Chromosomes segregate by interaction of spindle microtubules with kinetochores. In budding yeast the Dam1 complex forms rings around microtubules and recapitulates the functionality of a kinetochore–microtubule attachment. Mapping of subunits within the complex and their organization within the ring has made it possible to generate a model of Dam1 ring assembly.
Abstract
All eukaryotic cells must segregate their chromosomes equally between two daughter cells at each division. This process needs to be robust, as errors in the form of loss or gain of genetic material have catastrophic effects on viability. Chromosomes are captured, aligned, and segregated to daughter cells via interaction with spindle microtubules mediated by the kinetochore. In Saccharomyces cerevisiae one microtubule attaches to each kinetochore, requiring extreme processivity from this single connection. The yeast Dam1 complex, an essential component of the outer kinetochore, forms rings around microtubules and in vitro recapitulates much of the functionality of a kinetochore–microtubule attachment. To understand the mechanism of the Dam1 complex at the kinetochore, we must know how it binds to microtubules, how it assembles into rings, and how assembly is regulated. We used electron microscopy to map several subunits within the structure of the Dam1 complex and identify the organization of Dam1 complexes within the ring. Of importance, new data strongly support a more passive role for the microtubule in Dam1 ring formation. Integrating this information with previously published data, we generated a structural model for the Dam1 complex assembly that advances our understanding of its function and will direct future experiments.
INTRODUCTION
The accurate segregation of chromosomes between daughter cells is an essential step in cell division. Errors in this process lead to aneuploidy and can result in cell transformation or death (King, 2008). The kinetochore is a network of protein complexes that assembles on centromere regions of chromatin and acts as the connection point between chromatids and the spindle microtubules that segregate them into daughter cells (Westermann et al., 2007; Cheeseman and Desai, 2008). Kinetochores must sense microtubule attachment and determine whether this attachment of sister chromatids is to the same or different spindle poles (Li and Nicklas, 1995; Maresca and Salmon, 2009; Uchida et al., 2009). The stable engagement of a kinetochore is regulated by the Aurora B kinase, the activity of which has been shown to reduce the affinity of several kinetochore proteins for microtubules (Cheeseman et al., 2002; Ruchaud et al., 2007). A single unattached or incorrectly attached kinetochore is sufficient to trigger the spindle assembly checkpoint and halt progress into anaphase, preventing cell division.
Once all of the chromosomes in a cell preparing to divide are correctly bioriented and satisfy the spindle assembly checkpoint, chromosomes must be pulled apart into daughter cells (Westermann et al., 2007; Cheeseman and Desai, 2008). An essential function of the kinetochore is to couple chromosome movement to microtubule depolymerization. Kinetochores are able to track depolymerizing microtubule ends and harness the energy released during microtubule depolymerization to move chromosomes to opposite spindle poles (Koshland et al., 1988).
The molecular mechanisms by which chromosomes attach to kinetochores have remained a great mystery since the mitotic process was first visualized. The spindle microtubule is a dynamic polymer, and the mechanism by which a stable connection to an actively polymerizing/depolymerizing microtubule end can be maintained is still unclear. In budding yeast, in which there is a single microtubule attachment per kinetochore, the heterodecameric Dam1 complex has been shown to be an essential component of the yeast outer kinetochore and to form closed rings around microtubules in vitro (Cheeseman et al., 2001; Miranda et al., 2005; Westermann et al., 2005). The observation that Dam1 complexes can bind microtubules and assemble into rings (Hill, 1985; Miranda et al., 2005; Westermann et al., 2005) captured the imagination of kinetochore biologists and satisfied a prediction made two decades earlier that an encircling coupler could tether chromosomes to disassembling microtubules (Koshland et al., 1988). Functional, in vitro studies followed, showing that Dam1 could recapitulate many essential functions of the kinetochore: it tracked the plus end of a microtubule, generated a highly processive attachment, and could utilize the energy released by microtubule depolymerization to drag a load poleward (Asbury et al., 2006; Westermann et al., 2006). Taken together, these observations strongly point to a model in which the ring form of the Dam1 complex is used as a topological constraint to processively “surf” along the depolymerizing end of the microtubule as the protofilaments peel back (Hill, 1985; Miranda et al., 2005; Westermann et al., 2005), although the physical details of how this molecular coupling is achieved are still under debate (Asbury et al., 2006; Westermann et al., 2006; Grishchuk et al., 2008; for a recent review see Nogales and Ramey, 2009). The attachment of the Dam1 ring to other kinetochore proteins could then stably tether the chromosome to the end of a spindle microtubule.
The in vitro reconstitution of the 10-protein Dam1 complex made it possible to carry out electron microscopy (EM) studies of the complex. EM is ideally suited to visualize the Dam1 complex not only alone, but also, most important, self-assembled around microtubules into rings and spirals that interact in a novel manner with the underlying tubulin (Miranda et al., 2005; Westermann et al., 2005). Three-dimensional (3D) EM reconstructions provided the first structural views of the Dam1 complex and ring structure (Wang et al., 2007; Ramey et al., 2011). The 13-protofilament microtubules are surrounded by 16 repeats of the Dam1 complex oligomerized into a ring, or a turn of a spiral (Westermann et al., 2006), with the most proximal visualized mass of the Dam1 complex positioned ∼20 Å away from the ordered microtubule lattice (Westermann et al., 2005; Wang et al., 2007; Ramey et al., 2011). This distance and the accommodation of different repeats are achieved via interactions that are mediated by flexible elements in the Dam1 complex (likely in the proteins Duo1p and Dam1p; Hofmann et al., 1998; Miranda et al., 2007) and the disordered E-hook of tubulin (C-terminal tail of tubulin, highly charged with glutamic acid residues and involved in interaction with a number of microtubule-associated proteins) (Westermann et al., 2005; Ramey et al., 2011).
