A combination of bioinformatic and RNA interference analysis of Xenopus tropicalis RNA-seq data shows that the identification of microtubule-associated (MT) mRNAs can be used for discovering novel factors in the processes of spindle pole organization and centrosome structure. MT-RNAs are likely to contribute to spindle-localized mitotic translation.
Abstract
RNA localization is an important mechanism for achieving precise control of posttranscriptional gene expression. Previously, we demonstrated that a subset of cellular mRNAs copurify with mitotic microtubules in egg extracts of Xenopus laevis. Due to limited genomic sequence information available for X. laevis, we used RNA-seq to comprehensively identify the microtubule-interacting transcriptome of the related frog Xenopus tropicalis. We identified ∼450 mRNAs that showed significant enrichment on microtubules (MT-RNAs). In addition, we demonstrated that the MT-RNAs incenp, xrhamm, and tpx2 associate with spindle microtubules in vivo. MT-RNAs are enriched with transcripts associated with cell division, spindle formation, and chromosome function, demonstrating an overrepresentation of genes involved in mitotic regulation. To test whether uncharacterized MT-RNAs have a functional role in mitosis, we performed RNA interference and discovered that several MT-RNAs are required for normal spindle pole organization and γ-tubulin distribution. Together, these data demonstrate that microtubule association is one mechanism for compartmentalizing functionally related mRNAs within the nucleocytoplasmic space of mitotic cells and suggest that MT-RNAs are likely to contribute to spindle-localized mitotic translation.
INTRODUCTION
Localization of mRNAs to specific subcellular destinations is important for proper spatial and temporal control of gene expression. Examples of RNA localization are present in organisms ranging from bacteria to humans, indicating broad conservation of this type of posttranscriptional gene regulation (Holt and Bullock, 2009; Martin and Ephrussi, 2009; Montero et al., 2010). Furthermore, the incidence of RNA localization within any given transcriptome may be quite prevalent, as a survey of ∼3000 mRNAs in Drosophila embryos demonstrated 70% of transcripts showing discrete localization patterns (Lecuyer et al., 2007).
In the case of many well-studied examples, mRNA localization is critical for the spatial control of translation. The targeting of β-actin mRNA to lamellipodia of fibroblasts is important for translation of actin protein at the leading edge and cell motility (Kislauskis et al., 1997; Condeelis and Singer, 2005). The dendritic localization and expression of CaMKIIα mRNA affect synaptic and behavioral plasticity in mice (Miller et al., 2002). During early embryonic development, position-dependent localization and translation of the bicoid, nanos, and oskar mRNAs within the Drosophila embryo is essential for proper embryonic patterning (Johnstone and Lasko, 2001).
Several studies have shown the importance of the interphase microtubule cytoskeleton and microtubule-dependent motors in localizing specific mRNAs. For example, an intact microtubule cytoskeleton is essential for the distribution of bicoid and oskar mRNAs in Drosophila embryos (Cha et al., 2001; Weil et al., 2006; Zimyanin et al., 2008) and the transport of vg1 mRNA to the vegetal pole of Xenopus oocytes (Yisraeli et al., 1990; Messitt et al., 2008). Mitotic spindle microtubules are also sites for concentrating specific transcripts such as cyclin B (Raff et al., 1990; Groisman et al., 2000) and other mRNAs (Kingsley et al., 2007; Lecuyer et al., 2007). In addition, microtubules associate with polyribosomes and are sites of active translation, suggesting that some mRNAs targeted to mitotic microtubules undergo translation in close proximity to the spindle (Suprenant, 1993; Groisman et al., 2000; Blower et al., 2007).
Previously, we reported the identification of microtubule-associated mRNAs isolated from egg extracts of Xenopus laevis using a microarray-based approach (Blower et al., 2007). However, use of microarrays to interrogate the transcriptome is limited to analyzing the expression levels of mRNAs present on the arrays. In the case of X. laevis, the arrays are largely composed of unannotated expressed sequence tags and do not provide a complete picture of transcriptome complexity (i.e., no information on intronic sequences, untranslated regions, or noncoding RNAs). In this study, to take advantage of the sequenced genome and annotation resources of Xenopus tropicalis (Hellsten et al., 2010), we used RNA-seq to identify the complete microtubule-interacting transcriptome. Because RNA-seq has the ability to detect any transcribed sequence, this method provides an inherent advantage over microarray detection, which relies on hybridization to selected fragments of only known genomic sequences (Wang et al., 2009). Furthermore, the ability to align primary sequence data from RNA-seq experiments to annotated gene models provides much more information than X. laevis microarray datasets. Using a combination of bioinformatic and RNA interference (RNAi) analysis of X. tropicalis RNA-seq data, we demonstrate the identification of microtubule-associated mRNAs (MT-RNAs) as a resource for discovering novel factors involved in the processes of spindle pole organization and centrosome structure.
