field is the role of Cdc20 autoubiquitination in checkpoint silencing.
Pan and Chen (2004) first showed that Cdc20 underwent APC/C-dependent autoubiquitination and degradation during mitosis in yeast. This degradation required Mad2 and Mad3 (the yeast orthologue of BubR1), indicating that Cdc20 was ubiquitinated when it was bound to APC/C as part of the MCC. This mechanism was shown to be important for checkpoint-mediated inhibition of APC/C. Two related studies then showed that Cdc20 autoubiquitination was conserved in human cells, was stimulated by UbcH10 and p31comet, and was critical for MCC disassembly and checkpoint inactivation (Reddy et al., 2007; Stegmeier et al., 2007).
The role of Cdc20 autoubiquitination in checkpoint inactivation was later called into question, however. Nilsson et al. (2008) constructed a lysine-less Cdc20 mutant (Cdc20 K-less) that could not be ubiquitinated and showed that cells expressing this mutant established, but could not maintain, their spindle checkpoint in the presence of spindle poisons, consistent with Cdc20 ubiquitination being an APC/C-inhibitory checkpoint mechanism. Of interest, MCC containing Cdc20 K-less efficiently disassembled during checkpoint inactivation, leading Nilsson et al. to conclude that Cdc20 ubiquitination was not required for checkpoint inactivation.
In this study, we showed that p31comet is required for proper mitotic progression and for timely checkpoint inactivation in the absence of spindle poisons. The proteasome activity is required for MCC disassembly and APC/C activation following the recovery from nocodazole-mediated mitotic arrest. More important, we showed that p31comet depletion further delays MCC disassembly and checkpoint inactivation, even in the presence of proteasome inhibitors. Consistently, p31comet delays Mad2 dissociation from Cdc20 K-less and ΔIR (another Cdc20 mutant deficient for autoubiquitination) during checkpoint inactivation. Therefore, although p31comet can stimulate Cdc20 autoubiquitination and degradation, it has an additional function in promoting MCC disassembly that is independent of proteasomal degradation and Cdc20 ubiquitination.
We propose that p31comet and Cdc20 autoubiquitination act in partially redundant pathways to promote MCC disassembly and checkpoint inactivation (Figure 6). In this model, upon checkpoint activation, Cdc20 and the checkpoint proteins are recruited to unattached kinetochores in an Aurora B–dependent manner, enabling the conformational activation of Mad2 and increasing the rate of MCC formation (r1). Meanwhile, p31comet selectively binds to the Cdc20-bound, active conformer of Mad2 and, along with UbcH10, stimulates Cdc20 autoubiquitination and degradation (r2). Because Cdc20 bridges the interaction between BubR1 and Mad2, degradation of Cdc20 is expected to cause the complete disassembly of MCC. In a separate pathway, p31comet, together with unknown factors, promotes MCC disassembly (r3), independent of Cdc20 ubiquitination and degradation, creating free Cdc20. Finally, cap-independent, mitotic translation produces newly synthesized Cdc20 (r4). When the checkpoint is on, the rate of MCC formation is greater than the sum of the rates of MCC disassembly (i.e., r1 > r2 + r3), resulting in MCC accumulation (Figure 6).
Microtubule attachment during normal mitotic progression or inhibition of Aurora B by ZM447439 decreases the concentrations of checkpoint proteins at kinetochores, which is expected to reduce the rate of MCC formation. p31comet caps the Mad1–Mad2 core complex and blocks the conformational activation of Mad2, but p31comet binding to Mad1–Mad2 core complex does not appear to be regulated by the spindle checkpoint or Aurora B (data not shown). It likely acts as a constitutive mechanism to limit the rate of MCC formation. When the rate of MCC disassembly exceeds that of MCC formation (i.e., r2 + r3 > r1), the steady-state level of MCC drops. In addition, the sum of the rate of Cdc20 synthesis and the rate of nondegradative MCC disassembly is greater than that of MCC formation (i.e., r3 + r4 > r1), leading to an increase in the level of free Cdc20, APC/C activation, and checkpoint inactivation.
