The first PH domain of the myosin X cargo-binding domain is split into halves by insertion of another PH domain forming a PH1N-PH2-PH1C tandem. This tandem forms a rigid supramodule with the two lipid-binding pockets juxtaposed for cooperative binding to PI(3,4,5)P3-containing lipid membranes.
Abstract
Myosin X (MyoX) is an unconventional myosin that is known to induce the formation and elongation of filopodia in many cell types. MyoX-induced filopodial induction requires the three PH domains in its tail region, although with unknown underlying molecular mechanisms. MyoX's first PH domain is split into halves by its second PH domain. We show here that the PH1N-PH2-PH1C tandem allows MyoX to bind to phosphatidylinositol (3,4,5)-triphosphate [PI(3,4,5)P3] with high specificity and cooperativity. We further show that PH2 is responsible for the specificity of the PI(3,4,5)P3 interaction, whereas PH1 functions to enhance the lipid membrane–binding avidity of the tandem. The structure of the MyoX PH1N-PH2-PH1C tandem reveals that the split PH1, PH2, and the highly conserved interdomain linker sequences together form a rigid supramodule with two lipid-binding pockets positioned side by side for binding to phosphoinositide membrane bilayers with cooperativity. Finally, we demonstrate that disruption of PH2-mediated binding to PI(3,4,5)P3 abolishes MyoX's function in inducing filopodial formation and elongation.
INTRODUCTION
Myosin X (MyoX), encoded by Myo10, is an actin-based unconventional myosin localized at the tips of filopodia (Berg et al., 2000; Berg and Cheney, 2002; Tokuo and Ikebe, 2004; Zhang et al., 2004; reviewed in Sousa and Cheney, 2005). Overexpression of MyoX in heterologous cell lines, such as HeLa and COS7, induces the formation of large numbers of elongated filopodia (Bohil et al., 2006; Tokuo et al., 2007; Kerber et al., 2009; Watanabe et al., 2010). In endothelial cells, MyoX, in response to bone morphogenetic protein (BMP)–mediated up-regulation of expression, promotes filopodial extension and is required for BMP-dependent directed cell migration (Pi et al., 2007). The functional role of MyoX in directed cell migration has also been demonstrated in vivo in the cranial neural crest cells of Xenopus embryos (Hwang et al., 2009; Nie et al., 2009). The role that MyoX plays in filopodial formation is likely to be directly linked to the motor's function in directed cell migrations, as filopodia act as cell antenna that integrate various cellular signals to regulate actin cytoskeletal structures (Gupton and Gertler, 2007; Mattila and Lappalainen, 2008).
From its N- to C-termini, MyoX contains a motor domain, a neck region consisting of three IQ motifs, a single α-helix domain followed by a coiled-coil domain, three PH domains, a myosin tail homology (MyTH4) domain, and a band 4.1/ezrin/radixin/moesin (FERM) domain (Figure 1A). The motor domain binds to bundled actin filaments and powers the processive movement of MyoX (Nagy et al., 2008; Kerber et al., 2009). A GCN4-forced dimer of MyoX containing its head, neck, and predicted coiled-coil domains (known as MyoX-HMM) is sufficient for the filopodia tip localization and intrafilopodial movement of the motor (Berg and Cheney, 2002; Kerber et al., 2009). However, substrate-attached filopodial “buds” induced by MyoX-HMM often fail to elongate, and thus the PH repeats, MyTH4, and FERM domains of the motor are also required for the formation of elongated filopodia (Bohil et al., 2006; Tokuo et al., 2007; Plantard et al., 2010). Recently the MyTH4 and FERM domains were shown to act as a structural and functional supramodule in binding to deleted in colorectal cancer (DCC), a transmembrane MyoX cargo mediating directed axonal migrations (Zhu et al., 2007; Hirano et al., 2011; Wei et al., 2011).
FIGURE 1:
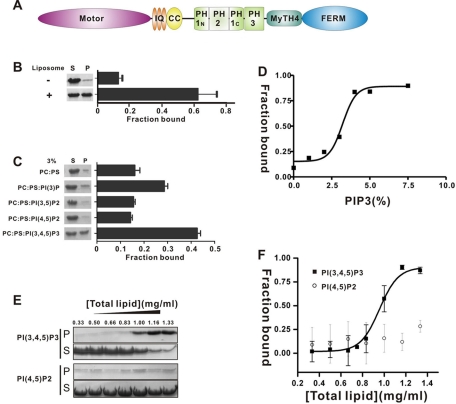
Characterization of the binding of MyoX PH1N-PH2-PH1C to lipid membranes. (A) Schematic diagram showing the domain organization of MyoX. (B) Cosedimentation-based assay of bindings of MyoX PH1N-PH2-PH1C to liposomes prepared from bovine brain lipid extract. After centrifugation, fractions labeled with S represent proteins in supernatants and fractions labeled with P represent proteins in pellets in the assays. The bar graph represents the fraction of the protein recovered in the liposome-bound pellet in each sedimentation assay. Values are mean ± SD of three different experiments. (C) Binding of MyoX PH1N-PH2-PH1C to defined, reconstituted PC/PS/PIP (77/20/3%) liposomes. Values are mean ± SD of three different experiments. (D) PI(3,4,5)P3 density-dependent binding between the PH1N-PH2-PH1C tandem and defined liposomes. All data points represent the cosedimentation assay between MyoX PH1N-PH2-PH1C and PI(3,4,5)P3-containing liposome. In the reconstituted liposome, PC concentration was fixed at 20%, and PS concentration was decreased from 80 to 72.5%,with the corresponding increase of the PI(3,4,5)P3 concentration from 0 to 7.5%. (E) Liposome concentration–dependent binding between the PH1N-PH2-PH1C tandem and the reconstituted PC/PS/PIPs (77/20/3%) liposomes. Total liposome concentration varied from 0.33 to 1.33 mg/ml in the cosedimentation assays. (F) The sigmoidal dose-dependent binding curve of PH1N-PH2-PH1C to the PC/PS/PI(3,4,5)P3 (77/20/3%) liposomes. Error bars, SD of liposome bindings from three different experiments.
Enrichment of phosphatidylinositol (3,4,5)-triphosphate [PI(3,4,5)P3] in the leading edges is important for the dynamics of filopodium (Luikart et al., 2008; Ketschek and Gallo, 2010). MyoX contains three consecutive PH domains immediately N-terminal to its MyTH4-FERM supramodule (Figure 1A). The PH domains of the motor couple with PI3 kinase activities by directly binding to the kinase product PI(3,4,5)P3 during MyoX-mediated cellular processes, including phagocytosis (Cox et al., 2002) and filopodial extension (Plantard et al., 2010). The organization of the MyoX PH domains is highly unusual: the second PH domain is inserted in the predicted β3/β4 loop of PH1, splitting PH1 into halves and forming a PH1N-PH2-PH1C tandem (Berg et al., 2000; Figure 1A). The amino acid sequences of PH1 and PH2, as well as all linker sequences connecting them, are highly conserved, and the splitting site of PH1 is identical in MyoX orthologues from different species, indicating that the PH1N-PH2-PH1C tandem is likely to have specific functions in addition to the individual functions of PH1 and PH2. In vitro biochemical assays, as well as in silico amino acid sequence analysis, demonstrated that the isolated PH2 is capable of specifically binding to PI(3,4,5)P3 (Isakoff et al., 1998; Plantard et al., 2010; Umeki et al., 2011). Using a loss-of-function approach, it was shown that the PH2 domain is required for filopodia induction as well as phagocytosis (Cox et al., 2002; Plantard et al., 2010). It was shown recently that the PH domains are involved in the direct binding to the motor head, thus maintaining MyoX in an autoinhibited conformation. Binding of PI(3,4,5)P3 to the PH domains can release the autoinhibited conformation of MyoX (Umeki et al., 2011). However, neither the biochemical properties nor the functional role of the PH1N-PH2-PH1C tandem of MyoX were directly tested.