RESULTS
Isolation and sequencing of MT-RNA from X. tropicalis eggs
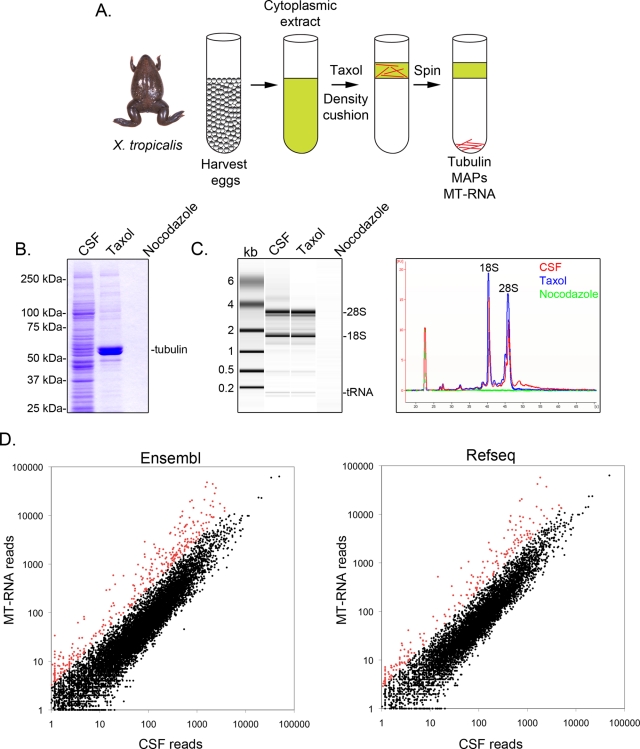
To identify X. tropicalis transcripts associated with microtubules, we prepared a cytosolic extract from unfertilized eggs arrested in metaphase (cytostatic factor [CSF]; Hannak and Heald, 2006; Brown et al., 2007). Treatment of this extract with Taxol allowed the formation of stable microtubules that could be purified by sedimentation through a glycerol cushion (Figure 1A). Coomassie gel analysis confirmed that α/β-tubulin sedimented in a Taxol-dependent manner, and represented the major protein species recovered in these preparations (Figure 1B). Lower levels of other proteins were also present in the Taxol pellet but not in preparations treated with the microtubule-depolymerizing drug nocodazole, indicating that proteins in the Taxol fraction specifically associated with microtubules.
FIGURE 1:
Purification and sequencing of MT-RNA. (A) Purification scheme to isolate MT-RNA. Eggs were harvested from female X. tropicalis frogs. After preparation of a cytoplasmic extract, Taxol was added to induce microtubule polymerization. Microtubules and MT-RNA were purified by sedimentation through a glycerol cushion. (B) Coomassie gel analysis of proteins isolated using the scheme described in A. Total CSF extract was compared with proteins sedimented in the presence of Taxol or nocodazole. (C) Bioanalyzer gel analysis of RNA isolated using the scheme described in A. RNA isolated from CSF extract was compared with RNA sedimented in the presence of Taxol or nocodazole. Both the gel projection and the line traces are shown. (D) RNA sequencing of CSF extract and MT-RNA. Sequences were aligned to the X. tropicalis genome and then compared with the Ensembl or RefSeq gene annotation databases to group sequences mapping to defined gene models. The scatter plots show read numbers per locus, plotted as a function of read number in CSF extract (x-axis) and read number in the MT-RNA fraction (y-axis). Data points that represent a value of 2 SDs above the mean log2(MT-RNA/CSF) ratio are plotted in red.
Next we examined general RNA composition in all X. tropicalis extract fractions using an Agilent Bioanalyzer (Figure 1C). Both rRNA and tRNA species were present in CSF extract and the microtubule-containing Taxol pellet, consistent with previous findings that translation occurs on microtubules and spindles in X. laevis egg extract (Groisman et al., 2000, Blower et al., 2007). A line trace of the gel projection revealed the mRNA signal was markedly lower in the microtubule-containing Taxol pellet, most notably in the region migrating above 28S rRNA, and suggested that a subset of mRNAs cosediment with microtubules in X. tropicalis.
To identify the mRNAs that associate with microtubules, we generated MT-RNA and CSF transcriptome libraries from poly-A purified RNA and performed Illumina platform sequencing (Materials and Methods). There are multiple incomplete annotation resources for the X. tropicalis genome, with only partial overlap between the gene models contained within each of these databases. To optimize our detection of annotated transcripts, we aligned library sequences to both the Ensembl and RefSeq databases (Materials and Methods; Supplemental Table S1). We detected transcribed sequences for 10,335 Ensembl gene models and 7169 RefSeq gene models, as depicted in Figure 1D and summarized in Supplemental Table S1.
To identify transcripts that show specific enrichment with microtubules, we compared reads per gene model in the MT-RNA fraction versus the whole transcriptome represented in CSF extract. The average log2(MT-RNA/CSF) read ratio was −0.40 (Ensembl) and −0.48 (RefSeq), confirming that most of the transcriptome is not preferentially associated with microtubules. To assign a quantitative definition for transcripts showing the strongest association with microtubules, we imposed a cutoff of two SDs above the average log2(MT-RNA/CSF) read ratio, reflecting a range of 3- to 36-fold microtubule enrichment. Approximately 3% of all RNAs detected within each database met this criterion (349 Ensembl and 218 RefSeq transcripts), consistent with the findings for X. laevis (Figure 1D; data points plotted in red; Blower et al., 2007). The sequence read number for transcripts qualifying as MT-RNAs ranged from a few copies to the tens of thousands, demonstrating that transcript abundance alone does not account for the observed enrichment of select transcripts with microtubules.
Inspection of reads aligned to the X. tropicalis genome revealed that 87% of sequences aligned to annotated exons of existing gene models (e.g., incenp, tpx2, and xkid; Figure 2, A– C). We also detected transcribed sequences occurring just proximal to gene models (97% of all reads aligned to or within 5 kb proximal to annotated gene models), likely indicating heterogeneity in untranslated regions (UTRs) of mRNAs (e.g., tpx2 and xkid; Figure 2, B and C). In addition, 2.5% of all reads aligned to annotated introns. These introns were positioned in the 5′ UTR, coding, and 3′ UTR regions at frequencies roughly proportional to their incidence in the genome. In one example, we detected retention and polysome association of an intron occurring between exons 8 and 9 in the coding region of the xkid chromokinesin, which predicts a rare protein-coding isoform (Figure 2C and Supplemental Figure S1; Funabiki and Murray, 2000). Using RT-PCR, we confirmed the presence of the intron-containing transcript isoforms for the xkid, set, pdcl3, cpsf7, gtf2f2, and wnt11 genes (Supplemental Figure S1 and unpublished data), thereby confirming the RNA-seq intron data.