In the cases of Cdc20 K-less and ΔIR, although the pathway of Cdc20 autoubiquitination and degradation is blocked (i.e., r2 ≈ 0), the p31comet-dependent and Cdc20 ubiquitination-independent pathway (r3) is still on and can promote MCC disassembly and checkpoint inactivation. The underlying reasons for the inability of Cdc20 K-less–expressing cells to maintain the spindle checkpoint are unclear but could not simply be attributed to total Cdc20 levels. One possibility is that Cdc20 K-less–expressing cells accumulate active APC/CCdc20 species.
In the cells treated with both MG132 and ZM, BubR1 dissociates from Cdc20 more slowly than Mad2 does (Figure 3B). Furthermore, BubR1 does not completely dissociate from Cdc20ΔIR in cells treated with ZM, and p31comet does not impede this partial dissociation of BubR1 from Cdc20 (Figure 5D). These findings suggest that Cdc20 autoubiquitination and the proteasome pathway might preferentially promote BubR1 dissociation from Cdc20. The proteasome-independent, p31comet-dependent pathway preferentially promotes Mad2 dissociation from Cdc20.
Why then does depletion of p31comet only cause a transient mitotic delay and not completely block checkpoint inactivation? We envision three possibilities. First, there are other, unidentified p31comet-independent and proteasome-independent pathways for MCC disassembly checkpoint inactivation. Second, the Cdc20 autoubiquitination pathway is not strictly p31comet-dependent. The basal rates of Cdc20 autoubiquitination and degradation in the absence of p31comet are sufficient to trigger MCC disassembly, albeit with a delay. Finally, trivial explanations, such as the incompleteness of p31comet depletion, cannot be ruled out.
Nonetheless, our findings strongly suggest that p31comet and Cdc20 autoubiquitination do not act in the same linear pathway. p31comet has a role in MCC disassembly that is independent of Cdc20 ubiquitination and degradation, providing a straightforward explanation for the disassembly of MCC containing Cdc20 K-less or ΔIR.
Purified p31comet is insufficient to promote the disassembly of recombinant MCC in vitro (Xia et al., 2004; data not shown), indicating that other factors are required. It is intriguing that Hershko and coworkers recently showed that p31comet-dependent MCC disassembly in HeLa cell extracts requires the hydrolysis of the β–γ bond of ATP but not the hydrolysis of the α–β bond (Teichner et al., 2011). Because hydrolysis of the α–β bond of ATP is sufficient to support ubiquitination, their results are consistent with the notion that p31comet has a role independent of Cdc20 ubiquitination in MCC disassembly. Identification of the ATP-dependent enzyme that cooperates with p31comet to promote MCC disassembly is an important task for the future.
MCC as a dynamic entity in mitosis
The steady-state levels of MCC are determined by the difference between the rate of MCC formation and the rate of disassembly. Three possible scenarios can explain the checkpoint-dependent accumulation of MCC in mitosis. In the first scenario, the rate of MCC formation increases during mitosis, whereas the rate of MCC disassembly remains constant during the cell cycle. In a second scenario, an increase in the rate of MCC formation during mitosis is accompanied by a concomitant drop in the rate of MCC disassembly. In a third scenario, both rates of MCC formation and disassembly increase during mitosis, with the former being greater than the latter. The fact that Cdc20 autoubiquitination requires Mad2 and BubR1 suggests the third scenario occurs in the cell. In other words, the spindle checkpoint elevates both the rates of MCC formation and disassembly.
Why then is the third scenario beneficial to the cell? The third scenario makes MCC a highly dynamic entity. As a consequence, the level of MCC is more responsive to the status of checkpoint signaling. Once all sister chromatids achieve proper attachment, MCC can quickly disassemble to allow APC/C activation and sister-chromatid separation. This scenario is reminiscent of elevated dynamic instability of microtubules during mitosis. By contrast, the first two scenarios produce a slower and undesirable process of MCC disassembly and checkpoint inactivation.