Here we demonstrate that the PH1N-PH2-PH1C tandem of MyoX specifically binds to PI(3,4,5)P3-containing lipid membranes. The high-resolution structure of the PH1N-PH2-PH1C tandem reveals that the two split halves of PH1 adopt an intact PH domain fold. The two PH domains form a structural supramodule with their phosphoinositide-binding pockets juxtaposed with each other for synergistic binding to lipid membranes. The structure of the PH1N-PH2-PH1C tandem, together with detailed biochemical analysis, demonstrates that PH2 provides the PI(3,4,5)P3-binding specificity of MyoX, whereas the split PH1 functions to enhance its lipid membrane–binding avidity via nonspecific binding to phosphoinositide. We further demonstrate that the lipid-binding properties of both PH1 and PH2 are required for MyoX to function in filopodial induction, indicating that the PH1N-PH2-PH1C tandem acts as a functional supramodule in sensing acute increases in PI(3,4,5)P3 concentration induced by PI3 kinase at the tip of filopodia.
RESULTS
The PH1N-PH2-PH1C tandem of MyoX specifically binds to PI(3,4,5)P3-containing liposomes with high affinity
We attempted to determine the lipid membrane–binding properties of PH1N-PH1C and PH2 individually by removing the PH2 sequence from the predicted β3/β4 loop of the split PH1, as our earlier studies of split PH domains from α-syntrophin, phospholipase Cγ, Rho kinase, and PI3 kinase enhancer showed that covalently fusing two halves of a split PH domain gives rise to a well-structured canonical PH domain fold (Yan et al., 2005, 2008; Wen et al., 2006, 2008). To our disappointment, neither PH1N-PH1C nor PH2 could be purified as soluble proteins despite extensive trials using different domain boundaries, implying the two domains in the tandem may not act as separately functional PH domains. In contrast, the PH1N-PH2-PH1C tandem can be expressed as a highly soluble protein that adopts a stable monomeric conformation at a wide range of concentrations (up to 0.5 mM tested; Supplemental Figure S1).
The purified PH1N-PH2-PH1C tandem robustly binds to liposomes prepared from total bovine brain lipid extracts in a cosedimentation-based lipid membrane–binding assay (Figure 1B). We tested the binding specificity of the PH1N-PH2-PH1C tandem for different phosphatidylinositol phosphates (PIPs) using reconstituted defined liposomes, each composed of phosphatidylcholine (PC), phosphatidylserine (PS), and one isoform of PIP at a ratio of 77:20:3 using the same assay. We kept the PIP concentration low (3%) in the defined liposomes to ensure assay stringency. The results demonstrate that the PH1N-PH2-PH1C tandem has a high selectivity for PI(3,4,5)P3 and a modest selectivity for PI(3)P (Figure 1C). Because PI(3)P is known to be present only in endosomes and multivesicular bodies at relatively constant levels (Lemmon, 2008), we decided not to further address its interaction with the PH1N-PH2-PH1C tandem. Instead, we focused on the interaction between PI(3,4,5)P3-containing lipid membranes with the PH1N-PH2-PH1C tandem, as PI3 kinase–mediated signaling is coupled with the functions of MyoX (Cox et al., 2002; Plantard et al., 2010; Umeki et al., 2011).
We performed a PI(3,4,5)P3 density-dependent binding assay between the PH1N-PH2-PH1C tandem and defined liposomes. In this experiment, the ratio of PI(3,4,5)P3 in the PC/PS/PI(3,4,5)P3 was increased incrementally from 0 to 7.5%, and the PC ratio was proportionally changed from 80 to 72.5%, with the PS concentration fixed at 20%. We observed a highly sigmoidal PI(3,4,5)P3 density-dependent binding curve, with a mid PI(3,4,5)P3 density point of ∼3% (Figure 1D). Only a basal level of membrane binding could be detected when the PI(3,4,5)P3 density was <2.5%, a condition corresponding to the low-activity state of PI3 kinase. In contrast, the PH1N-PH2-PH1C tandem is essentially saturated when PI(3,4,5)P3 density reaches ∼5%, indicating that the PH1N-PH2-PH1C tandem can efficiently respond to acute increases in plasma membrane PI(3,4,5)P3 concentration in response to PI3 kinase activation (Papayannopoulos et al., 2005). Taken together, these biochemical data indicate that the PH1N-PH2-PH1C tandem of MyoX is ideal as a sensor for cellular PI3 kinase activity.
We then performed a liposome concentration–dependent PH1N-PH2-PH1C tandem binding assay. In this assay, we fixed the PIP concentration in the defined liposome at 3% (i.e., a PC:PS:PIP ratio of 77:20:3) and varied total liposome concentration. We found that the PH1N-PH2-PH1C tandem binds to PI(3,4,5)P3-containing liposomes in a dose-dependent manner (Figure 1, E and F). The sigmoidal dose-dependent binding curve in Figure 1F indicates that the interaction between the PH1N-PH2-PH1C tandem and the PI(3,4,5)P3-containing liposomes is cooperative, and this binding cooperativity is the result of a synergistic action of both PH domains in the PH1N-PH2-PH1C tandem (see later discussion for details). In contrast, only a background level of binding could be detected between the PH1N-PH2-PH1C tandem and PI(4,5)P2-containing liposomes (Figure 1, E and F), further demonstrating the specificity of the PH1N-PH2-PH1C tandem/PI(3,4,5)P3 interaction.
The PH1N-PH2-PH1C tandem forms a structural supramodule
To elucidate the mechanistic basis governing the specific and cooperative binding of the PH1N-PH2-PH1C tandem to PI(3,4,5)P3-containing membranes, we tried to determine the atomic structure of the PH1N-PH2-PH1C tandem using NMR spectroscopy and x-ray crystallography. The poor quality of NMR spectra of the wild-type PH1N-PH2-PH1C indicated that the protein is not amenable to NMR-based structure determination. Despite numerous trials, we failed to obtain crystals of the wild-type PH1N-PH2-PH1C with or without the PI(3,4,5)P3 head group bound to the protein. However, we were able to obtain high-quality crystals of a PH1N-PH2-PH1C mutant in which three positively charged residues (K1187 in the β1/β2 loop of PH1 and K1223/K1224 in the β1/β2 loop of PH2) were neutralized with Asn. The structure of the PH1N-PH2-PH1C mutant was solved and refined to a 2.5-Å resolution (Figure 2A and Table 1).