FIGURE 2:
RNA-seq data mapped to the incenp, tpx2, xkid, and stag1 loci. Peaks representing accumulated sequences (red) were plotted relative to existing gene models (blue). (A) The incenp gene on scaffold 306: 113565–123005. Peak scale represents 0–5547 reads. (B) The tpx2 gene on scaffold 2319: 3641–9330. Peak scale represents 0–4751 reads. (C) The xkid gene on scaffold 1489: 21516–28810. Peak scale represents 0–425 reads. (D) The stag1 gene in an unannotated region of scaffold 10: 2160151–2183813. Peak scale represents 0–109 reads.
In cases in which sequences aligned to regions of the genome with no apparent gene models, we used the program TopHat to identify unannotated, multiexonic transcripts present in our data set (Trapnell et al., 2009). First, we identified splice junctions between transcriptional islands in our data set. We then subtracted all predicted splice junctions that overlapped with annotated exons and linked nongenic transcriptional islands using TopHat splice junction predictions (Materials and Methods). Using this approach, we identified an additional 83 protein-coding genes not identified in any X. tropicalis gene annotations (e.g., stag1; Figure 2D and Supplemental Table S1).
Together, these data demonstrate the power of RNA-seq to identify complete transcriptomes and indicate that most sequences present in the MT-RNA fraction represent full-length, spliced, protein-coding mRNAs.
X. tropicalis MT-RNAs are enriched with transcripts implicated in mitotic and cell cycle function
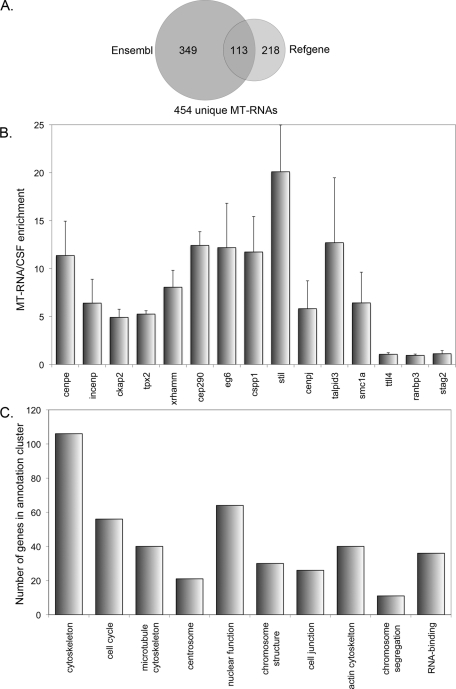
To determine the degree of overlap between gene models for Ensembl and RefSeq MT-RNAs, we used scaffold coordinates and BLAST to generate a combined list of MT-RNAs (Supplemental Table S2). This approach identified MT-RNA transcripts arising from 454 independent loci (Figure 3A). Furthermore, comparison of genes encoding X. tropicalis MT-RNAs to other vertebrate genomes revealed a high degree of conservation, with 95% of transcripts having human and mouse orthologues (Supplemental Table S2).
FIGURE 3:
Bioinformatic and real-time PCR analysis of MT-RNA. (A) Venn diagram showing the degree of overlap for MT-RNAs identified from the Ensembl and RefSeq databases. A total of 454 unique MT-RNAs were identified by RNA-seq. (B) Quantitative real-time RT-PCR was performed on selected transcripts (cenpe, incenp, ckap2, tpx2, xrhamm, cep290, eg6, cspp1, stil, cenpj, talpid3, and smc1a) identified as MT-RNAs by RNA-seq to confirm microtubule association. MT-RNA was isolated from X. tropicalis eggs as described in Figure 1 and was compared with total RNA from CSF extract. Several transcripts that did not show selective enrichment on microtubules (ttll4, ranbp3, stag2) served as controls. Error bars represent the SE from the mean of three independently prepared extracts. (C) GO analysis was performed to determine whether any GO terms were dominant among the transcripts identified as MT-RNAs. Shown are the top 10–ranked GO terms using the annotation clustering function of the NIH DAVID database.
Next we used quantitative RT-PCR to verify that transcripts identified as MT-RNAs were enriched on microtubules in independent extract preparations (Figure 3B). The MT-RNA transcripts cenpe, incenp, ckap2, tpx2, xrhamm, cep290, eg6, cspp1, stil, cenpj, talpid3, and smc1a all showed quantitative enrichment in Taxol-stabilized, microtubule-containing fractions relative to their abundance in CSF extract. In contrast, the transcripts ttll4, ranbp3, and stag2 were approximately equally represented in the MT-RNA fraction and CSF extract. These data confirm that specific transcripts show quantitative enrichment on microtubules and validate the accuracy of MT-RNAs identified using RNA-seq.
To test whether transcripts with related Gene Ontology (GO) terms were enriched in the MT-RNA library, we used the National Institutes of Health (NIH) Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource (Huang et al., 2009). This database enabled us to extract GO terms for the X. tropicalis MT-RNAs, using the National Center for Biotechnology Information (NCBI) Gene ID for the human homologues as identifiers. This approach resulted in a more comprehensive GO term list than any X. tropicalis annotation resource currently in existence. In addition, we performed functional annotation clustering to identify gene lists with related GO terms. Figure 3C shows the results of the top 10–ranked annotation clusters identified, which reflect DAVID enrichment scores of >3. Six of the 10 annotation clusters were composed of several genes that function in organizing the cell cycle, microtubule cytoskeleton, centrosomes, and chromosome structure/segregation (Figure 3C and Supplemental Table S3). However, MT-RNAs were not found to be solely composed of transcripts functioning in mitosis, as there were groups of genes identified that acted in distinct processes such as cell junction formation or RNA binding. From this analysis, we conclude that transcripts that regulate the cell cycle and mitotic apparatus are overrepresented in the pool of microtubule-associated RNAs versus the genome at large, which suggests that MT-RNAs may play a role in controlling mitosis.
