PTTG1 is associated with the cis face of the Golgi apparatus and the centrosome, forming a complex with proteins involved in microtubule nucleation. PTTG1 depletion produces a delay in centrosomal and noncentrosomal microtubule nucleation and causes defects in both cell polarization and migration.
Abstract
Pituitary tumor transforming gene 1 (PTTG1), also known as securin, has been implicated in many biological functions, including inhibition of sister chromatid separation, DNA repair, organ development, and regulation of the expression and secretion of angiogenic and metastatic factors. Although most of these functions of securin seem to depend on the localization of PTTG1 in the nucleus of the cell, a fraction of the protein has been also detected in the cytoplasm. Here we demonstrate that, in different cell types, a portion of cytoplasmic PTTG1 is associated with the cis face of the Golgi apparatus and that this localization depends on PTTG1 phosphorylation status. In this organelle, PTTG1 forms a complex with proteins involved in microtubule nucleation, including GM130, AKAP450, and γ-tubulin. RNA interference–mediated depletion of PTTG1 produces a delay in centrosomal and noncentrosomal microtubule nucleation. Cells lacking PTTG1 show severe defects in both cell polarization and migration in wound-healing assays. To our knowledge, this is the first study reporting the role of PTTG1 in microtubule nucleation and cell polarization, two processes directly involved in cell migration. We believe that these findings will contribute to understanding the mechanisms underlying PTTG1-mediated biological functions.
INTRODUCTION
Pituitary tumor-transforming gene 1 (PTTG1), originally isolated from rat pituitary tumor cells, was subsequently identified as a member of the securin family proteins (Zou et al., 1999). PTTG1 is a multifunctional protein with roles in the control of mitosis, cell transformation, DNA repair, and gene regulation (Pei and Melmed, 1997; Ramos Morales et al., 2000; Tong et al., 2007; Bernal et al., 2008). PTTG1 is expressed at high levels in multiple tumors of different etiologies (Saez et al., 1999, 2006; Heaney et al., 2000; Shibata et al., 2002). The mechanisms by which PTTG1 promotes tumorigenesis are unclear. The fact that PTTG1 regulates sister chromatid separation during cell division suggests that anomalous levels of PTTG1 may lead to aneuploidy by defective sister chromatid separation, resulting in tumor development (Lengauer et al., 1997; Jallepalli et al., 2001). However, other, nonexcluding mechanisms might participate in PTTG1 tumorigenic function. PTTG1 binds p53 and modulates its transcriptional activity. This interaction between PTTG1 and p53 could lead directly to the accumulation of DNA damage and subsequent development of malignant tumors (Bernal et al., 2002). PTTG1 was also shown to possess transactivation ability, which correlated with its transforming properties. Well-established targets of PTTG1 are the oncogenic gene c-myc and the mitogenic and angiogenic factors basic fibroblast growth factor and vascular endothelial growth factor, which sustain tumor growth and contribute to the tumorigenic microenvironment (Vlotides et al., 2007). These physiological roles of PTTG1 correlate with its nuclear localization into the cell; nevertheless, PTTG1 has been also localized in the cytoplasm, but there is little information concerning the role of the cytoplasmic PTTG1 fraction. It has been reported that some cytoplasmic PTTG1 localizes in the Golgi apparatus (GA) in human pituitary adenomas (Minematsu et al., 2007). However, whether this localization of PTTG1 is also observed in other cells and the potential role of PTTG1 in the GA remain to be investigated.
In mammalian cells, the GA is an organelle in which individual stacks of flattened cisternae are joined laterally by tubular connections to form a single compact reticulum localized in close proximity to the centrosome and adjacent to the nucleus (Sutterlin and Colanzi, 2010). Several biological functions are associated with the GA being the central organelle along the secretory pathway. The GA is involved in other physiological processes, such as cell signaling and differentiation, cell polarity, migration, control of cell division, and microtubule (MT) nucleation (Chabin-Brion et al., 2001; Colanzi and Corda, 2007; Efimov et al., 2007; Vinogradova et al., 2009). Furthermore, the centrosome is the major microtubule-organizing center (MTOC) in proliferating and migrating culture animal cells, leading to the formation of radial MT arrays in which MT minus ends are anchored at the centrosomes and plus ends extend to the cell periphery. It has been shown that a number of cytoplasmic MTs originate from the GA and that Golgi-associated MT nucleation depends on γ-tubulin and cytoplasmic linker-associated proteins (CLASPs; Chabin-Brion et al., 2001; Efimov et al., 2007). More recently, MT nucleation at the GA was shown to require AKAP450, a centrosomal γ-TuRC–interacting protein that also forms a distinct network associated with the Golgi (Rivero et al., 2009). A role for Golgi-nucleated MTs in cell polarization and motility has been proposed (Efimov et al., 2007). In fact, depletion of either CLASPs or AKAP450 proteins abolished MT formation from the Golgi, and these cells were defective in cell migration (Rivero et al., 2009). Moreover, it has been reported that PTTG1 overexpression promotes cell motility and lymph node metastasis and PTTG1 down-regulation significantly reduces cell motility (Ito et al., 2008).
On the basis of these findings, we sought to clarify the subcellular localization of PTTG1 and its possible involvement in MT nucleation. In this work, we show that PTTG1 is associated with the cis side of the GA and is a component of the MT nucleating complexes containing AKAP450, GM130, and γ-Tub, which may influence MT nucleation and cell migration.