FIGURE 2:
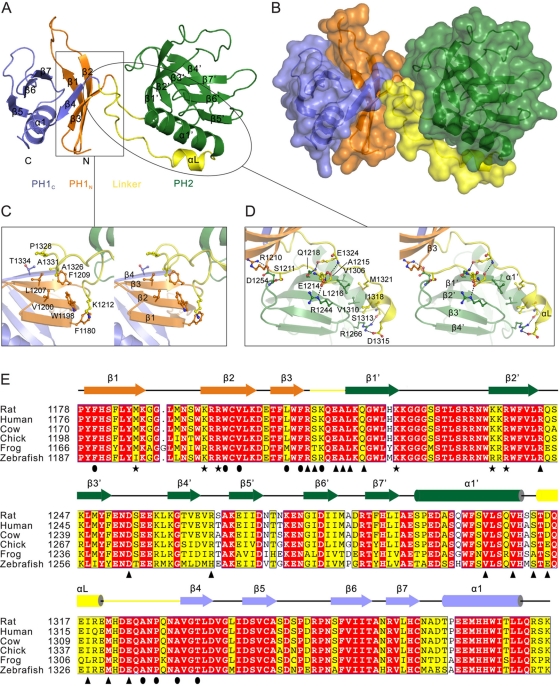
Crystal structure of the MyoX PH1N-PH2-PH1C tandem. (A) Ribbon diagram representation of the overall structure of the MyoX PH1N-PH2-PH1C tandem. PH1N and PH1C are orange and blue, respectively; PH2 is green; and the PH1 and PH2 linker is yellow. This coloring scheme is followed throughout. (B) Translucent surface diagram of the PH1N-PH2-PH1C tandem with the same orientation as in A. (C) Stereo view of the interface between PH1 and the linker region (boxed region in A). (D) Stereo view of the interface between PH2 domain and the linker region (oval in A). (E) Sequence alignment of PH1N-PH2-PH1C of MyoX from different species. The secondary structural elements are indicated above the alignment and colored as in A. Residues in the signature motif predicted to be critical for phosphoinositide recognition are highlighted with stars. Residues involved in the interface between the PH1 and the linker region are indicated with ovals, and residues forming the PH2 and the linker interface are highlighted with triangles.
TABLE 1:
Data collection and refinement statistics for MyoX PH12_3KN.
| A. Diffraction dataa | |
|---|---|
| Space group | P41 |
| Unit cell (Å) | a = 36.8 |
| b = 36.8 | |
| c = 154.1 | |
| α = β = γ = 90º | |
| Resolution (Å) | 30–2.53 (2.60–2.53) |
| Observed reflections | 18,309 |
| Unique reflections | 6630 |
| Rmergeb (%) | 6.6 (33.3) |
| I/σ | 10.0 (2.3) |
| Average redundancy | 2.8 (2.6) |
| Completeness (%) | 96.7 (96.7) |
| B. Refinementa | |
| Rcrystc (last shell) | 21.5 (33.6) |
| Rfreed (last shell) | 27.8 (38.3) |
| Mean B factors (Å2) | 43.7 |
| Bond lengthe (Å) | 0.012 |
| Bond angle (deg) | 1.1 |
| Ramachandran plot (%) | |
| Most favored | 90.1 |
| Additionally allowed | 9.9 |
aThe values in parentheses refer to the highest-resolution shell.
bRmerge =∑|i − Im|/∑Ii, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
cRcryst = ∑‖Fobs| − |Fcalc‖/∑|Fobs|, where Fobs and Fcalc are observed and calculated structure factors, respectively.
dRfree = ∑T‖Fobs| − |Fcalc‖/∑T|Fobs|, where T is a test data set of ∼10% of the total reflections randomly chosen and set aside prior to refinement.
eRoot-mean-square deviation from ideal values.
The structure of the PH1N-PH2-PH1C tandem shows that both the split PH1 and PH2 adopt canonical PH domain folds, consisting of a conserved core composed of two-sheeted β-barrel with one end capped by a C-terminal amphipathic α-helix (Figure 2A). PH1 is split between β3 and β4 by the insertion of the PH2 domain. Like all other known split PH domains (Teo et al., 2006; Yan et al., 2005, 2008; Wen et al., 2006, 2008), the two halves of MyoX PH1 (PH1N and PH1C) interact with each other to form an intact PH domain fold.
The unique and most important feature of the PH1N-PH2-PH1C tandem with respect to the other split PH domains is that the split PH1 domain and the inserting PH2 domain are physically integrated into a structural supramodule by two highly conserved linking sequences at the PH2 insertion site (i.e., the segment linking β3 of PH1 and β1′ of PH2, and the stretch connecting α1′ of PH2 and β4 of PH1; see Figure 2, A, B, and E). The relative orientations of the two PH domains of MyoX are fixed, although the two PH domains make minimal contact with each other, with the exception of the Arg-1210–Asp-1254 salt bridge (Figure 2D; see later discussion for more details). In sharp contrast, the orientations of the split PH domains are highly flexible with respect to the inserting domains in other split PH domains (Yan et al., 2005, 2008; Wen et al., 2006, 2008).
The interface between PH1 and the linker region is primarily hydrophobic, with a total buried surface area of ∼420 Å2 (Figure 2C). A number of highly conserved hydrophobic residues from the first β-sheet of PH1 (β1–β4) with their side chains facing outside of the PH domain β-barrel (e.g., Phe-1180, Trp-1198, Val-1200, Leu-1207, and Phe-1209) pack with conserved Ala-1326, Pro-1328, Ala-1331, and Lys-1212 in the linker region (Figure 2, C and E). The interface between PH2 and the linker region is more extensive and involves residues from the first β-sheet of PH2 (β1′–β4′) and the α1′ helix, burying ∼680 Å2 of surface area through hydrophobic interactions, hydrogen bonding, and two salt bridges between Asp-1254 and Arg-1210 and between Asp-1315 and Arg-1266 (Figure 2, D and E).
Mutation of any single hydrophobic residue in the PH1/linker interface (Figure 2C) invariably leads to heavy aggregation of the PH1N-PH2-PH1C tandem mutant protein (unpublished data). We also tested the role of the salt bridge between Asp-1254 and Arg-1210 in the PH2/linker interface by substituting Asp-1254 with an Arg. Although the Asp-1254–Arg-1210 salt bridge is at the edge of the PH2/linker interface, the D1254R-PH1N-PH2-PH1C mutant was eluted with a highly broadened low–molecular weight peak and a heavy aggregated high–molecular weight peak on an analytical gel filtration column (Supplemental Figure S1), indicating that the mutant protein is conformationally heterogeneous, presumably due to the destabilized PH1/PH2 interface caused by the D1254R substitution. The foregoing biochemical data are consistent with the structure-based prediction that the highly conserved residues in the PH1/PH2 interface maintain the conformation of the PH1N-PH2-PH1C supramodule.