MT-RNAs interact with spindle microtubules in vivo
To investigate how MT-RNAs are spatially distributed with respect to spindle microtubules in vivo, we performed fluorescence in situ hybridization (FISH) for select MT-RNAs in conjunction with tubulin immunofluorescence in X. laevis XL177 tissue culture cells (Figure 4). The MT-RNAs incenp, xrhamm, and tpx2 have known roles in regulating spindle assembly (Wittman et al., 2000; Groen et al., 2004; Sampath et al., 2004) and were chosen for analysis since these transcripts showed a high degree of microtubule enrichment in both the RNA-seq and quantitative RT-PCR assays. In contrast, β-actin mRNA did not show selective biochemical enrichment on microtubules, consistent with several studies showing the importance of the actin cytoskeleton for localization of β-actin mRNA (Sundell and Singer, 1991; Latham et al., 2001; Oleynikov and Singer, 2003), and was used as a negative control.
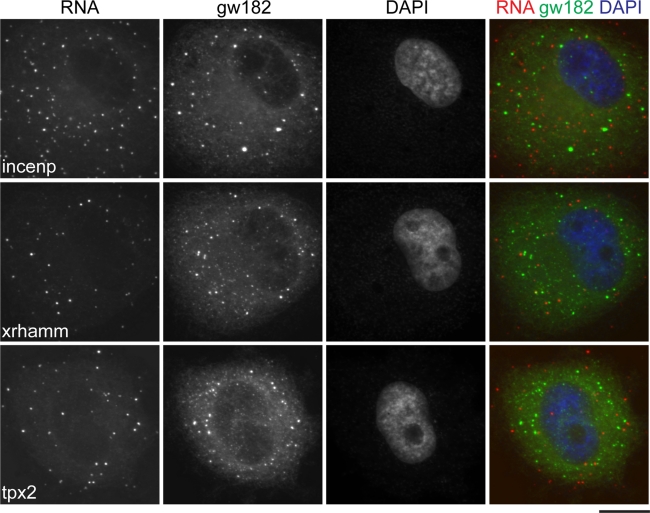
FIGURE 4:
MT-RNAs associate with spindle microtubules in vivo. X. laevis XL177 cells were fixed and processed for combined immunofluorescence for tubulin and FISH as described in the Materials and Methods. (A) The fluorescence hybridization signal for the MT-RNAs incenp, xrhamm, and tpx2. β-actin was used as a control. (B) The RNA signal merged with tubulin immunofluorescence and DAPI staining to visualize spindle microtubules and chromatin. Scale bar, 10 μm. (C) A selected area of the panels in B were enlarged four times. Scale bar, 2.5 μm.
We observed that incenp RNA showed two general staining patterns within metaphase cells. In the first pattern, incenp RNA localized throughout the spindle body and along astral microtubules. In the second pattern, incenp RNA particles were still observed on astral microtubules; however, incenp RNA within the spindle body was preferentially distributed on chromosomes, similar to the distribution of incenp protein (Cooke et al., 1987; Mackay et al., 1993). Both xrhamm and tpx2 RNAs showed localization within the spindle body and along astral microtubules, with little staining of chromosomes.
In contrast, β-actin mRNA showed a markedly different staining pattern. Specifically, the majority of the β-actin RNA signal was present in the form of small particles distributed throughout the nucleocytoplasmic volume with no apparent selective association with microtubules. Because metaphase cells in XL177 cultures lack lamellipodia, these data are consistent with the diffuse localization pattern for β-actin RNA present in ∼70% of fibroblast cells (Latham et al., 1994; Shestakova et al., 2001). Larger β-actin particles were also present but tended to accumulate at the cell periphery rather than overlap astral microtubules. These data demonstrate that the MT-RNAs incenp, xrhamm, and tpx2 are associated with spindle microtubules in vivo. Furthermore, because XL177 cells are derived from tadpole-stage frogs (Ellison et al., 1985), these data argue that the incenp, xrhamm, and tpx2 RNAs associate with microtubules in somatic cell types as well as oocytes.
The appearance of incenp, xrhamm, and tpx2 mRNAs as discrete particles led us to consider whether these RNAs were components of P-bodies. P-Bodies are cytoplasmic ribonucleoprotein structures harboring proteins involved in the processes of mRNA decay and translational repression (Moser and Fritzler, 2010). P-Body motion in the cytoplasm is microtubule dependent (Aizer et al., 2008), suggesting the possibility that some RNAs could associate with microtubules via P-body association. We therefore tested whether the incenp, xrhamm, and tpx2 mRNA FISH signal overlapped with the integral P-body component gw182 (Eystathioy et al., 2003). Because P-bodies disassemble prior to mitosis, we were limited to examining interphase cells for these experiments (Yang et al., 2004; unpublished data). As depicted in Figure 5, we observed that the incenp, xrhamm, and tpx2 mRNAs were observed in cytoplasmic particles, the vast majority of which were spatially distinct from gw182/P-bodies. We conclude the MT-RNAs incenp, xrhamm, and tpx2 are not sequestered in translationally repressive cytoplasmic domains during interphase and associate with spindle microtubules independent of P-bodies.
FIGURE 5:
MT-RNAs are distinct from P-body domains. XL177 cells were labeled with antibodies to gw182 and fluorescent probes to detect the incenp, xrhamm, and tpx2 mRNAs. Scale bar, 10 μm.
RNAi of human MT-RNA orthologues identifies novel genes that contribute to spindle pole morphology and γ-tubulin distribution
Our GO analysis indicated that the MT-RNA population was enriched with transcripts encoding proteins that contribute to mitosis, which suggested that uncharacterized genes in the data set might also encode proteins with mitotic functions. To test for a mitotic function of uncharacterized MT-RNAs, we identified the human orthologues for 10 MT-RNA transcripts and performed RNAi in HeLa cells using endoribonuclease-prepared short interfering RNAs (esiRNAs; Figure 6; Yang et al., 2002; Kittler et al., 2007). We chose to test several transcripts encoding coiled-coil domain-containing proteins (CCDC18, CCDC52, CCDC66, CCDC100, CCDC104, KIAA1731, TRIM36), as the coiled-coil motif was present in 100 of the 454 MT-RNAs (Supplemental Table S2) and is a domain associated with several proteins known to be involved in spindle pole and centrosome function (Compton et al., 1992; Yang et al., 1992; Doxsey et al., 1994; Bouckson-Castaing et al., 1996; Gergeley et al., 2000). In addition, because CASC1/LAS1 and TALPID3 have been suggested to interact with tubulin and centrosomes, respectively (Liu et al., 2007; Yin et al., 2009), these genes were also chosen for functional analysis.