RESULTS
Subcellular localization of PTTG1
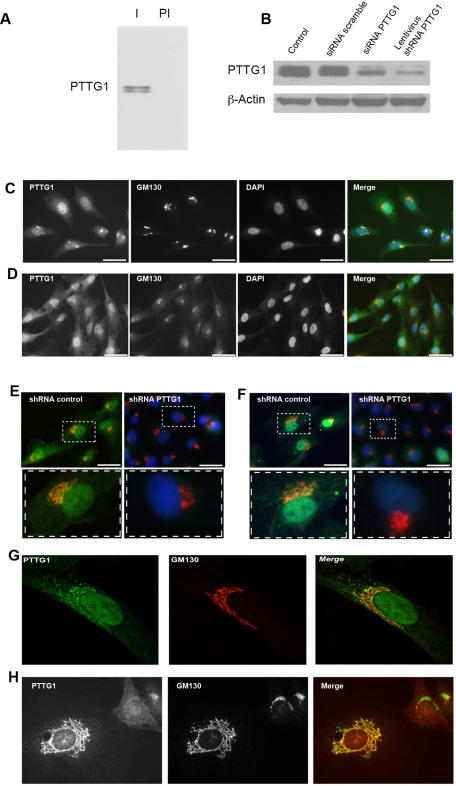
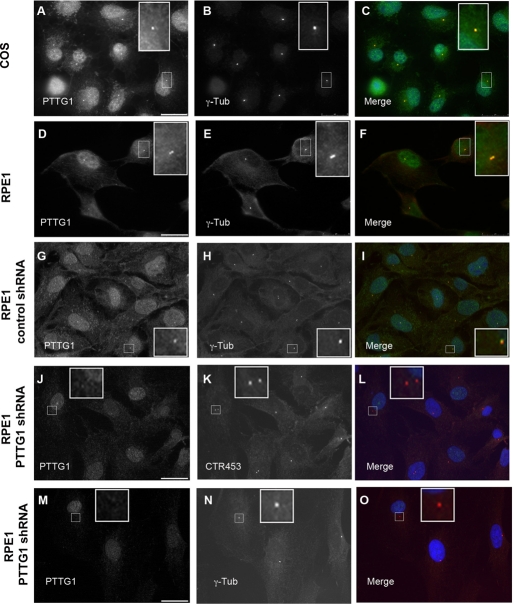
We generated a polyclonal antibody against a carboxy-terminal fragment of PTTG1 protein containing residues 108–202. The affinity-purified antibody recognized in Western blot a double band of ∼29 kDa corresponding to phosphorylated and unphosphorylated PTTG1 protein (Ramos Morales et al., 2000; Figure 1A). The specificity of this antibody was confirmed by immunoblotting using protein extracts from RPE1 cells transfected with scrambled small interfering RNA (siRNA) or knockdown PTTG1 siRNA or infected with Lentivirus expressing short hairpin PTTG1 RNA (Figure 1B). Immunostaining of HeLa and RPE1 cells with the affinity-purified antibody revealed that PTTG1 was associated with the GA and the nucleus. In addition, a dispersed signal in the cytoplasm was also observed (Figure 1, C, D, and G). The staining pattern given by the PTTG1 antibody could be diminished either by short hairpin RNA (shRNA)–mediated PTTG1 depletion (Figure 1, E and F) or by preincubation of the antibody with the recombinant protein used as antigen for the immunizations. Identical subcellular localization was observed in the tumor cell line Cos-7 and in the immortalized cell line NIH3T3 (Figure 1, F and H), confirming that remarkable Golgi localization is not cell-type dependent.
FIGURE 1:
PTTG1 protein is associated to the GA. (A) Cell extracts from RPE1 cells were analyzed by Western blot using purified anti-PTTG1 antibody (I) and preimmune serum (PI). (B) RPE1 cells were transfected with scrambled or PTTG1 siRNA duplexes or infected with Lentivirus expressing short hairpin PTTG1 RNA. Levels of PTTG1 protein were detected by Western blot analysis. β-Actin was used as loading control. (C) HeLa and (D) RPE1 cells were analyzed by immunofluorescence with antibodies to PTTG1 (green) and GM130 (red) and with the dye DAPI to visualize DNA (blue). (E) RPE1 and (F) Cos-7 cells were infected with Lentivirus (MOI = 10) expressing scrambled short hairpin (left) or PTTG1 shRNA (right). Cells were stained with antibodies to PTTG1 (green) and GM130 (red) and with the dye DAPI to visualize DNA (blue). Boxes in E and F are enlarged at the bottom of each figure. (G) RPE1 cells were stained with antibodies to PTTG1 (green) and GM130 (red). (H) NIH3T3 cells were stained with antibodies to PTTG1 (red) and GM130 (green).
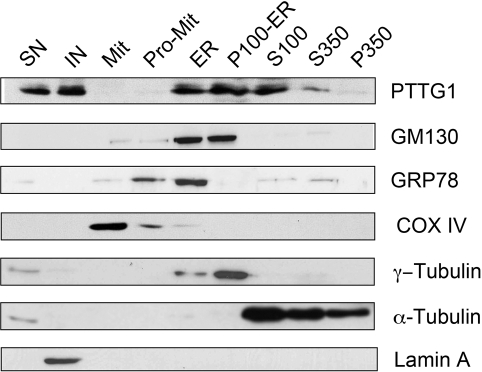
We explored in detail the distribution of PTTG1 protein in subcellular fractions in human promyelocytic HL60 cells. Purity of fractions was assayed by Western blotting using antibodies against specific proteins considered as markers of the different organelles. Enrichment of PTTG1 was detected in soluble and insoluble nuclear fractions and high-speed membrane fractions corresponding to endoplasmic reticulum (ER) and GA (P100-ER). Significant levels of PTTG1 were also found in the cytosolic fraction S100. However, it was virtually absent from mitochondria-enriched fractions (Figure 2).
FIGURE 2:
Detection of PTTG1 in subcellular fractions of HL60 cells. Purification procedures of subcellular fractions were performed as described in Materials and Methods. Proteins were separated by SDS–PAGE, transferred to nitrocellulose membrane, and then probed with anti-PTTG1 polyclonal antibody. To control the purity of the different fractions, the same filter was probed with the following specific antibodies: GM130 and GRP78 against Golgi apparatus and endoplasmic reticulum fractions, respectively, COX IV against mitochondria and protomitochondria fractions, γ-tubulin against centrosomal and Golgi apparatus fractions, α-tubulin against cytoplasmic fraction, and lamin A against nuclear fraction. ER, endoplasmic reticulum; IN, insoluble nuclear fraction; Mit, mitochondria; P100-ER, Golgi apparatus fraction; P350, pellet after 350,000 × g centrifugation; Pro-Mit, protomitochondria; S100 and S350, cytosolic fractions after 100,000 and 350,000 × g ultracentrifugation, respectively; SN, soluble nuclear fraction. Blot is representative of two different experiments.
PTTG1 is targeted to the cis-Golgi, and its localization is independent of microtubule integrity
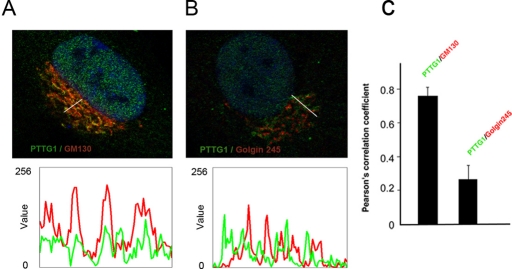
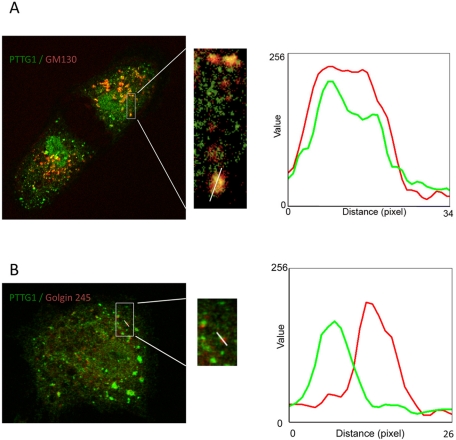
To precisely determine the distribution of PTTG1 network in the GA, we analyzed by confocal microscopy RPE1 cells costained with PTTG1 antibody and different Golgi compartment markers. As shown in Figure 3A, PTTG1 showed more extensive signal overlap with the cis-Golgi protein GM130 than with golgin-245 marker (Figure 3B). Fluorescence intensity profiles along lines drawn over the Golgi area showed the degree of correlation between the two staining patterns (Figure 3, A and B, bottom). Image correlation analysis, which is a pixel-to-pixel comparison based on the Pearson correlation coefficient, is shown in Figure 3C. Whereas PTTG1 showed a high degree of colocalization with GM130 (r = 0.7598) the correlation with golgin-245 (r = 0.2593) was significantly lower. These results indicate that PTTG1 is associated with the cis side of the GA. Nocodazole (NZ) treatment depolymerizes MTs, resulting in fragmentation of the pericentrosomal GA into small stacks that are dispersed through the cytosol. After NZ treatment, PTTG1 remains associated with Golgi ministacks, showing similar correlation with the markers mentioned (Figure 4), indicating that targeting of PTTG1 to the GA is independent of MT and GA integrity.