PH2 is responsible for PI(3,4,5)P3-binding specificity, whereas the split PH1 increases general membrane-binding avidity
A number of PH domains—for example, PH domains from general receptors for phosphoinositides (Grp1), Bruton's tyrosine kinase (Btk), a dual specificity for PI(3,4,5)P3 and PI(3,4)P2 to PH domains of the dual adaptor for phosphotyrosine and 3-phosphoinositides (DAPP1), and protein kinase B (PKB/Akt)—are known to bind selectively to PI(3,4,5)P3 (Lemmon, 2008). The structures of these PH domains in complex with Ins(1,3,4,5)P4, the soluble head group of PI(3,4,5)P3, are known. We performed a structure-based amino acid sequence alignment analysis of the split PH1 and PH2 with these PI(3,4,5)P3-selective PH domains (Supplemental Figure S2). It is known that all of these PI(3,4,5)P3-binding PH domains contain a signature motif with conserved positively charged amino acid residues, K-Xn-(K/R)-X-R, in which the first Lys is located at the penultimate position of the β1 strand and the (K/R)-X-R sequence corresponds to the residues 2–4 of the β2 strand (Lemmon, 2008). The PH2 domain of MyoX PH1N-PH2-PH1C contains this signature motif, whereas the split PH1 lacks the first Lys (Supplemental Figure S2), suggesting that PH2 is primarily responsible for selectively binding to PI(3,4,5)P3. In addition, the structure of the DAPP1 PH domain is very similar to that of MyoX PH2, and the positions of the residues responsible for binding to PI(3,4,5)P3 in DAPP1 PH are well matched with corresponding residues in the sequence and the structure of MyoX PH2 (Figure 3, A and B, and Supplemental Figure S2). Using the DAPP1 PH/Ins(1,3,4,5)P4 complex structure as a template (Ferguson et al., 2000), we built a MyoX PH2/Ins(1,3,4,5)P4 complex structural model (Figure 3A). Analogous to the DAPP1 PH/Ins(1,3,4,5)P4 complex (Figure 3B), the MyoX PH2/Ins(1,3,4,5)P4 complex structural model shows that Arg-1239 in β2′ forms salt bridge(s) with the 3-phosphate, Tyr-1250 in β3′ forms a hydrogen bond with the 4-phosphate, Lys-1223 in β1′ forms salt bridges with the 3- and 4-phosphates, and Arg-1288 immediately preceding β7′ forms a salt bridge with the 4-phosphate of Ins(1,3,4,5)P4 (Figure 3A).
FIGURE 3:
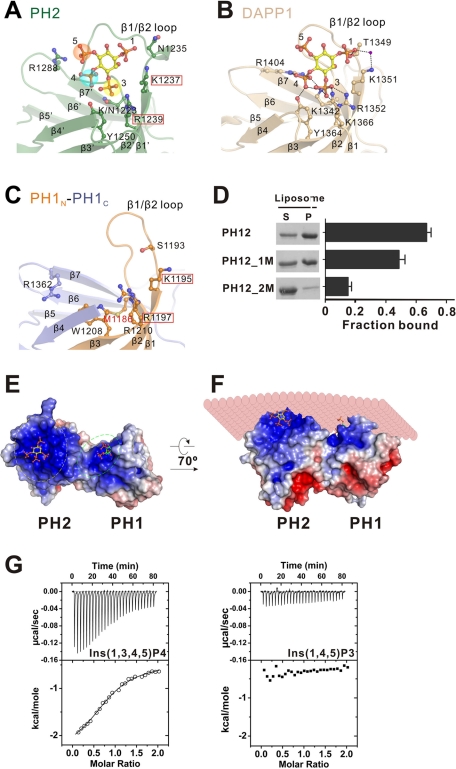
Structural basis of the specific binding between PH1N-PH2-PH1C and PI(3,4,5)P3. (A) Ribbon diagram representation of the docked MyoX PH2/Ins(1,3,4,5)P4 complex structural model. The residues predicted to be critical for Ins(1,3,4,5)P4 binding are shown in stick model. The residues substituted with Ala to test their role in binding to defined PIP-containing liposome are indicated with red boxes. (B) Ribbon diagram structure of the Ins(1,3,4,5)P4-bound DAPP1 PH (PDB ID: 1FAO), which was used to model the MyoX PH2/Ins(1,3,4,5)P4 complex structure. (C) Ribbon diagram representation of the MyoX PH1 domain showing that the canonical lipid-binding pocket of the domain contains several positively charged residues. The two positively charged residues chosen to be substituted with Ala to test the domain's membrane binding are boxed in red. (D) Sedimentation-based assay of bindings of the wild-type PH12 and its two mutants, PH12_1M and PH12_2M, to defined liposomes containing 3% PI(3,4,5)P3. (E, F) Surface electrostatic potential representation of the PH12 domain (blue to red, +5 to −5 times kBTec−1, where k is the Boltzmann constant, T is temperature, and ec is the elementary charge constant), showing two prominent positively charged lipid head group–binding pockets in the tandem. Ins(1,3,4,5)P4 (shown in the stick model) is modeled into the PH2 lipid-binding pocket, and Ins(1,4,5)P3 is modeled into the lipid-binding pocket of PH1. It is also shown in F that the two PIP lipid-binding pockets of the PH12 tandem are juxtaposed to lipid membrane bilayers. (G) ITC curves showing that MyoX PH12 can specifically interact with Ins(1,3,4,5)P4 with a KD of ∼32.9 ± 4.2 μM [the error represents the fitting of the experimental data with each PH12 containing one Ins(1,3,4,5)P4 binding-site model]. No detectable binding can be found between PH12 and Ins(1,4,5)P3.
It is well established that the first lysine in the signature motif at the penultimate position of the β1 strand is conserved in phosphoinositide-binding PH domains (Supplemental Figure S2). Removal of this positive charge by amino acid substitution is known to greatly decrease or eliminate the lipid-binding capacities of PH domains (Lemmon, 2008). In the split PH1 of MyoX, the corresponding residue is a Met (Met-1186; Figure 3C), suggesting that PH1 is not likely to bind specifically to phosphoinositides. Nonetheless, PH1 contains a prominent positively charged cavity corresponding to the canonical PIP-binding pocket (Figure 3C), suggesting that the domain may interact nonspecifically with the negatively charged membrane surface.
Figure 3E shows a surface charge potential representation of the PH1N-PH2-PH1C tandem. The structural rigidity of the PH1N-PH2-PH1C supramodule holds the lipid-binding pockets on the same face of the tandem. This allows the two PH domains to bind synergistically to planar phosphatidylinositol-containing membrane bilayers. In other words, the relative orientations of the two PH domains are fixed in a conformation in which PH2 can recognize and bind to PI(3,4,5)P3 while PH1 nonspecifically binds to negatively charged lipid head groups. This interaction model is highly consistent with the cooperative, PI(3,4,5)P3-dependent interaction between the PH1N-PH2-PH1C tandem and phosphatidylinositol membranes shown in Figure 1, D–F.