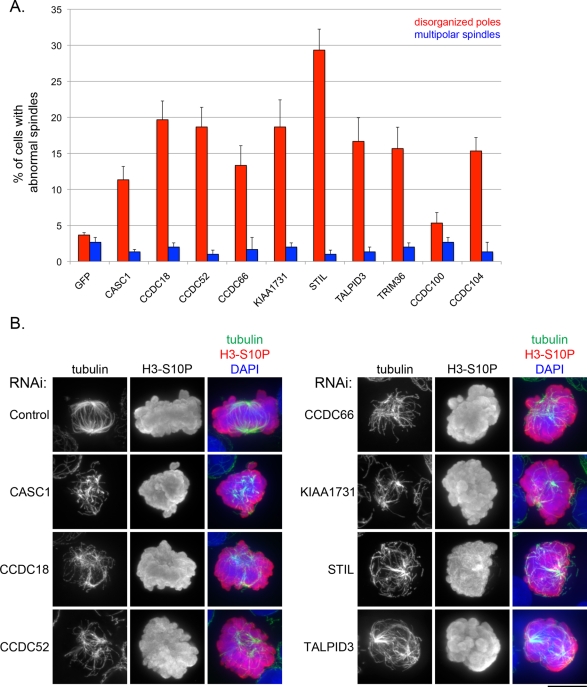
FIGURE 6:
RNAi of MT-RNAs results in defective spindle pole organization. (A) RNAi was performed in HeLa cells to deplete transcripts for uncharacterized MT-RNAs. In control transfections, siRNAs for green fluorescent protein were used. Bar graph showing the percentage of HeLa cells displaying either abnormal spindle pole organization (red) or the formation of supernumerary spindle poles (blue). The error bars represent the SE from three independent experiments. (B) Images showing examples of spindles observed in RNAi experiments. HeLa cells were stained with an anti-tubulin antibody to label spindle microtubules, an anti–histone H3-S10P antibody to label chromatin of mitotic cells, and DAPI to label DNA for both interphase and mitotic cells. Scale bar, 10 μm.
Real-time PCR analysis of transcript levels confirmed that all MT-RNAs were effectively depleted (∼70–97% knockdown) in our experiments (Supplemental Figure S2). We performed immunofluorescent staining for tubulin and the mitotic marker histone H3-S10P and scored the appearance of spindle morphology in mitotic figures with condensed chromosomes. RNAi of the STIL gene served as a positive control in these experiments since depletion of this MT-RNA transcript was reported to result in defective mitotic spindles in HeLa cells (Pfaff et al., 2007).
In 9 of 10 experimental RNAi transfections to deplete MT-RNAs (CASC1, CCDC18, CCDC52, CCDC66, KIAA1731, STIL, TALPID3, TRIM36, and CCDC104), we observed that MT-RNA depletion resulted in a significantly increased frequency of abnormal spindle pole morphology (Figure 6A). Abnormal spindles in knockdown cell populations often appeared as a random array of microtubule fragments or bundles, with varying degrees of defective pole coalescence (Figure 6B). This phenotype occurred in an average ∼11–29% of cells in experimental transfections, compared with a 3.6% frequency in control transfections. In contrast, the frequency of multipolar spindles in both control and experimental transfections was similar (Figure 6A), showing that the effect of depleting MT-RNAs was specific to spindle pole organization.
In addition, we performed immunofluorescence for γ-tubulin to test whether centrosomes were affected by MT-RNA depletion (Figure 7). In control transfections and six of the experimental transfections, the vast majority of interphase cells had one to two γ-tubulin foci per cell, consistent with normal centrosome number. Of interest, we observed that knockdown of CASC1, KIAA1731, STIL, and TALPID3 resulted in a significant and reproducible increase in the number of cells showing a dispersed cytoplasmic cloud of γ-tubulin (11–18% of cells; Figure 7A). In the case of the KIAA1731 RNAi data, we note that the observation of dispersed γ-tubulin is consistent with a recent report describing the loss of two other centrosome markers in cells depleted of the KIAA1731 transcript (Knorz et al., 2010). On the basis of our results, we conclude that the CASC1, KIAA1731, STIL, and TALPID3 genes all contribute to maintaining normal cytoplasmic concentrations of γ-tubulin in the vicinity of the centrosome.
FIGURE 7:
RNAi of MT-RNAs results in abnormal γ-tubulin distribution. (A) RNAi was performed in HeLa cells to deplete transcripts for the same MT-RNAs analyzed in Figure 4. Bar graph showing the percentage of HeLa cells with γ-tubulin dispersed throughout the cytoplasm. The error bars represent the SE from three independent experiments. (B) Images showing examples of γ-tubulin distribution observed in RNAi experiments. HeLa cells were stained with an anti-γ-tubulin antibody and DAPI to stain DNA. Scale bar, 10 μm.
DISCUSSION
In sum, we used transcriptome sequencing to comprehensively survey the fraction of mRNAs associated with microtubules in X. tropicalis eggs and identified hundreds of mRNAs associated with microtubules. The MT-RNAs incenp, tpx2, and xrhamm were distributed throughout the spindle region as well as on astral microtubules in vivo, suggesting that some MT-RNAs are targeted where the protein product is functionally required. We found that X. tropicalis MT-RNAs are enriched in genes known to be involved in cell division, cytoskeletal and centrosome structure, and chromosome function. In addition, we demonstrated the utility of identifying MT-RNAs in discovering novel factors involved in the processes of spindle pole organization and centrosome structure. We conclude that mitotic microtubules are sites for sequestering mRNAs that encode functionally related proteins.