FIGURE 3:
PTTG1 is associated with the cis face of the GA. RPE1 cells were double labeled for (A) PTTG1 (green) and GM130 (red) and (B) PTTG1 (green) and golgin-245 (red) and analyzed by confocal microscopy. Merged images are shown. Bottom, fluorescence intensity profiles of lines drawn over Golgi membranes in images A and B. (C) Quantitative analysis of colocalization was based on Pearson's correlation coefficient. Values are means of five lines drawn at random positions in 20 different cells; each line contained 280 pixels on average.
FIGURE 4:
PTTG1 is associated with GA in the absence of microtubules. RPE1 cells were treated with 10 μM nocodazole for 2 h and labeled for (A) PTTG1 (green) and GM130 (red) or (B) PTTG1 (green) and golgin-245 (red). Merged images are shown. Enlarged images showing this association are demarcated in boxed areas to the right of each image. Fluorescence intensity profiles of lines drawn over ministacks into the boxed areas are shown at right.
Phosphorylation of PTTG1 by Cdk1 reduces its localization to Golgi membranes
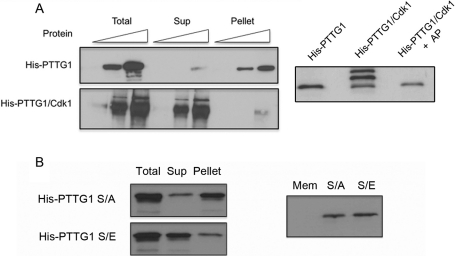
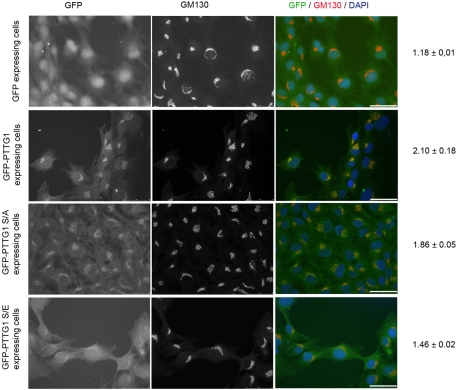
In previous work, we showed that PTTG1 is phosphorylated at the Ser-165 residue during mitosis by Cdk1 (Ramos Morales et al., 2000). The Golgi localization of PTTG1 and its phosphorylation by Cdk1 suggest that PTTG1 could be dissociated from Golgi ministacks in mitosis, as occurs in other GA proteins (Lowe et al., 1998; Litvak et al., 2004). To demonstrate this possibility, we isolated Golgi membranes from interphase HL60 cells and incubated them with increasing concentrations of histidine (His)-PTTG1 fusion protein. The recombinant His-PTTG1 fusion protein was detected by immunoblotting with anti-His antibody, and it was found mainly in the pellet fraction, associated with the Golgi membranes (Figure 5A, top). However, phosphorylation of His-PTTG1 by Cdk1 reduced notably its ability to bind the purified Golgi membranes, and most of the phosphorylated His-PTTG1 protein was found in the supernatant (Figure 5A, bottom). We next decided to test whether mitotic phosphorylation of PTTG1 would affect its interaction with Golgi membranes by using glutamate substitution, which most closely mimics the properties of phosphoserine. Site-directed mutagenesis was performed to generate phosphorylation-deficient (PTTG1S165A) and phosphomimetic (PTTG1S165E) mutants. Binding experiments showed that the binding capacity of PTTG1 to Golgi membranes was unaffected after substituting the serine with an alanine; however, the PTTG1S165E phosphomimetic mutant exhibited a decreased binding capacity to Golgi membranes (Figure 5B). In vivo experiments were carried out with RPE1 cells expressing wild-type PTTG1 and PTTG1 S/A and PTTG1 S/E mutants fused to green fluorescent protein (GFP) to determine their localization. The recruitment level was calculated as the ratio of fluorescence intensity between the GA and the cytoplasm. As Figure 6 shows, GFP-PTTG1 and GFP-PTTG1 S/A proteins were preferentially recruited to GA (ratio, 2.10 ± 0.18 and 1.86 ± 0.05, respectively). The GFP-PTTG1 S/E phosphomimetic protein was less accumulated in GA, and it was also present as a diffusely dispersed protein in the cytoplasm (ratio, 1.46 ± 0.02). Overall, these results suggest that in mitosis the most of the phosphorylated PTTG1 protein is absent from the Golgi ministacks and that its phosphorylation by Cdk1 could facilitate this dissociation.
FIGURE 5:
Binding of PTTG1 to Golgi membranes. (A) Phosphorylation of PTTG1 by Cdk1 kinase facilitates its dissociation from Golgi membranes. Isolated Golgi membranes (10 μg) were incubated with increasing concentration of purified His-PTTG1 or His-PTTG1 that was phosphorylated by Cdk1 in vitro. The association of the recombinant protein with the Golgi membranes (pellet) was determined by immunoblotting with anti-His antibody. Right, Western blot of in vitro phosphorylated PTTG1 by Cdk1 and dephosphorylated PTTG1 by alkaline phosphatase (AP). (B) Binding of PTTG1 mutants to Golgi membranes. Golgi membranes were incubated in vitro with the phosphorylation-deficient mutant His-PTTG1 S/A and with the phosphomimetic mutant His-PTTG1 S/E (700 ng). The association of the recombinant protein with the Golgi membranes (pellet) was determined by immunoblotting with anti-His antibody. Right, Western blot of membranes (10 μg) and mutant fusion proteins using anti-PTTG1 antibody.
FIGURE 6:
Replacement of serine 165 affects the recruitment of intact PTTG1 to the Golgi apparatus. RPE1 cells were transfected with GFP-PTTG1, GFP-PTTG1-S/A, and GFP-PTTG1-S/E. The cells were stained with anti-GM130 antibody and DAPI. Right, merged images. Relative GA recruitment (mean ± SD) of each construction is shown on the right. N = 35–50 cells scored per construction from two independent experiments. Scale bars, 50 μm.
To identify the region of PTTG1 involved in the association with the GA, we generated the glutathione S-transferase (GST)–fused truncated forms GST-PTTG1ΔC and GST-PTTG1ΔN, which lack the 82 C-terminal residues and 108 N-terminal residues, respectively. Incubation of Golgi membranes with these truncated forms showed that GST-PTTG1ΔC still associated with Golgi membranes (Supplemental Figure S1), indicating that the interaction was mediated through the N-terminal region of the PTTG1 protein.