We directly tested the roles of the two PH domains in the PH1N-PH2-PH1C tandem in binding to liposomes containing 3% PI(3,4,5)P3. We substituted the positively charged residues at the second and fourth positions of the β2 strand of each PH domain (i.e., the last two positively charged residues in the K-Xn-(K/R)-X-R signature motif) and tested the lipid membrane–binding capacities of the mutant tandems. Substitutions of Lys-1195 and Arg-1197 of PH1 with Ala (referred to as PH12_1M) mildly decreased the lipid membrane–binding capacity of the PH1N-PH2-PH1C tandem (Figure 3D), consistent with the predicted nonspecific avidity-enhancing role of the split PH1 in lipid membrane binding. In contrast, replacements of Lys-1237 and Arg-1239 of PH2 with Ala (denoted PH12_2M) essentially eliminated the lipid membrane–binding capacity of the PH1N-PH2-PH1C tandem (Figure 3D), supporting our structure-based prediction that PH2 is chiefly responsible for specific binding to PI(3,4,5)P3-containing membranes. The direct and specific binding of PI(3,4,5)P3 to the PH2 domain in the PH1N-PH2-PH1C tandem was further probed using an isothermal titration calorimetry (ITC)–based binding assay. We found that a water-soluble head group of phosphoinositide, Ins(1,3,4,5)P4, binds to PH1N-PH2-PH1C with a KD of ∼32.9 ± 4.2 μM (Figure 3G). No binding could be detected between Ins(1,4,5)P3 and PH1N-PH2-PH1C (Figure 3G). In addition, we could not detect any binding between the Lys1237Ala/Arg1239Ala mutant of PH1N-PH2-PH1C and Ins(1,3,4,5)P4 (unpublished data).
The PH domains of MyoX are localized to PI(3,4,5)P3-enriched basolateral membranes in polarized MDCK cells
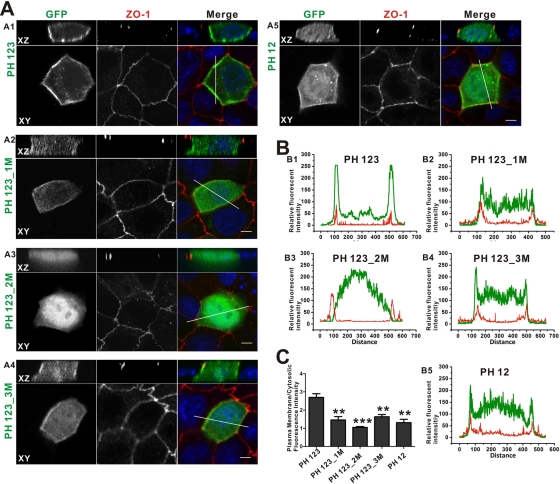
We used polarized MDCK cells as a model to assess the lipid membrane–binding properties of the MyoX PH domains under cellular conditions. It is known that PI(3,4,5)P3 and PI(4,5)P2 are restricted to the basolateral and apical membranes, respectively, of polarized MDCK cells (Gassama-Diagne et al., 2006). Confocal images of the MyoX PH repeats show that the wild-type PH123 is selectively targeted to basolateral membranes with no detectable apical membrane localization (Figure 4, A1 and B1). A PH123 mutant with Lys-1237 and Arg-1239 in PH2 substituted with Ala (PH123_2M) is almost completely diffused in cytosol (Figure 4, A3, B3, and C), indicating that the MyoX PH repeats selectively bind to PI(3,4,5)P3-containing phosphatidylinositol membranes, and PH2 in the PH123 repeats plays an essential role for the basolateral membrane localization in MDCK cells. Substitutions of Lys-1195 and Arg-1197 in the split PH1 domain with Ala weaken the membrane localization of PH123, and a significant proportion of the mutants are diffused in cytoplasm (Figure 4, A2, B2, and C). It is noted that, compared with PH123_2M, the PH123_1M mutant is still partially enriched at the basolateral membranes, consistent with the idea that the split PH1 plays an accessory role in the lipid membrane binding of the MyoX PH1N-PH2-PH1C tandem. We also tested the role of potential lipid-binding properties in PH3 in mediating basolateral localization of MyoX PH123 by substituting the two positively charged residues in the predicted second and fourth residues of the domain's β2 strand (K1417A and R1419A-, denoted PH123_3M). The PH123_3M mutant is also partially enriched at the basolateral membranes (Figure 4, A4, B4, and C), suggesting that the PH3 domain plays at most a minor role in the lipid membrane binding of MyoX. Because the MyoX PH1N-PH2-PH1C tandem is more diffused in polarized MDCK cells than the PH123_3M mutant (Figure 4, A5), we reasoned that the PH3 domain might facilitate basal lateral membrane binding via a lipid-unrelated mechanism (e.g., possibly by binding to a protein target). Consistent with this hypothesis, a recent study showed that MyoX PH3 does not bind to PIP lipids (Cox et al., 2002; Plantard et al., 2010).
FIGURE 4:
The MyoX PH repeats target to PI(3,4,5)P3-enriched basolateral membranes in polarized MDCK cells. (A) Confocal image of polarized MDCK cells transfected with GFP-tagged PH123 (A1), PH123_1M (A2), PH123_2M (A3), PH123_3M (A4), and PH12 (A5). An XZ section is shown at the top of each column, and a single optical XY section of the corresponding image is shown below the XZ section. Anti–ZO-1 antibody (red) and 4′,6-diamidino-2-phenylindole (DAPI; blue) are used to stain tight junctions and nuclei of polarized MDCK cells, respectively. Scale bar, 5 μm. (B) Fluorescence-intensity profiles represent the area marked by the white lines in A. Green curve represents the fluorescence intensity of GFP-tagged MyoX PH domains: PH123 (B1), PH123_1M (B2), PH123_2M (B3), PH123_3M (B4), and PH12 (B5). Red curve represents the fluorescence intensity of ZO-1 used to mark membranes of MDCK cells. (C) Quantitative analysis of membrane and cytosolic distributions of GFP-tagged MyoX PH repeats and its mutants. Values are means ± SE from three independent experiments (20 cells per time), analyzed with unpaired t test; **p < 0.01, ***p < 0.001.
We also tested the membrane localization properties of the PH123-MyTH4-FERM fragment of MyoX (i.e., the entire cargo-binding tail of the motor) using MDCK cells. Like the PH123 repeats of the motor, the wild-type PH123-MyTH4-FERM is membrane enriched (Supplemental Figure S3A). Disruption of the lipid-binding capacity of PH2 or the split PH1 eliminates or weakens the membrane localization of PH123-MyTH4-FERM, respectively (Supplemental Figure S3, B and C), indicating that PH repeat–mediated lipid membrane binding is essential for proper membrane localization of the motor.