Our data suggest that one purpose for spindle localization of mRNAs is to translate proteins that function in mitosis in close proximity to the spindle and chromosomes. In the case of the cyclin B and xkid transcripts, microtubule targeting by CPEB facilitates polyadenylation-induced translation (Raff et al., 1990; Groisman et al., 2000; Eliscovich et al., 2008). Determining whether spindle microtubules and mitotic signaling pathways act to modulate the translational capacity for other MT-RNAs identified in this study will be an important goal for future study. Localized translation is well known to play an important role in efficient protein function in large cells such as oocytes and neurons (Kindler et al., 2005). However, our FISH data demonstrating that MT-RNAs are also enriched near the site of protein action in tissue culture cells suggest that localized translation is also important in smaller, somatic cells.
Of interest, the global rate of translation is dramatically decreased during mitosis in higher eukaryotes to ∼25% of the rate of translation of interphase cells (Prescott and Bender, 1962; Pyronnet et al., 2001). The decrease in translation rate is caused by an inhibition of cap-dependent translation (Bonneau and Sonenberg, 1987), which results in an up-regulation of internal ribosome entry site (IRES)–dependent translation during mitosis. Of note, two mRNAs with known cell cycle–regulated IRES elements, odc1 and cdk11 (Cornelis et al., 2000; Pyronnet et al., 2000), were found to be modestly enriched on microtubules in our data set (1.2- and 3.1-fold, respectively). Several studies have shown that mitotic translation (Groisman et al., 2000) and inhibition of cap-dependent translation (Wilker et al., 2007) are important for proper mitotic progression. Our data set of MT-RNAs likely contains many new candidate mRNAs that are translated during mitosis, possibly by an IRES-dependent mechanism.
The mechanism of how MT-RNAs are transported to spindle microtubules remains to be determined. One possibility would be that ribosomes complexed with MT-RNAs are targeted to the spindle apparatus, perhaps through an interaction made possible by nascent peptide synthesis. However, we previously demonstrated that active translation is not required for mRNA localization to microtubules (Blower et al., 2007). Instead, we favor the hypothesis that mRNAs are actively transported to microtubules in a translationally repressed state (Besse and Ephrussi, 2008) using microtubule motor proteins.
Taken together, the RNAi data indicate that several MT-RNA transcripts are required to maintain normal spindle pole organization and centrosome structure. These MT-RNAs could be of particular importance in maintaining pole structure for the centrosome-free meiotic spindle present in the oocyte. However, because these data were acquired in HeLa cells, which have centrosomes, we envision that these MT-RNAs also participate in centrosome-driven mitotic spindle pole organization after fertilization of the oocyte. This general role for MT-RNAs in spindle pole organization is likely to contribute to the accurate segregation of genetic material during the rapid cell divisions taking place in the early embryo. Four of nine MT-RNAs that showed a spindle pole organization phenotype upon RNAi-mediated depletion also showed an abnormal cytoplasmic distribution of γ-tubulin, suggesting that these MT-RNAs could act in part to maintain pole structure through enforcing proper localization of γ-tubulin. This function could be of particular importance in the context of the fertilized oocyte, in which the sperm centrosome needs to be significantly remodeled in order to form the spindle for the first mitotic division of the zygote (Felix et al., 1994; Stearns and Kirschner, 1994; Fry et al., 2000). We note MT-RNAs are likely to make additional contributions to pole organization since only a subset of genes tested showed an abnormal γ-tubulin distribution upon RNAi-mediated depletion. In sum, our study demonstrates that hundreds of mRNAs are localized to mitotic spindles and suggests that spindle-localized translation is an important and novel mechanism of mitotic control.
MATERIALS AND METHODS
X. tropicalis MT-RNA purification and library preparation
X. tropicalis frogs were obtained from Nasco (Fort Atkinson, WI) and housed in accordance with the Massachusetts General Hospital Animal Facility Guidelines. CSF extract was prepared as described (Brown et al., 2007) except for a modification to increase egg yield. To induce egg laying, frogs were injected with 20 U of human chorionic gonadotropin (hCG; Sigma-Aldrich, St. Louis, MO) on two successive days, followed by a final injection of 200 U hCG on the third day.
To induce microtubule polymerization, Taxol was added to CSF extract at a final concentration of 10 μM. In control reactions, nocodazole was added at a final concentration of 10 μM. After incubation for 30 min at room temperature, reactions were diluted with 10 volumes BRB-80 (80 mM 1,4-piperazinediethanesulfonic acid [PIPES], pH 6.8, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid) plus 30% glycerol. Diluted extract was layered over a 10-ml cushion of BRB-80 plus 60% glycerol and centrifuged for 10 min at 17,000 × g. The supernatant was aspirated, and the interface was washed twice with water. The Taxol pellet containing MT-RNA was then resuspended in TRIzol (Invitrogen, Carlsbad, CA) for RNA isolation. For comparison, an aliquot of total CSF extract was incubated similarly and resuspended directly in TRIzol.
To generate CSF and MT-RNA libraries for Illumina sequencing, we prepared poly-A RNA from 25 μg of total RNA using an oligo-dT LNA (Exiqon, Vedbaek, Denmark) according to the manufacturer's instructions. Approximately 0.5 μg of poly-A RNA was fragmented using 1× RNA fragmentation buffer (Applied Biosystems/Ambion, Austin, TX) for 5 min at 70°C to optimize gene coverage. Fragmentation reactions were quenched and precipitated in ethanol. To generate end-tagged cDNA libraries, we used the reverse transcription reaction conditions described previously (Cloonan et al., 2008). The oligonucleotide 5′-CAAGCAGAAGACGGCATACGANNNNNN-3′ was used to attach the first linker sequence through random-primed first-strand cDNA synthesis. The oligonucleotide 5′-ACAGGTTCAGAGTTCTACAGTCCGACGATCGGG-3′ was used to as a G-rich anchor to append the second linker sequence to first-strand cDNA using the template-switching activity of MMLV reverse transcriptase (Clontech, Mountain View, CA). cDNAs were amplified using the Expand PCR Kit (Roche, Indianapolis, IN) and gel purified prior to precipitation in ethanol.