PTTG1 associates with microtubule nucleation complexes, and its absence attenuates MT repolymerization after NZ challenge
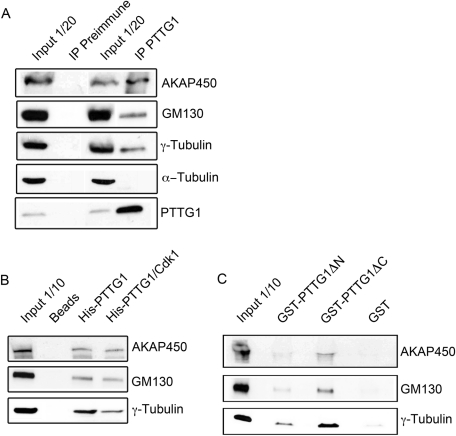
The colocalization of PTTG1 and GM130 observed by confocal microscopy led us to investigate whether PTTG1 interacts with this protein and/or with some of its binding partners. Endogenous GM130 was coimmunoprecipitated with endogenous PTTG1 from RPE1 cell extracts using the anti-PTTG1 antibody (Figure 7A). This interaction was also observed between exogenously expressed GM130 and PTTG1 (data not shown). AKAP450, a centrosomal and Golgi-associated γ−TURC-interacting protein that specifically binds the cis side of the Golgi in a GM130-dependent manner, also coimmunoprecipitated with PTTG1. AKAP450 indirectly associates with γ-tubulin through binding with γ-tubulin complex protein 2 (GCP2) and/or GCP3 and GM130. As expected, γ-tubulin was also coimmunoprecipitated with anti-PTTG1 antibody (Figure 7A). Whether these interactions are direct or not remains to be determined. We also tested whether PTTG1 could bind these proteins in a pull-down assay and whether these interactions were affected by phosphorylation of the Ser-165 residue. Whole-cell extracts from RPE1 cells were incubated with beads coated with phosphorylated and unphosphorylated His-tagged PTTG1. Both versions of PTTG1 bound to GM130, AKAP450, and γ-tubulin proteins, although these associations slightly decreased with the phosphorylated PTTG1, particularly in the case of the γ-tubulin interaction (Figure 7B). The interaction of these proteins with the GST-fused truncated forms GST-PTTG1ΔC and GST-PTTG1ΔN was also tested in a pull-down assay; as shown Figure 7C, AKAP450, GM130, and γ-tubulin associate preferentially with the N-terminal region of the PTTG1 protein.
FIGURE 7:
In vivo and in vitro interactions of PTTG1 with proteins involved in microtubule nucleation. (A) Coimmunoprecipitation of PTTG1, AKAP450, GM130, and γ-tubulin from RPE1 cells using anti-PTTG1 antibody. α-Tubulin was used as negative control. An amount of 1/20 of the input proteins used in the coimmunoprecipitation experiment was analyzed by Western blotting with antibodies against AKAP450, GM130, and γ-tubulin. IP, immunoprecipitates. (B) Interaction of AKAP450, GM130, and γ-tubulin with unphosphorylated PTTG1 and Cdk1-phosphorylated PTTG1. His-tagged fusion proteins were incubated with RPE1 extracts, and their association with AKAP450, GM130, and γ-tubulin was determined by immunoblotting. An amount of 1/10 of the input proteins used in the pull-down experiment was analyzed by Western blotting with antibodies against each protein. (C) Interaction of AKAP450, GM130, and γ-tubulin with GST-fused truncated forms of PTTG1. These experiments were performed as described in B.
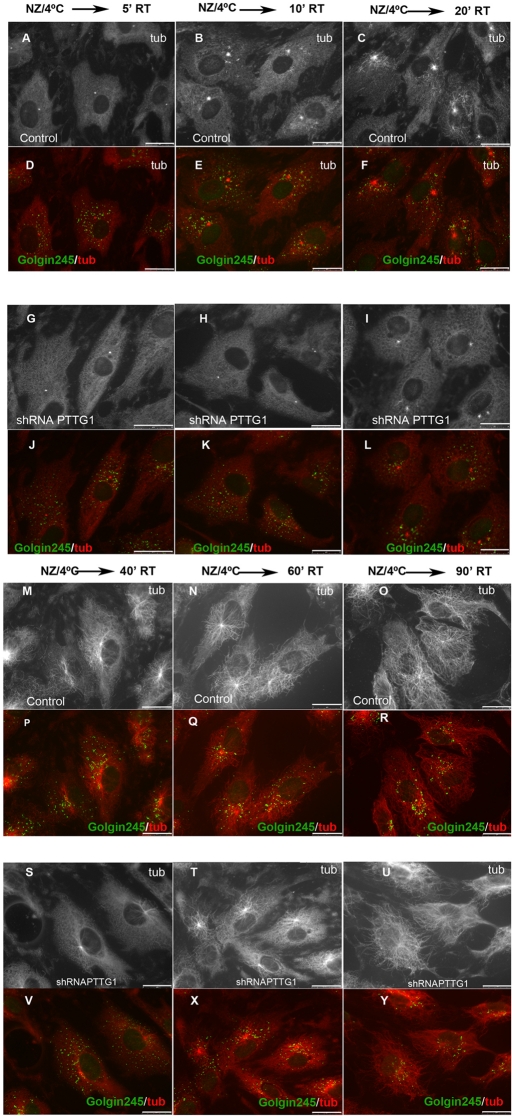
Taking into account that AKAP450 and γ-tubulin provide MT nucleation sites to the centrosome and that GM130, by anchoring AKAP450, participates in MT nucleation at the GA, we analyzed whether the absence of PTTG1 affected the MT nucleation process. To depolymerize acetylated and nonacetylated microtubules, PTTG1-depleted and control RPE1 cells were treated with 10 μM NZ for 2 h at 37°C and then incubated for 40 min at 4°C; the MT nucleation was analyzed at different times following NZ washout and incubation at 37°C. A notable difference was observed between control and PTTG1-depleted cells. In control cells, noncentrosomal MTs started to appear 10 min after NZ washout, becoming more evident after 20 min in fresh media (Figure 8, B, C, and M). However, in PTTG1-depleted cells, the nucleation of centrosomal and noncentrosomal MTs was delayed and the number of MTs was notably reduced. In fact, centrosomal and noncentrosomal MTs were absent in PTTG1-depleted cells 10 min after NZ washout and hardly evident 10 min later (Figure 8, H and I). Differences in nucleation were still observed 40 min after NZ washout (Figure 8, M and S), and they were reduced at longer times (60 min; Figure 8, N and T). Control and PTTG1-depleted cells showed a similar microtubule network 90 min after NZ washout (Figure 8, O and U).