PH repeat–mediated membrane binding is essential for MyoX to induce filopodial formation
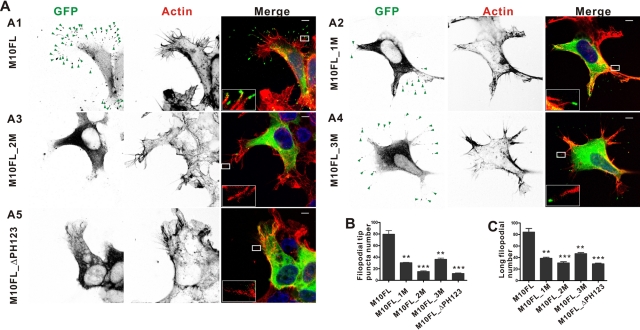
MyoX is known to induce filopodia formation, and both endogenous and overexpressed MyoX localize at the tips of filopodia (Berg and Cheney, 2002; Bohil et al., 2006; Tokuo et al., 2007; Plantard et al., 2010). We tested the role of PH repeat–mediated PI(3,4,5)P3-membrane binding on MyoX's function in filopodial formation. Consistent with reported findings (Berg and Cheney, 2002; Bohil et al., 2006; Tokuo et al., 2007), HEK293T cells expressing the wild-type. full-length MyoX display a large number of green fluorescent protein (GFP) puncta at filopodial tips and massive numbers of substrate-attached elongated filopodia (Figure 5, A1, B1, and C). In line with a recent report, removal of all three PH repeats eliminated the filopodia tip localization and filopodial induction capabilities of MyoX (Plantard et al., 2010). Of importance, disruption of the PH2-mediated PI(3,4,5)P3 binding of MyoX (i.e., the Lys1237Ala/Arg1239Ala mutant of the full-length MyoX) also eliminated the filopodia tip localization and filopodial-inducing activities of MyoX (Figure 5, A3, B3, and C), indicating that PH2-mediated binding to PI(3,4,5)P3-containing membranes is absolutely required for the cellular functions of the motor. Consistent with our in vitro lipid membrane–binding experiments, the lipid binding–deficient mutations in the split PH1 also have significant effects on the filopodial-inducing activities of MyoX (Figure 5, A2, B2, and C), although the impact of the mutations is not as large as that caused by the mutations in PH2. The M10FL_3M mutant has an even smaller effect on the filopodia induction activity of the motor (Figure 5, A4, B4, and C). We repeated the MyoX-induced filopodia formation assay of the wild-type and various mutants of the full-length MyoX shown in Figure 5 in COS7 cells. Strikingly similar filopodia induction patterns were found for these MyoX constructs in COS7 cells (Supplemental Figure S4) and in HEK293T cells (Figure 5). In an earlier study, it was shown that mutation of one amino acid residue in the predicted lipid binding pocket of PH1 (K1179A, equivalent of K1187 in our study, which is the last instead of the penultimate residue in the β1 strand of PH1) or PH3 (K1395A, equivalent of K1403 in our study, which is the penultimate residue in the β1 strand of PH3) had no detectable impact on cellular localization of MyoX (i.e., both MyoX mutants are localized at filopodia tips) (Plantard et al., 2010). In contrast, a large portion of cytoplasm localized M10FL_1M or M10FL_3M was detected in the current study (Figure 5, A2 and A4). A likely explanation for this discrepancy is that the PH1 and PH3 mutations used in our study (K1195A/R1197A in PH1 and K1417A/R1419A in PH3) are expected to completely disrupt their lipid-binding capacities. The single point mutation in PH1 used in the Plantard et al. (2010) study is the last amino acid residue of the β1 strand, and this residue is known not to be critical for lipid binding of PH domains (Lemmon, 2008). The single point mutation of the penultimate residue in the β1 strand of PH3 used by Plantard et al. (2010) is likely to have a smaller effect than the double mutation used in our study. Therefore the PH1 and PH3 mutants of MyoX described by Plantard et al. (2010) are not likely to be significantly different from the wild-type MyoX. It is also possible that different experimental conditions between the two studies (e.g., different protein expression levels) might account for the observed differences. To minimize such variations possibly introduced by experimental conditions, we chose to compare both total amount of filopodia and the amount of elongated filopodia induced by different forms of MyoX expressed at a comparable level in our study.
FIGURE 5:
PH domains are essential for MyoX's filopodial induction activity. (A) Confocal images of HEK293T cells transfected with GFP-tagged M10FL (A1), M10FL _1M (A2), M10FL _2M (A3), M10FL _3M (A4), and M10FL_ΔPH123 (A5). Inverted contrast images are shown for GFP-tagged MyoX and actin filaments. Green triangles point to MyoX puncta localized at the tip of filopodia. Right, the distribution pattern of GFP-tagged MyoX. Actin filaments are stained with phalloidin (red) and nuclei are stained with DAPI (blue). Scale bar, 5 μm. (B, C) Quantitative analysis of GFP puncta (B) and long filopodia (C) (filopodial length, >0.5 μm) numbers per transfected cell. Values are means ± SE from three independent experiments (by counting 20 cells per experiment), analyzed with unpaired t test; **p < 0.01, ***p < 0.001.
DISCUSSION
Split PH domains constitute a unique subfamily among the hundreds of PH domains in eukaryotic genomes (Lemmon, 2008). With the determination of the MyoX split PH domain structure in this study, the atomic structures of all six known split PH domains have been solved (i.e., those in phospholipase Cγ, syntrophins, Rho kinases, the Vps36 subunit of the yeast ESCRT-II protein sorting complex, the neuronal GTPase PI3 kinase enhancer [PIKE], and MyoX; Yan et al., 2005, 2008; Alam et al., 2006; Hirano et al., 2006; Teo et al., 2006; Wen et al., 2006, 2008). Several features are common to all split PH domains. First, PH domain splitting invariably occurs at the flexible loops connecting two consecutive β-strands of the PH domain β-barrel. As a result, the insertion of one or more additional domains does not alter the overall fold of the split PH domains (Supplemental Figure S5). Second, the splitting site varies in different split PH domains. The PH domains of PLCγ, syntrophin, and MyoX are all split into two halves at their respective β3/β4 loops. The two zinc fingers of Vps36 and the Cys-rich C1 domain of Rho kinase insert into the β6/β7 loop of their PH domains, respectively. The nuclear localization sequence of PIKE is located at the β5/β6 loop of the PH domain (Supplemental Figure S5). Third, although different split PH domains contain different insertions, split PH domains invariably function together with their insertions to form multidomain supramodules with functions distinct from the simple sum of the functions of the individual domains. For example, in PLCγ, the split PH domain and the inserted SH2-SH2-SH3 domains play an autoinhibitory role in regulating the lipase activity of PLCγ (DeBell et al., 2007). The split PH domain of Vps36 and its two inserted zinc fingers function together to tether the ESCRT-II complex to the ESCRT-I complex via zinc finger 1 and to endosome membranes via the split PH/PI(3)P interaction and the binding of zinc finger 2 to ubiquitinated cargos on the endosome membranes (Alam et al., 2006; Hirano et al., 2006; Teo et al., 2006). The split PH domains of syntrophins, PIKE, and Rho kinase function with their insertions sequences to enhance lipid membrane–binding avidities (Yan et al., 2005, 2008; Wen et al., 2008). In addition, it should be noted that the split PH domain family does not represent the only case in which intact domains are split into halves by the insertion of other domains. Statistical analysis shows that ∼9% of nonredundant proteins structures deposited at the Protein Data Bank (www.rcsb.org/pdb) contain domain insertions (Selvam and Sasidharan, 2004). We believe that domain splitting is likely to be a general strategy for multiple protein domains to assemble into functional supramodules with functions distinct from the individual or simple sum of individual domains.