In summary, the current evidence indicates that MCC assembly and disassembly are highly dynamic processes in mitosis. The differences between the rates of multiple competing reactions determine the steady-state levels of MCC and active APC/C and hence influence the status of the spindle checkpoint. p31comet and ubiquitination of Cdc20 function somewhat redundantly to promote checkpoint inactivation, ensuring robust and timely anaphase initiation.
MATERIALS AND METHODS
Cell culture and transfection
HeLa Tet-on cells were maintained in DMEM (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (Invitrogen) and 100 μg/ml penicillin and streptomycin (Invitrogen). To arrest cells at G1/S, cells were treated with 2.5 mM thymidine for 18 h. To arrest cells in mitosis, cells were treated with 100 nM Taxol or 330 nM nocodazole for 12–16 h.
The Cdc20 siRNA is Silencer Select Pre-designed siRNA (Ambion, Austin, TX) against Cdc20 (ID s2748) with the following sequence: sense strand, CGAAAUGACUAUUACCUGAtt; antisense strand, UCAGGUAAUAGUCAUUUCGga. The p31comet siRNAs were synthesized by Dharmacon (Lafayette, CO) and have the following sequences: siRNA1, GGCUGCUGUCAGUUUACUUtt; siRNA2, GAAGAUUGGUUUCGACCCAtt; siRNA3, UUUAAAGGCUUCCGCGAGUtt; siRNA4, CGAAGAAACUCACGCCUCCtt; and siRNA5, GCACCGUCGUCAUGGCACAtt. Transfection of the siRNAs was performed with Lipofectamine RNAiMAX (Invitrogen) following manufacturer's instructions. An siRNA with the random sequence of GGCAUCUUAGCCCGUACUUtt was used as control. The final concentrations of siRNAs used in the transfections were 2–10 nM.
The Myc-Cdc20 WT, K-less, and ΔIR stable cell lines were made by transfecting HeLa Tet-on cells with pTRE2-Myc-Cdc20 WT, K-less, or ΔIR plasmids using Effectene transfection reagent (Qiagen, Valencia, CA). Resistant colonies were selected using 400 μg/ml hygromycin (Clontech, Mountain View, CA). Single colonies were picked and screened for inducible expression in the absence and presence of 1.5 μg/ml doxycycline (Clontech). For actual experiments, Myc-Cdc20 WT–expressing cells (clone 11) were cultured in the presence of doxycycline. Myc-Cdc20 ΔIR–expressing cells (clone 6) were also cultured in the presence of doxycycline. Myc-Cdc20 K-less–expressing cells (clone 9) were cultured in the absence of doxycycline to keep the expression levels of Cdc20 K-less low.
Antibodies, immunoblotting, and immunoprecipitation
Antibodies against BubR1, Cdc20, APC2, Mad2, and p31comet were described previously (Tang et al., 2001; Xia et al., 2004). The mouse anti-Myc (9E10) antibody was purchased from Roche (Indianapolis, IN). For quantitative immunoblotting, anti–rabbit immunoglobulin G (IgG) (H+L) (Dylight 800 conjugates) and anti–mouse IgG (H+L) (Dylight 680 conjugates) (Cell Signaling, Beverly, MA) were used as secondary antibodies. The membranes were scanned with the Odyssey Infrared Imaging System (LI-COR, Lincoln, NE).
For immunoprecipitation, the cell pellets were lysed with lysis buffer (50 mM Tris-HCl, pH 7.7, 120 mM KCl, 0.1% NP40, 1 mM dithiothreitol) supplemented with protease inhibitor tablets (Sigma-Aldrich, St. Louis, MO). After centrifuging for 30 min at 4°C at top speed in a microcentrifuge, the supernatants were incubated with protein A beads coupled to anti-Myc antibody or other antibodies for 2 h at 4°C. After washing, the beads were boiled in SDS sample buffer and analyzed by SDS–PAGE, followed by immunoblotting.