FIGURE 8:
PTTG1 depletion interferes with microtubule repolymerization. RPE1 control (A–F, M–R) or PTTG1-depleted (G–L, S–Y) cells were treated with NZ (10 μM) for 2 h at 37°C and then incubated for 40 min at 4°C, fixed, and stained for α-tubulin (red) and golgin-245 (green) after 5 min (A, D, G, J), 10 min (B, E, H, K), 20 min (C, F, I, L), 40 min (M, P, S, V), 60 min (N, Q, T, X), or 90 min (O, R, U, Y) of NZ washout. Inverted images of tubulin staining are shown in A–C, G–I, M–O, and S–U. Merged images are shown in D–F, J–L, P–R, and V–Y. Scale bars, 25 μm.
The effect of PTTG1 on centrosomal MT nucleation, together with its interaction with γ-tubulin and AKAP450, suggests a role of PTTG1 at the centrosome. Actually, the intracellular location of GFP-PTTG1 fusion protein in the centrosome was reported in HCT116 cells (Tong et al., 2008); in addition, using our PTTG1 antibody, we localized endogenous PTTG1 in the centrosome in both Cos-7 and RPE1 cells (Figure 9). This intracellular location supports an effective interaction of PTTG1 with γ-tubulin and AKAP450 centrosomal proteins.
FIGURE 9:
Centrosomal localization of PTTG1. Antibodies against endogenous PTTG1 and γ-tubulin or CTR453 proteins were used in COS-7 (A–C) and RPE1 (D–O) cells to show centrosomal colocalization. To test the specificity of centrosomal labeling, RPE1 cells were infected with scramble shRNA (G–I) or PTTG1 shRNA (J–O) lentiviral vectors (MOI = 10). After 72 h, cells were fixed in methanol and processed for immunofluorescence. Left and middle, inverted images of PTTG1 and γ-tubulin or CTR453 staining, respectively. Right, merged images with PTTG1 in green and CTR453 and γ-tubulin in red. The small inset corresponds to the boxed region in each figure. Scale bars, 25 μm.
Given that GM130 anchors AKAP450 to the cis-Golgi (Rivero et al., 2009), we investigated whether PTTG1 is needed for this recruitment. Depletion of PTTG1 did not apparently affect the interaction between AKAP450 and GM130 at the GA (Supplemental Figure S2). Furthermore, Western blot analysis indicated that levels of GM130, γ-tubulin, and AKAP450 are not affected significantly after PTTG1 depletion (Supplemental Figure S3). We conclude that, although it seems to be dispensable, PTTG1 is involved in MT nucleation probably by facilitating, as a chaperone, the nucleation of centrosomal and noncentrosomal microtubules.
Migration defects in PTTG1-depleted RPE1 cells
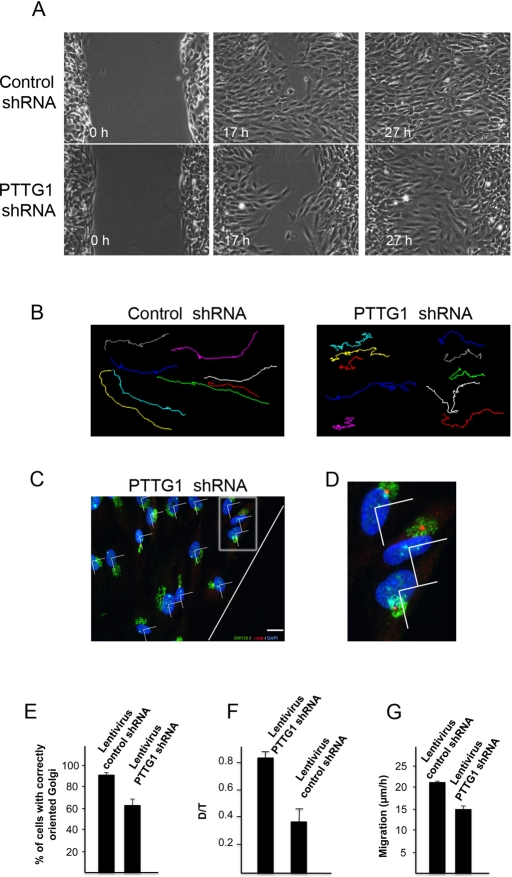
Given that MTs are indispensable for polarized cell migration and PTTG1 depletion delays the nucleation process of MTs, we tested whether PTTG1 depletion affected cell migration. In a wound-healing assay, cells infected with control shRNA Lentivirus moved into the wound area, filling the gap created by the scratch within 21 h. In contrast, cells infected with PTTG1 shRNA Lentivirus needed at least 27 h to close the gap (Figure 10A). The analysis of the movement of individual cells showed that RPE1 control cells migrate in a directionally persistent manner (Figure 10B, left). In contrast PTTG1-depleted cells showed random migration (Figure 10B, right), suggesting that PTTG1 is involved in the cell migration process. To investigate whether this defect in migration was due to GA/centrosome misorientation, we analyzed the reorientation of these organelles in scramble and PTTG1-depleted cells 7 h after wounding. Immunostaining with antibodies to GM130 and γ-tubulin and nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI), followed by quantitative analysis, confirmed that 38% of PTTG1-depleted cells showed a misorientated GA/centrosome compared with 10% of scrambled control cells (Figure 10, C–E). PTTG1 depletion affects the Golgi/centrosome reorientation and therefore directed cell migration, reducing the effective migration by a factor of 2.2 (Figure 10F). Furthermore, an estimate of migration speed indicated a 30% reduction in rate for the knockdown cells (Figure 10G). Therefore the impairment in wound healing was the consequence of a loss in directionally persistent migration and a loss of general cell motility.
FIGURE 10:
Migration defects in PTTG1-depleted cells. (A) Living RPE1 cells infected with control or shRNA lentiviruses (MOI = 10) were imaged at 7-min intervals for 27 h. The frames of the movie at 0 and 17 h are presented and used to estimate the percentage of surface covered by the cells. The last frame at 27 h is also shown. (B) Migration tracks of randomly picked cells are presented in different colors. (C) Representative immunofluorescence experiment of PTTG1-depleted cells at 7 h after wounding. Cells were stained with anti-GM130 (green), γ-tubulin (red), and DAPI. Cells were considered as correctly oriented if the Golgi and centrosome were contained inside the quadrant in front of the nucleus and facing the scratch (white line). Scale bar, 10 μm. (D) Enlarged view of the boxed area in C. (E) Percentage of control and PTTG1-depleted cells with Golgi and centrosome correctly reoriented. More than 90 cells were measured from each condition. (F) Directionally persistent migration distance of control and PTTG1-depleted cells. The directionally persistent migration (D/T) is the ratio of direct distance from start to end point (D) divided by the total track distance (T). Twenty-two cells were measured for each genotype. (G) Migration speed (μm/h) of control and PTTG1-depleted cells. Error bars, ±SD.
DISCUSSION
In this work, we report a novel function of PTTG1 and previously unrecognized PTTG1-binding partners. Using new, highly specific polyclonal antiserum against PTTG1, we confirmed binding of PTTG1 to AKAP450, GM130, and γ-tubulin and demonstrated that depletion of PTTG1 notably attenuates MT repolymerization and affects cell migration.