The split PH domain of MyoX is unique in two ways. First, its PH1 domain is split by another PH domain (PH2), forming a PH1N-PH2-PH1C tandem. Second, the two PH domains in the PH1N-PH2-PH1C tandem interact with each other to form a structurally rigid supramodule. The relative orientation of the two lipid-binding pockets allows the MyoX PH repeats to bind to PI(3,4,5)P3-containing membranes with high specificity and cooperativity (Figure 1). We demonstrate in this study that both PH1 and PH2 are required for the proper cellular localization as well as filopodial induction activities of MyoX, although PH2-mediated selective binding to PI(3,4,5)P3 plays a more prominent role in both processes (Figures 4 and 5).
The cooperative nature of the PH1N-PH2-PH1C/PI(3,4,5)P3 interaction allows the supramodule to respond sharply to relatively small changes in PI(3,4,5)P3 concentration at a specific threshold, making it an ideal sensor for PI3 kinase activity. This is reflected in the sigmoidal shape of the PH1N-PH2-PH1C/PI(3,4,5)P3 binding curve; the slope is steepest at ∼3.0 ± 0.5% PI(3,4,5)P3, meaning that the bound fraction of PH1N-PH2-PH1C is most sensitive to concentration changes about this point (Figure 1D). In contrast, PI(3,4,5)P3-specific PHs domains that possess high binding affinities but lack cooperativity are unable to respond as sharply to concentration changes at a specific threshold due to the hyperbolic shape of the general, noncooperative ligand-binding curve.
It is likely that the cooperative PI(3,4,5)P3 binding of its PH repeats plays a critical role in determining the localization of MyoX at the tips of filopodia (Sousa and Cheney, 2005). We envision that the activation of PI3 kinase at the tips of filopodia leads to local and sustained elevation of PI(3,4,5)P3, consequently inducing PH repeat–mediated MyoX enrichment at filopodial tips (Di Paolo and De Camilli, 2006; Kolsch et al., 2008; Lemmon, 2008). Decreases in PI(3,4,5)P3 concentration at membranes distal from the filopodial tips, by simple diffusion-mediated dilution and/or by metabolic turnover, would lead to sharp decreases in MyoX association to these plasma membranes owing to the sigmoidal PI(3,4,5)P3 membrane-binding curve of the MyoX PH repeats. It is possible that the PH repeats and the MyTH4-FERM tandem of the MyoX tail might function synergistically in interpreting PI3 kinase activation signals and signals for other transmembrane receptors (e.g., netrin receptor DCC, integrin) in regulating the cellular localization and activities of the motor (Zhang et al., 2004; Zhu et al., 2007; Hirano et al., 2011; Plantard et al., 2010; Wei et al., 2011). A recent study showed that binding of PI(3,4,5)P3 to the PH domains directly regulates the motor activity by releasing the PH repeat–mediated autoinhibition of MyoX (Umeki et al., 2011). Elucidating the detailed molecular mechanism governing the interaction between the PH repeats and the motor head of MyoX will certainly be highly valuable in understanding this unusual PH repeat–mediated motor activity regulation.
MATERIALS AND METHODS
Constructs and protein expression
The coding sequence of MyoX PH1N-PH2-PH1C (XP_226874, residues 1170–1317) and PH123 (XP_226874, residues 1170–1500) were PCR amplified from rat Myo10 and cloned into a pET32a vector. The GFP-tagged full-length MyoX expression plasmid (NP_036466.2, 1–2058), was a gift from Wencheng Xiong. Point mutations, including PH12_1M (K1195A/R1197A), PH12_2M (K1237A/R1239A), PH12_3M (K1417A/R1419A), PH12_3KN (K1187N/K1224N/K1225N), PH12_D1254R, PH123_1M (K1195A/R1197A), PH123_2M (K1237A/R1239A), PH123_3M (K1417A/R1419A), FL_1M (K1193A/R1195A), FL_2M (K1235A/R1237A), and FL_3M (K1415A/R1417A), were created by PCR-based mutagenesis. For truncation mutant FL_ΔPH123, the coding sequence of PH123 (NP_036466.2, residues 1168x1498) was deleted from the full-length MyoX. Protein was expressed in BL21(DE3) Escherichia coli cells. The His6-tagged proteins were purified using Ni2+ nitrilotriacetic acid–agarose column followed by a size-exclusion chromatography.
Lipid-binding assays
Defined liposomes were reconstituted from synthetic PC and PS (Avanti Polar Lipids, Alabaster, AL) with or without certain concentrations of PIPs (Echelon Biosciences, Salt Lake City, UT). Lipids dissolved in chloroform were mixed in a glass tube at an appropriate ratio, and the solvent was evaporated under a stream of N2 gas at 4°C. An appropriate amount of buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 100 mM NaCl, 1 mM dithiothreitol [DTT]) was added to bring the final lipid concentration to 5 mg/ml. The lipid solution was rigorously vortexed for 5 min, and then the mixture was hydrated by 10 cycles of freeze and thaw with liquid N2. Brain lipid extracts (Folch fraction I, B1502; Sigma-Aldrich, St. Louis, MO) were resuspended in a buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, and 1 mM DTT. The protein sample (5 μM final concentration) was incubated with liposomes in 50 μl of buffer for 15 min at room temperature and then spun at 80,000 × g for 60 min at 4°C in a Beckman TLA100.1 rotor. The supernatants were removed for determination of proteins not bound to liposomes. The pellets were washed twice with the same buffer and brought up to the same volume as the supernatant. The supernatant and the pellet proteins were subjected to SDS–PAGE and visualized by Coomassie blue staining.
Crystallization, data collection and process
Crystals of PH12_3KN mutant (10.0 mg/ml in 50 mM sodium phosphate buffer, 100 mM NaCl, pH 7.0) were grown using the hanging-drop method by mixing 1 μl of protein sample with equal volume of 20% (wt/vol) PEG8000 and 0.05 M potassium phosphate monobasic at 16°C. Crystals were transferred to the reservoir solution containing 15% (vol/vol) glycerol as the cryoprotectant and flash cooled with liquid nitrogen. Diffraction data were collected at 110 K on a RAXIS IV++ imaging-plate system with a MicroMax-007 copper rotating-anode generator (Rigaku, The Woodlands, TX). The diffraction data were processed and scaled using MOSFLM and SCALA in the CCP4 suite (CCP4, 1994).
Structure determination and refinement
The PH12_3KN mutant structure was solved by the molecular replacement method with the program Phaser (McCoy et al., 2007) using the structure of DAPP1 PH domain (Protein Data Bank code 1FAO) as the search model. Structures were fitted and rebuilt with the program COOT (Emsley and Cowtan, 2004) and refined with REFMAC5 (Vagin et al., 2004) and CNS (Brunger et al., 1998). The overall qualities of the structural models were assessed using PROCHECK (Laskowski et al., 1993). The coordinates of PH12_3KN were deposited in the Protein Data Bank (www.rcsb.org/pdb) under the access code of 3TFM. Data collection and refinement statistics of the PH12_3KN structures are summarized in Table 1.