RNA-seq and alignment
X. tropicalis end-tagged cDNA libraries were sequenced at Massachusetts General Hospital on an Illumina GA2 sequencer. Sequences consisting of 12 base pairs and longer were aligned to the X. tropicalis JGI4.1/xenTro2 genome assembly (http://genome.ucsc.edu), allowing for one mismatch or one insert/deletion per read. Each read was divided by the number of incidences in the genome, with reads aligning greater than five times removed to facilitate analysis. Reads were grouped by alignment to individual gene models according to the scaffold coordinates specified in the Ensembl (www.ensembl.org) and RefSeq (www.ncbi.nlm.nih.gov/RefSeq) databases. Visualization of reads mapped to the X. tropicalis genome was performed using IGV 2.0 software (Robinson et al., 2011). The aligned sequence read numbers totaled 2,004,856 and 2,455,598 for the CSF and MT-RNA libraries, respectively. After normalization between library samples, gene models with less than one read were subtracted from the data set, as were reads occurring antisense to gene models. The annotated sequencing data are listed in Supplementary Table S1.
Identification of unannotated genes
MT-RNA and CSF reads were pooled and used as input for the TopHat splice junction prediction program. We then wrote a custom Perl script to remove all predicted splice junctions that overlapped with annotated exons (RefSeq or Ensembl). We then searched for predicted splice junctions that mapped on the same strand of the same scaffold within 819 bases of one another, which is two SDs greater than the average exon length. We then captured groups of greater than two junction pairs that could be clustered in this manner, which represents a transcript of at least three exons. Junction clusters were then manually inspected using the UCSC Genome Browser (http://genome.ucsc.edu) and BLASTX to identify protein-coding genes.
Gene Ontology
Identification of the human homologues to X. tropicalis MT-RNAs by BLAST analysis allowed us to assign an NCBI Gene ID to each MT-RNA. We then used the NIH DAVID Bioinformatics Database 6.7 (http://david.abcc.ncifcrf.gov) to identify the GO terms for 412 of 454 MT-RNAs. Functional annotation clustering of the Gene ID list resulted in identifying 153 total annotation clusters (Supplemental Table S3). The top-ranked annotation clusters, which were assigned DAVID enrichment scores of >3, were depicted as shown in Figure 2C.
Immunofluorescence and FISH
X. laevis XL177 cells (Ellison et al., 1985) were cultured in Leibowitz's media containing 15% fetal bovine serum (FBS). Cells were fixed in BRB-80 containing 4% paraformaldehyde and 0.05% glutaraldehyde for 10 min and were extracted in BRB-80 plus 0.5% Triton for 20 min. Microtubules were stained using the mouse anti-tubulin antibody DM1A (Sigma-Aldrich) at a 1:500 dilution, followed by an Alexa 488–conjugated anti–mouse antibody (Invitrogen) at 1:500 dilution. P-Bodies were stained using human antisera that recognize X. laevis gw182 (Eystathioy et al., 2002; Lund et al., 2011), followed by an Alexa 488–conjugated anti–human antibody (Invitrogen) at 1:500 dilution. Cells were postfixed in 2% paraformaldehyde for 10 min to preserve antibody staining prior to FISH hybridization.
For RNAi experiments, HeLa S3 cells (a gift of A. Fischer) were cultured in DMEM media containing 10% FBS, 1× nonessential amino acids, and 2 mM l-glutamine. After transfection, cells were permeabilized in CSK buffer (10 mM PIPES, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2) plus 0.5% Triton for 30 s and fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde for 10 min at room temperature. Immunofluorescence for tubulin was as described earlier. In parallel, cells were stained for γ-tubulin using the mouse GTU88 monoclonal antibody (Sigma-Aldrich) at 1:1000 dilution, followed by the Alexa 488–conjugated anti–mouse antibody (Invitrogen) at 1:500 dilution. Coverslips were washed in PBS containing 0.2% Tween, stained with 4′,6-diamidino-2-phenylindole (DAPI), and mounted in Vectashield (Vector Labs, Burlingame, CA).
Probes for FISH analysis were generated from the full-length cDNA clones for X. laevis incenp, xrhamm, tpx2, and β-actin (Open Biosystems, Thermo Biosystems, Huntsville, AL). Approximately 0.5 μg of plasmid DNA was labeled with Cy3-dCTP (GE Healthcare, Piscataway, NJ), using a Klenow Labeling Kit (Invitrogen). Unincorporated nucleotide was removed using Microspin-S200 columns (GE Healthcare). Probes were precipitated in ethanol and then reprecipitated with the addition of tRNA, Cot-1 DNA, and salmon sperm single-stranded DNA. Probes were then resuspended in hybridization buffer (2× saline–sodium citrate [SSC], 10% dextran sulfate, 50% formamide) and denatured before hybridization.
Prior to FISH analysis, fixed cells were dehydrated sequentially in 70% ethanol, 85% ethanol, 95% ethanol, and 100% ethanol. FISH probes were applied directly to air-dried coverslips and incubated overnight in a humid chamber at 37°C. Coverslips were washed twice in 2× SSC/50% formamide, once in 2× SSC, and three times in 1× SSC. All washes were performed at 39°C for 5 min each. DAPI was used at 100 ng/ml in 1× SSC to stain nuclei, and coverslips were mounted in Vectashield.
Images were captured on an Olympus (Center Valley, PA) BX61 microscope equipped with a Hamamatsu charge-coupled device camera (ORCA; Hamamatsu, Japan). Stacks of confocal images spaced 0.2 μm apart were rendered as maximum Z-projections using either Slidebook or Metamorph software.