PTTG1 possesses transcriptional activity. A previous chromatin immunoprecipitation-on-chip study revealed that PTTG1 is a global transcription factor as well as an inhibitor of premature sister chromatid separation (Zou et al., 1999). These functions logically require a nuclear localization of PTTG1. However, PTTG1 can be also found in the cytoplasm; however, to date, the physiological role of this cytoplasmic PTTG1 fraction remains unclear.
Here we show that PTTG1 localizes at the GA in different types of cells. To prove this result, purified polyclonal antibodies were used both in immunofluorescence microscopy experiments and Western blot assays on subcellular protein fractions. These results confirmed and extended a previous report showing the localization of PTTG1 at the GA and vesicles of the mouse pituitary cell line AtT-20 (Minematsu et al., 2007). The present study analyzed several aspects of PTTG1 localization in the Golgi. We showed that PTTG1 is specifically associated with the cis side of the GA. This association seems to be independent of a correct polymerization and assembly of MTs and GA integrity. Other proteins, such as GM130, AKAP450, and the vesicle docking protein p115, remain associated with Golgi fragments generated after treating the cells with NZ, a drug that depolymerizes MTs and converts the Golgi ribbon into ministacks dispersed throughout the cytoplasm (Lowe et al., 2000; Rivero et al., 2009).
Association of PTTG1 with GA membranes seems to depend on its phosphorylation state. Our in vitro experiments show that phosphorylation of His-PTTG1 fusion protein by Cdk1 kinase prevents its association to membranes of GA. Furthermore, in vivo experiments show that GFP-PTTG1 fusion protein is recruited to GA, whereas the GFP-PTTG1 S/E phosphomimetic form is more weakly accumulated in GA, being also present as a diffusely dispersed protein in the cytoplasm. During prophase, as the Golgi complex starts to break down, PTTG1 is phosphorylated at Ser-165 by Cdk1 kinase and remains phosphorylated during further breakdown and partitioning of the Golgi fragments. In contrast, as the Golgi fragments start to reassemble in telophase, PTTG1 is dephosphorylated by PP2A phosphatase. It is important to highlight the similarities between the timing of the kinase and phosphatase involved in the phosphorylation and dephosphorylation of PTTG1 and those of other Golgi proteins involved in the mitotic fragmentation and reassembling of the GA during mitosis whose localization is also regulated by phosphorylation (Nakamura et al., 1997; Lowe et al., 1998, 2000; Litvak et al., 2004). Taken together, these findings point to a putative novel role of PTTG1 during mitosis different from its well-characterized function as a securin.
Furthermore, coimmunoprecipitation experiments showed that PTTG1 interacts with the proteins GM130, AKAP450, and γ-tubulin. These proteins are present at the cis side of the GA and form a complex involved in MT nucleation from the GA (Rivero et al., 2009). PTTG1-depleted cells showed a notable attenuation of MT repolymerization, suggesting that PTTG1 is a new member of the multiprotein complex associated with this process. Although centrosomes serve as the principal MT-organizing center, GA can also operate as an MTOC in a centrosome-independent manner (Efimov et al., 2007). It was reported that the absence of the centrosomal proteins such as CENP-F or Hook2 delays dramatically the MT repolymerization. This effect was demonstrated to be centrosome specific because Golgi membranes retain the capability to nucleate MTs, whereas the centrosome does not (Moynihan et al., 2009). Depletion of the cis-Golgi protein GM130 prevents the recruitment of AKAP450 to the pericentrosomal GA, leading to the disorganization of AKAP450 network and impairment of noncentrosomal nucleating activity (Rivero et al., 2009). Here we show that PTTG1 depletion delays both centrosomal and noncentrosomal MT repolymerization, providing additional evidence for the presence of PTTG1 in both MTOCs. In addition, a function as chaperone was previously described for PTTG1 (Zou et al., 1999). Accordingly, although ours results indicate that PTTG1 is not essential for the MT repolymerization, it could play a role as a chaperone in both centrosomal and noncentrosomal MTOCs, facilitating the formation and stability of these complexes.
Cell migration requires polarization of the cell into a leading edge and a trailing edge. The importance of Golgi-derived and centrosomal MTs in cell polarity and migration has been reported (Efimov et al., 2007; Miller et al., 2009; Rivero et al., 2009; Yadav et al., 2009). We proved that PTTG1-depleted cells show a severe impairment in migration ability. Two factors at least could cause this effect: the repolymerization delay of the MTs and the misorientation of the centrosome and the Golgi complex. A previous study described functional differences between Golgi- and centrosome-nucleated MTs with regard to cell polarization and migration. Cells depleted of AKAP450 at the Golgi, but not at the centrosome, were defective in cell migration in a wound-healing assay but retained the ability to reorient their centrosome and Golgi membranes toward the leading edge of the cell (Rivero et al., 2009). In contrast, GM130-depleted cells were also unable to migrate but contained aberrant centrosomes and were unable to reorient, suggesting that GM130 is required for cell migration through its regulatory role on the centrosome and possibly also through effects on MT assembly and dynamics (Kodani and Sutterlin, 2008). The localization of PTTG1 at the centrosome and Golgi and its interaction with essential MTOC proteins suggest that PTTG1 plays a role in the functional interactions between GA and the centrosome, which in turn could affect fundamental cellular processes such as cell polarization and cell migration. However, we cannot rule out the involvement of PTTG1 in other mechanisms that modulate some of these processes. In fact, a recent report showed that p53 regulates polarity of cell division in mammary stem cells; the loss of p53 favors symmetric divisions contributing to initiation and growth of tumors (Cicalese et al., 2009). Considering that PTTG1 is a modulator of p53 activity, it will be interesting to investigate in a future work a potential link between cell fate determination by p53 and the involvement of PTTG1 in cell polarity.
MATERIALS AND METHODS
Cell culture and antibodies
Immortalized human retinal pigment epithelial cells hTer-RPE1 (Clontech, Mountain View, CA) were maintained in DMEM/F12 with 10% fetal bovine serum (FBS). HeLa, HL-60, and Cos-7 cells (American Type Culture Collection, Manassas, VA) were grown in DMEM with 10% FBS; NIH3T3 cells were grown under identical conditions, except that the medium was supplemented with 10% calf born serum. To generate anti-PTTG1 antibody, the recombinant GST-fused, C-terminus of PTTG1 (amino acid positions 108–202) was produced in Escherichia coli and purified on glutathione–agarose beads (Sigma-Aldrich, St. Louis, MO). Rabbit polyclonal antibody against GST-fused PTTG1 C-terminus was affinity purified by absorption of the sera to His-fused PTTG1 protein immobilized onto CNBr-activated Sepharose (GE Healthcare, Piscataway, NJ) and elution at high pH. Monoclonal anti–α-tubulin, anti–γ-tubulin (clone GTU-88), and monoclonal anti-His antibodies were purchased from Sigma-Aldrich; monoclonal anti-GM130 and monoclonal anti-AKAP450 were from BD Biosciences (San Diego, CA). Rabbit polyclonal anti-GRP78, rabbit monoclonal anti-GM130, and mouse monoclonal anti–lamin A were from Abcam (Cambridge, MA). Autoimmune serum recognizing golgin-245 was previously characterized (Infante et al., 1999). Monoclonal anti-Cdk1, monoclonal anti-GST, monoclonal anti-PCNA, goat anti-CoxIV, and goat anti-AKAP450 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The monoclonal anti-CTR453 antibody recognizes a pericentriolar antigen and was a gift from M. Bornens.