Cellular localization and filopodia quantification
HEK293T cells were transiently transfected with 0.5 μg of each plasmid per well using a Lipofectamine PLUS Kit (Invitrogen, Carlsbad, CA) in 12-well plates, and cells were cultured 24 h in DMEM containing 10% fetal bovine serum (FBS) in 10% CO2 before fixation. MDCK cells were transfected using a nucleotransfection kit (Amaxa, Köln, Germany) by electroporation, and cells were cultured 48 h in MEM containing 10% FBS in 10% CO2 before fixation. Cells were imaged with an LSM 510 META laser scanning confocal microscope (Zeiss, Wetzlar, Germany) or a TE2000E inverted fluorescent microscope (Nikon, Melville, NY). Images from the Nikon microscope were used for quantification. Anti–ZO-1 antibody was used as the marker for tight junctions. Fluorescence intensities of plasma membrane and cytosolic region were quantitatively measured by MetaMorph image processing software (Molecular Devices, Sunnyvale, CA). For HEK293T cells, actin was stained by rhodamine-conjugated phalloidin (Invitrogen) to visualize filopodia. The numbers of filopodia (crossing the cell edge, with length >0.5 μm) and GFP puncta were tracked and quantified by MetaMorph. Data were analyzed using a Student's t test. At least 20 cells were quantified for each construct, and values (means ± SD) were calculated from three independent experiments.
Supplementary Material
Acknowledgments
We thank Richard Cheney and Wencheng Xiong for providing the myosin X cDNA construct, Ling-Nga Chan for helping in cell biology experiments, Yanxiang Zhao for the x-ray machine time, and Anthony Zhang for editing the manuscript. This work was supported by grants from the Research Grants Council of Hong Kong to M.Z. (663808, 664009, 660709, 663610, HKUST6/CRF/10, SEG_HKUST06, and AoE/B-15/01-II) and to Z.W. (662710).
Abbreviations used:
- BMP
bone morphogenetic protein
- DCC
deleted in colorectal cancer
- FERM
band 4.1/ezrin/radixin/moesin
- HMM
heavy meromyosin
- MyoX
myosin X
- MyTH4
myosin tail homology
- PC
phosphatidylcholine
- PS
phosphatidylserine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0354) on September 30, 2011.
REFERENCES
- Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol. 2006;13:1029–1030. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cox D, Berg JS, Cammer M, Chinegwundoh JO, Dale BM, Cheney RE, Greenberg S. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat Cell Biol. 2002;4:469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- DeBell K, Graham L, Reischl I, Serrano C, Bonvini E, Rellahan B. Intramolecular regulation of phospholipase c-gamma1 by its C-terminal Src homology 2 domain. Mol Cell Biol. 2007;27:854–863. doi: 10.1128/MCB.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Gassama-Diagne A, Yu W, ter Beest M, Martin-Belmonte F, Kierbel A, Engel J, Mostov K. Phosphatidylinositol-3,4,5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat Cell Biol. 2006;8:963–970. doi: 10.1038/ncb1461. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci STKE. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, Kato R, Stenmark H, Wakatsuki S. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol. 2006;13:1031–1032. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hatano T, Takahashi A, Toriyama M, Inagaki N, Hakoshima T. Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain. EMBO J. 2011;30:2734–2747. doi: 10.1038/emboj.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YS, Luo T, Xu Y, Sargent TD. Myosin-X is required for cranial neural crest cell migration in Xenopus laevis. Dev Dyn. 2009;238:2522–2529. doi: 10.1002/dvdy.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ML, Jacobs DT, Campagnola L, Dunn BD, Yin T, Sousa AD, Quintero OA, Cheney RE. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketschek A, Gallo G. Nerve growth factor induces axonal filopodia through localized microdomains of phosphoinositide 3-kinase activity that drive the formation of cytoskeletal precursors to filopodia. J Neurosci. 2010;30:12185–12197. doi: 10.1523/JNEUROSCI.1740-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Luikart BW, Zhang W, Wayman GA, Kwon C-H, Westbrook GL, Parada LF. Neurotrophin-dependent dendritic filopodial motility: a convergence on PI3K signaling. J Neurosci. 2008;28:7006–7012. doi: 10.1523/JNEUROSCI.0195-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, Rock RS. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Kee Y, Bronner-Fraser M. Myosin-X is critical for migratory ability of Xenopus cranial neural crest cells. Dev Biol. 2009;335:132–142. doi: 10.1016/j.ydbio.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Pi X, Ren R, Kelley R, Zhang C, Moser M, Bohil AB, Divito M, Cheney RE, Patterson C. Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J Cell Biol. 2007;179:1569–1582. doi: 10.1083/jcb.200704010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantard L, Arjonen A, Lock JG, Nurani G, Ivaska J, Stromblad S. PtdIns(3,4,5)P3 is a regulator of myosin-X localization and filopodia formation. J Cell Sci. 2010;123:3525–3534. doi: 10.1242/jcs.069609. [DOI] [PubMed] [Google Scholar]
- Selvam RA, Sasidharan R. DomIns: a Web resource for domain insertions in known protein structures. Nucleic Acids Res. 2004;32:D193–D195. doi: 10.1093/nar/gkh047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AD, Cheney RE. Myosin-X: a molecular motor at the cell's fingertips. Trends Cell Biol. 2005;15:533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Tokuo H, Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem Biophys Res Commun. 2004;319:214–220. doi: 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- Tokuo H, Mabuchi K, Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J Cell Biol. 2007;179:229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeki N, Jung HS, Sakai T, Sato O, Ikebe R, Ikebe M. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18:783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Watanabe TM, Tokuo H, Gonda K, Higuchi H, Ikebe M. Myosin-X induces filopodia by multiple elongation mechanism. J Biol Chem. 2010;285:19605–19614. doi: 10.1074/jbc.M109.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yan J, Lu Q, Pan L, Zhang M. Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc Natl Acad Sci USA. 2011;108:3572–3577. doi: 10.1073/pnas.1016567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Liu W, Yan J, Zhang M. Structure basis and unconventional lipid membrane binding properties of the PH-C1 tandem of Rho kinases. J Biol Chem. 2008;283:26263–26273. doi: 10.1074/jbc.M803417200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Yan J, Zhang M. Structural characterization of the split pleckstrin homology domain in phospholipase C-gamma1 and its interaction with TRPC3. J Biol Chem. 2006;281:12060–12068. doi: 10.1074/jbc.M600336200. [DOI] [PubMed] [Google Scholar]
- Yan J, Wen W, Chan LN, Zhang M. Split pleckstrin homology domain-mediated cytoplasmic-nuclear localization of PI3-kinase enhancer GTPase. J Mol Biol. 2008;378:425–435. doi: 10.1016/j.jmb.2008.02.052. [DOI] [PubMed] [Google Scholar]
- Yan J, Wen W, Xu W, Long JF, Adams ME, Froehner SC, Zhang M. Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin. EMBO J. 2005;24:3985–3995. doi: 10.1038/sj.emboj.7600858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Berg JS, Li Z, Wang Y, Lang P, Sousa AD, Bhaskar A, Cheney RE, Stromblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- Zhu XJ, Wang CZ, Dai PG, Xie Y, Song NN, Liu Y, Du QS, Mei L, Ding YQ, Xiong WC. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat Cell Biol. 2007;9:184–192. doi: 10.1038/ncb1535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.