RNAi
The human genes CASC1, CCDC18, CCDC52, CCDC66, CCDC100, CCDC104, KIAA1731, STIL, TALPID3, and TRIM36 were identified from the X. tropicalis orthologues using BLAST (NCBI). We used DEQOR software (Henschel et al., 2004) to identify optimal sequences for RNAi targeting and designed oligonucleotides complementary to those sequences for PCR amplification. PCR fragments were then used as templates for generation of double-stranded RNA (dsRNA), and short RNA duplexes were produced from dsRNA using RNase III (Yang et al., 2002). RNase III digestions were subsequently purified using Invitrogen PureLink miRNA spin columns. Approximately 5 pmol of siRNAs were transfected into HeLa S3 cells using the transfection reagent Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. After 48 h, cells were processed for immunofluorescence or RNA content analysis.
Supplementary Material
Acknowledgments
We thank members of the Blower laboratory, Tom Fazzio, and Paul Kaufman for helpful discussions and Marvin Fritzler for the GW182 antibodies. J.A.S. was supported by a Pfizer Fellowship from the Life Science Research Foundation and a Tosteson Award from the Massachusetts Biomedical Research Corporation. M.D.B. was supported by a Burroughs Wellcome Fund Career Award.
Abbreviations used:
- CSF
cytostatic factor
- FISH
fluorescence in situ hybridization
- GO
Gene Ontology
- IRES
internal ribosome entry site
- MT-RNA
microtubule-associated mRNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0629) on September 21, 2011.
REFERENCES
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–80. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau AM, Sonenberg N. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J Biol Chem. 1987;262:11134–11139. [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J Cell Sci. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald RJ. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol. 2007;176:765–770. doi: 10.1083/jcb.200610043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106:35–46. doi: 10.1016/s0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Compton DA, Szilak I, Cleveland DW. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Singer RH. How and why does β-actin mRNA target. Biol Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol. 1987;105:2053–2067. doi: 10.1083/jcb.105.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Eliscovich C, Peset I, Vernos I, Mendez R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat Cell Biol. 2008;10:858–865. doi: 10.1038/ncb1746. [DOI] [PubMed] [Google Scholar]
- Ellison TR, Mathisen PM, Miller L. Developmental changes in keratin patterns during epidermal maturation. Dev Biol. 1985;112:329–337. doi: 10.1016/0012-1606(85)90403-8. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Chan EKL, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLsm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of gamma-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Descombes P, Twomey C, Bacchieri R, Nigg EA. The NIMA-related kinase X-Nek2B is required for efficient assembly of the zygotic centrosome in Xenopus laevis. J Cell Sci. 2000;113:1973–1984. doi: 10.1242/jcs.113.11.1973. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Gergeley F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA. 2000;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr Biol. 2004;14:1801–1811. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–437. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschel A, Buchholz F, Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113-W120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley EP, Chan XY, Duan Y, Lambert JD. Widespread RNA segregation in a spiralian embryo. Evol Dev. 2007;9:527–539. doi: 10.1111/j.1525-142X.2007.00194.x. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH. β-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4:337–44. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- Knorz VJ, Spalluto C, Lessard M, Purvis TL, Adigun FF, Collin GB, Hanley NA, Wilson DI, Hearn T. Centriolar association of ALMS1 and likely centrosomal functions of the ALMS-motif-containing proteins C10orf90 and KIAA1731. Mol Biol Cell. 2010;21:3617–3629. doi: 10.1091/mbc.E10-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham VM, Kislauskis EH, Singer RH, Ross AF. Beta-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham VM, Yu EH, Tullio AN, Adelstein RS, Singer RH. A Rho-dependent signaling pathway operating through myosin localizes beta-actin mRNA in fibroblasts. Curr Biol. 2001;11:1010–1016. doi: 10.1016/s0960-9822(01)00291-3. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Liu Y, Vikis HG, Yi Y, Futamura M, Wang Y, You M. Degradation of lung adenoma susceptibility 1, a major candidate mouse lung tumor modifier, is required for cell cycle progression. Cancer Res. 2007;67:10207–10213. doi: 10.1158/0008-5472.CAN-07-2574. [DOI] [PubMed] [Google Scholar]
- Lund E, Sheets MD, Imboden SB, Dahlberg JE. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 2011;25:1121–1131. doi: 10.1101/gad.2038811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay AM, Eckley DM, Chue C, Earnshaw WC. Molecular analysis of the INCENPs (inner centromere proteins): separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. J Cell Biol. 1993;123:373–385. doi: 10.1083/jcb.123.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KM. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15:426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIa impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Montero LP, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser JJ, Fritzler MJ. Cytoplasmic ribonucleoprotein (RNP bodies and their relationship to GW/P bodies. Int J Biochem Cell Biol. 2010;42:828–843. doi: 10.1016/j.biocel.2009.11.018. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13:199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. The Zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol. 2007;27:5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- Raff JW, Whitfield WG, Glover DM. Two distinct mechanisms localise cyclin B transcripts in syncytial Drosophila embryos. Development. 1990;110:1249–1261. doi: 10.1242/dev.110.4.1249. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, Condeelis J. The physiological significance of beta-actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci USA. 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Suprenant KA. Microtubules, ribosomes, and RNA: evidence for cytoplasmic localization and translational regulation. Cell Motil Cytoskeleton. 1993;179:1365–1373. doi: 10.1002/cm.970250102. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Wilker EW, et al. 14–3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- Wittman T, Wilm M, Karsenti E, Vernos I. TPX2, a novel Xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen C, Brodsky FM, Bishop JM. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EKL. Gw182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- Yin Y, et al. The Talpid3 gene (KIAA0586) encodes a centrosomal protein that is essential for primary cilia formation. Development. 2009;136:655–664. doi: 10.1242/dev.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli JK, Sokol S, Melton DA. A two-step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development. 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux P, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.