Cell fractionation, fusion proteins, and binding experiments
HL-60 human cells (108) were recovered by centrifugation at 500 × g for 5 min at 4°C and washed twice with ice-cold PBS. Purification of different subcellular fractions was performed essentially as previously described (Fernandez-Ayala et al., 2005). Interphase Golgi membranes were isolated from HL-60 cells according to the described protocol (Balch et al., 1984). To produce a six-His fusion protein, PTTG1 cDNA was cloned into pRSETB vector (Invitrogen, Carlsbad, CA). Mutations Ser-165 to Ala (S165A) and Ser-165 to Glu (S165E) in PTTG1 were made in the pRSETB expression vector using the Transformer Site-Directed Mutagenesis Kit from Clontech. The mutagenic primers for S165A and S165E were 5′- G CTG GGC CCC CCT GCG CCT GTG AAG ATG-3′ and 5′-G CTG GGC CCC CCT GAA CCT GTGAAG ATG-3′, respectively (the mutated codons are underlined). Fusion proteins were induced in E. coli BL21 (DE3) by addition of 1 mM isopropyl-b-d-thiogalactopyranoside, and the fusion proteins were isolated from bacterial lysates with Talon beads (Clontech), eluted with 50 mM EDTA, and dialyzed against 50 mM NaHCO3, 150 mM NaCl, pH 7. The GST Gene Fusion System (GE Healthcare) was used to produce GST-PTTG1 protein and GST-PTTG1ΔN and GST-PTTG1-ΔC mutant forms. These PTTG1 mutant forms lack the 82 C-terminal residues and the 108 N-terminal residues, respectively. To generate the GFP version of PTTG1 mutants, corresponding sequences were amplified by PCR and cloned in pEGFP-C1 vector (Clontech). Binding to Golgi membranes was performed by incubation of dialyzed GST-PTTG1 or His-PTTG1 fusion proteins with 5–10 μg of purified Golgi membranes for 30 min at 4°C. The Golgi membranes were recovered by centrifugation, and the pellet and the supernatants were analyzed by Western blotting. Binding and dialysis of the fusion proteins was performed in a buffer containing 40 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.4), 0.15 M KCl, 1 mM MgCl2, and 0.2 M sucrose.
Coimmunoprecipitation, in vitro phosphorylation, and His pull-down assays
RPE1 cells were lysed in NP40 lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 1% NP-40) with protease and phosphatase inhibitors. Cellular lysates from (1–1.5) × 107 cells were incubated with magnetic Dynabeads protein A for 1 h at 4°C. After magnetic separation, beads were discarded and supernatants incubated overnight at 4°C with polyclonal anti-hPTTG or preimmune serum, followed by beads for 2 h. Beads were washed five times with lysis buffer, and bound proteins were dissolved into SDS sample buffer at 95°C for 5 min and subjected to SDS–PAGE and analyzed by Western blot according to standard protocols. Immunoprecipitation of Cdk1 was carried out by incubating lysates from HeLa cells synchronized in mitosis phase with anti-Cdk1 antibody, as described earlier. In vitro phosphorylation of His-PTTG1 was performed as described (Ramos Morales et al., 2000). Dephosphorylation assay was performed by incubating the in vitro phosphorylated PTTG1 for 30 min at 37°C with alkaline phosphatase (Roche, Indianapolis, IN). For six-His pull-down assays, fusion proteins (100–500 ng) were adsorbed to Dynabeads TALON and incubated with whole-cell extracts (200–500 μg) from RPE1 cells for 2 h. Beads were washed six times in lysis buffer, and bound proteins were eluted by the addition of SDS-sample buffer heated at 95°C for 5 min. Finally, the samples were subjected to SDS–PAGE and analyzed by Western blot.
siRNAs, constructs, and lentiviral production and infection
Transient PTTG1 down-regulation was achieved using the oligonucleotide targeting of the coding region 5′-GTCTGTAAAGACCAAGGGA-3′. Scramble siRNA (5′- CGUACGCGGAAUACUUCGATT-3′) was used as control. Interference and control oligonucleotides were subcloned in pSUPER as described (Brummelkamp et al., 2002). The H1-shRNA cassette from pSUPER-PTTG1 and pSUPER-scramble was excised after digestion with EcoRI and XhoI and subcloned into the pHRSIN-DUAL-GFP vector, kindly provided by Mary K. Collins (Windeyer Institute, London, United Kingdom). Insertion and orientation of the DNA fragment were confirmed by sequencing. For production of Lentivirus, 3 × 106 293T cells were seeded onto a 10-cm Petri dish and transfected after 24 h with Lipofectamine 2000 (Invitrogen) using 13.5 μg of transfer vector pHRSIN carrying shRNA PTTG1 and 9 and 4.5 μg of the HIV packaging plasmids pCMVDR8.91 and pMDG, respectively. Lentivirus was harvested 48 h posttransfection, passed through a 0.45-μm filter, and concentrated by ultracentrifugation at 100,000 × g for 90 min. Virus particles were resuspended in serum-free DMEM-F12 (Invitrogen), snap frozen in liquid nitrogen, and stored at −80°C. Lentiviral particle titers were determined by flow cytometry of RPE-1–infected cells. For infection with the lentiviral stock, RPE-1 cells were seeded at 105 cells in a six-well plate 24 h prior to infection, and then lentiviral particles (multiplicity of infection [MOI] = 10) were added and the culture medium was replaced 24 h after infection.
MT regrowth assay
Cell grown on coverslips were treated with 10 μM NZ for 2 h at 37°C and then incubated for 40 min at 4°C. After NZ removal by washing five times with ice-cold medium, cells were moved to a dish with warm medium (37°C) and were fixed at various times for immunostaining.
Immunofluorescence microscopy
Cells grown on coverslips were fixed with methanol at −20°C for 6 min or 4% paraformaldehyde/phosphate-buffered saline (PBS) at room temperature for 15 min and then permeabilized by incubation in 0.01% Triton X-100 in PBS. After fixation, the coverslips were incubated in blocking buffer (PBS containing 0.01% Tween-20, 3% bovine serum albumin) for 45 min and then with primary antibodies diluted in blocking buffer for 60 min. Secondary antibodies were conjugated with Alexa Fluor 488, Alexa Fluor 546, and Alexa Fluor 633 (Invitrogen). Nuclei were labeled with DAPI (200 ng/ml). Relative recruitment of construction to GA was calculated by dividing the mean pixel density measured in the GA by that measured in a region of the cytoplasm outside the GA. Wide-field fluorescence imaging was performed using a Leica DM6000B microscope (Wetzlar, Germany). Confocal images were captured by a confocal Leica TCS SP5 using HCX PL APO Lambda blue 63× 1.4 oil objective. Images were analyzed using Leica Application Suite software. Adobe Photoshop CS4 (Adobe Systems, Mountain View, CA) was used to process all images. Quantization of images was performed with the Metamorph Offline 7.5.1.0 (Molecular Devices, Sunnyvale, CA), using the command linescan. Values of Pearson's correlation coefficient were processed using KaleidaGraph 4.0.