Live-cell imaging
For live-cell imaging, ∼25,000 HeLa Tet-on cells were seeded into each well of a four-well chambered coverglass. At 24 h after seeding, cells were transfected with the control siRNA, p31comet siRNA1, or p31comet siRNA5 at 5 nM concentration. At 36 h after siRNA transfection, live-cell imaging was performed with a DeltaVision microscope (Applied Precision, Issaquah, WA). For each sample, 10 fields were chosen at random. Differential interference contrast images were taken with a 40× objective lens at 4-min intervals for a total duration of 21 h. Four 5-um optical slices were acquired for each time point. Images were processed with ImageJ (National Institutes of Health, Bethesda, MD) and Photoshop (Adobe, San Jose, CA).
Flow cytometry
HeLa Tet-on cells or the Myc-Cdc20–expressing stable cell lines were transfected with p31comet siRNA1 or both p31comet siRNA1 and the Cdc20 siRNA. Thymidine was added at 6 h after transfection. The cells were released into fresh media after the 18-h thymidine treatment. At 7 h after release, Taxol was added for another 7 h before ZM447439 (Selleck Chemicals, Houston, TX) addition. Cells were harvested at different time points after adding ZM447439 and were fixed with 70% ethanol. Propidium iodide was used to stain DNA. To stain mitotic cells, fixed cells were incubated with anti–phospho-MPM2 antibody (Millipore, Billerica, MA) followed by Alexa Fluor 488 donkey anti–mouse secondary antibody (Invitrogen). Cells were analyzed by a flow cytometer, and the data were processed with FlowJo (FloJo, Ashland, OR).
APC/C ubiquitination assays
APC/C ubiquitination assays using Xenopus or human APC/C were performed essentially as described (Tang and Yu, 2004).
Supplementary Material
Acknowledgments
We thank Jon Pines for providing the Cdc20 K-less expression plasmid and Laura Diaz-Martinez for reading the manuscript critically. This work was supported by the National Institutes of Health (GM61542) and the Welch Foundation (I-1441). H.Y. is an Investigator at the Howard Hughes Medical Institute.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- Bub1/3
budding uninhibited by benomyl 1/3
- FACS
fluorescence-activated cell sorting
- Mad1–3
mitotic arrest deficiency 1–3
- MCC
mitotic checkpoint complex
- NEBD
nuclear envelope breakdown
- RNAi
RNA interference
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0389) on September 21, 2011.
REFERENCES
- Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Shi J, Orth JD, Mitchison TJ. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 2009;16:347–358. doi: 10.1016/j.ccr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HT, Poon RY. Orderly inactivation of the key checkpoint protein mitotic arrest deficient 2 (MAD2) during mitotic progression. J Biol Chem. 2011;286:13052–13059. doi: 10.1074/jbc.M110.201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Mapelli M, Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr Opin Struct Biol. 2007;17:716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tang Z, Yu H. Functional analysis of the spindle-checkpoint proteins using an in vitro ubiquitination assay. Methods Mol Biol. 2004;281:227–242. doi: 10.1385/1-59259-811-0:227. [DOI] [PubMed] [Google Scholar]
- Teichner A, Eytan E, Sitry-Shevah D, Miniowitz-Shemtov S, Dumin E, Gromis J, Hershko A. p31comet promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci USA. 2011;108:3187–3192. doi: 10.1073/pnas.1100023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti R, Palazzo L, Grieco D. Requirement for proteolysis in spindle assembly checkpoint silencing. Cell Cycle. 2010;9:564–569. doi: 10.4161/cc.9.3.10581. [DOI] [PubMed] [Google Scholar]
- Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31comet antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Liu CJ, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 2008;6:e50. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr Opin Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.