Wound migration assay
Cell migration was measured by using Culture-Inserts (Ibidi, Martinsried, Germany). The Culture-Inserts were transferred to six-well plates, and RPE1 cells were seeded at a density of 7 × 104/ml in each well of Culture-Inserts. Cells were cultured in phenol red–free complete medium. After a 24-h incubation, the Culture-Inserts were removed, and a 400-μm cell-free gap was created. Cell migration was observed with a Leica DMI6000 inverted microscope equipped with a Hamamatsu ORCA-ER camera (Hamamatsu, Japan) and using LEICA N PLAN 10×, 20×/numerical aperture 0.25 objectives. Processing, quantification, and analysis were carried out using a Metamorph Offline 7.5.1.0 program. For GA reorientation assays, cells were fixed 7 h after wounding with 100% methanol at −20°C for 6 min, and the GA and centrosome were visualized by staining for G130 and γ-tubulin, respectively. DAPI (Sigma-Aldrich) was used as a nuclear counterstain. Golgi/centrosomes located in front of the nucleus, within the quadrant facing the wound, were scored as correctly oriented.
Supplementary Material
Acknowledgments
We thank C. Méndez-Vidal for critical reading of the manuscript. J.A.P.-T. and R.M.R. were supported by grants from the Ministerio de Educación y Cultura of Spain and the Dirección General de Universidades e Investigación of the Junta de Andalucía. M.A.M. was a recipient of a postdoctoral contract from the Junta de Andalucía.
Abbreviations used:
- GA
Golgi apparatus
- MT
microtubule
- MTOC
microtubule-organizing center
- PTTG1
pituitary tumor transforming gene 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0838) on September 21, 2011.
REFERENCES
- Balch WE, Dunphy WG, Braell WA, Rothman JE. Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell. 1984;39:405–416. doi: 10.1016/0092-8674(84)90019-9. [DOI] [PubMed] [Google Scholar]
- Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet. 2002;32:306–311. doi: 10.1038/ng997. [DOI] [PubMed] [Google Scholar]
- Bernal JA, Roche M, Mendez-Vidal C, Espina A, Tortolero M, Pintor-Toro JA. Proliferative potential after DNA damage and non-homologous end joining are affected by loss of securin. Cell Death Differ. 2008;15:202–212. doi: 10.1038/sj.cdd.4402254. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr Opin Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Efimov A, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ayala DJ, Brea-Calvo G, Lopez-Lluch G, Navas P. Coenzyme Q distribution in HL-60 human cells depends on the endomembrane system. Biochim Biophys Acta. 2005;1713:129–137. doi: 10.1016/j.bbamem.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet. 2000;355:716–719. doi: 10.1016/S0140-6736(99)10238-1. [DOI] [PubMed] [Google Scholar]
- Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios RM. GMAP-210, A cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J Cell Biol. 1999;145:83–98. doi: 10.1083/jcb.145.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, et al. Pituitary tumor-transforming 1 increases cell motility and promotes lymph node metastasis in esophageal squamous cell carcinoma. Cancer Res. 2008;68:3214–3224. doi: 10.1158/0008-5472.CAN-07-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- Kodani A, Sutterlin C. The Golgi protein GM130 regulates centrosome morphology and function. Mol Biol Cell. 2008;19:745–753. doi: 10.1091/mbc.E07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Litvak V, Argov R, Dahan N, Ramachandran S, Amarilio R, Shainskaya A, Lev S. Mitotic phosphorylation of the peripheral Golgi protein Nir2 by Cdk1 provides a docking mechanism for Plk1 and affects cytokinesis completion. Mol Cell. 2004;14:319–330. doi: 10.1016/s1097-2765(04)00214-x. [DOI] [PubMed] [Google Scholar]
- Lowe M, Gonatas NK, Warren G. The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J Cell Biol. 2000;149:341–356. doi: 10.1083/jcb.149.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minematsu T, et al. PTTG is a secretory protein in human pituitary adenomas and in mouse pituitary tumor cell lines. Endocr Pathol. 2007;18:8–15. doi: 10.1007/s12022-007-0005-9. [DOI] [PubMed] [Google Scholar]
- Moynihan KL, Pooley R, Miller PM, Kaverina I, Bader DM. Murine CENP-F regulates centrosomal microtubule nucleation and interacts with Hook2 at the centrosome. Mol Biol Cell. 2009;20:4790–4803. doi: 10.1091/mbc.E09-07-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG) Mol Endocrinol. 1997;11:433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- Ramos Morales F, Dominguez A, Romero F, Luna R, Multon MC, Pintor Toro JA, Tortolero M. Cell cycle regulated expression and phosphorylation of hpttg proto-oncogene product. Oncogene. 2000;19:403–409. doi: 10.1038/sj.onc.1203320. [DOI] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez C, Japon MA, Ramos Morales F, Romero F, Segura DI, Tortolero M, Pintor Toro JA. hpttg is over-expressed in pituitary adenomas and other primary epithelial neoplasias. Oncogene. 1999;18:5473–5476. doi: 10.1038/sj.onc.1202914. [DOI] [PubMed] [Google Scholar]
- Saez C, Martinez-Brocca MA, Castilla C, Soto A, Navarro E, Tortolero M, Pintor-Toro JA, Japon MA. Prognostic significance of human pituitary tumor-transforming gene immunohistochemical expression in differentiated thyroid cancer. J Clin Endocrinol Metab. 2006;91:1404–1409. doi: 10.1210/jc.2005-2532. [DOI] [PubMed] [Google Scholar]
- Shibata Y, et al. Expression of PTTG (pituitary tumor transforming gene) in esophageal cancer. Jpn J Clin Oncol. 2002;32:233–237. doi: 10.1093/jjco/hyf058. [DOI] [PubMed] [Google Scholar]
- Sutterlin C, Colanzi A. The Golgi and the centrosome: building a functional partnership. J Cell Biol. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Ben-Shlomo A, Zhou C, Wawrowsky K, Melmed S. Pituitary tumor transforming gene 1 regulates Aurora kinase A activity. Oncogene. 2008;27:6385–6395. doi: 10.1038/onc.2008.234. [DOI] [PubMed] [Google Scholar]
- Tong Y, Tan Y, Zhou C, Melmed S. Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene. 2007;26:5596–5605. doi: 10.1038/sj.onc.1210339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova T, Miller PM, Kaverina I. Microtubule network asymmetry in motile cells: role of Golgi-derived array. Cell Cycle. 2009;8:2168–2174. doi: 10.4161/cc.8.14.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev. 2007;28:165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.