A number of studies have provided information on the protein interactions within the Dam1 complex. A two-hybrid and targeted binding assay approach to identifying pairwise binding partners within the complex, as well as binding partners of Dam1p in the rest of the kinetochore, showed Dam1p, Duo1p, and Spc34p to be central interaction hubs in the complex (Shang et al., 2003). A mutational and selective expression approach also shed light on how the complex is organized (Miranda et al., 2007). Absence of Hsk3 results in two subcomplexes—Ask1p-Dad2p-Dad4p (which does not bind to microtubules) and Dam1p-Duo1p-Spc34p-Spc19p-Dad1p-Dad3p (which does bind to microtubules but does not form rings). In the absence of Dam1p, the complex that forms is also missing Duo1p and the Dad1p–Dad3p dimer (which can be expressed and purified independently and forms a stable structural module). Thus Dam1p and Duo1p have been proposed to form a structural unit (Miranda et al., 2007). Recently the Dam1 complex was used as a proof of concept for a novel high-throughput domain interaction mapping technique (Ikeuchi et al., 2010). In this study, two-hybrid interactions between proteins were monitored after generating many truncations of each protein through a selective PCR step. The process favors the minimal PCR product and, therefore, the smallest protein fragment that still supports binding between the two proteins. Using this powerful tool, the authors were able to map the minimal domains needed for binding in a complete map of the complex.
Defining the architecture of the Dam1 complex and its self-assembly into a ring structure is essential for understanding the mechanisms by which rings may contribute to the end-on attachment of spindle microtubules to chromosomes (Shimogawa et al., 2006; Tanaka and Desai, 2008) and how the complex couples microtubule disassembly to processive chromosome movement. It is also crucial for determining how the assembly of the ring could be regulated and how the ring could attach to other components of the kinetochore. To understand the assembly of Dam1 into rings, it is necessary to address two major structural questions: How are the 10 proteins that make up each Dam1 complex organized within the structure of the complex? And how do Dam1 complexes associate when they oligomerize around the microtubule?
RESULTS AND DISCUSSION
Localization of Dam1 complex subunits by maltose-binding protein labeling
Although a wealth of biochemical information has been generated concerning the connectivity between the proteins within the Dam1 complex, structural markers do not yet exist to fit this interaction map onto the structure of the complex. Several regions of the complex are of particular importance. Knowing which proteins exist at the interfaces between complexes within the Dam1 ring could direct genetic and biochemical experiments to further our understanding of the biological importance of Dam1 complex assembly in vivo. In addition, identifying proteins at the Dam1–microtubule interface or those exposed at surfaces available for connection(s) with other elements of the kinetochore is vital to understanding the activities of the Dam1 complex.
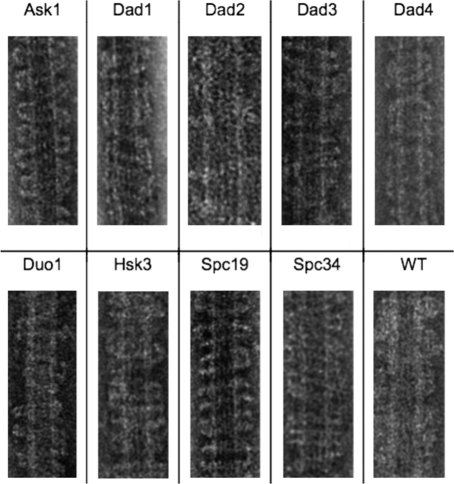
To locate individual protein subunits within the Dam1 complex, we constructed an N-terminal maltose-binding protein (MBP) fusion protein of each of the 10 Dam1 complex proteins and, in each case, purified each fusion protein associated with the respective other nine wild-type subunits of the complex. Our strategy was to image these 10 different tagged complexes by negative-stain EM, to carry out two-dimensional (2D) classification and alignment of particle images, and to generate high–signal-to-noise class averages. Detection of additional density in a class average generated from images of tagged complexes of a particular subunit, compared with wild-type complexes, would indicate the location of the MBP-labeled subunit. This type of strategy was used successfully for other complexes (Bertin et al., 2008; Chen et al., 2008). Ten different complexes were expressed, each with one tagged and nine wild-type subunits. Each was purified through a hexahistidine (6xHis) tag on the MBP using nickel bead chromatography followed by gel filtration. Initially we assumed that some of the tags would be in locations that would interfere with complex formation. Surprisingly, all of our tagged Dam1 complexes expressed well and were purified as complete 10-subunit complexes (Supplemental Figure S1), with one exception: the complex containing the MBP–Dam1p construct failed to express and could not be purified. Each of the other nine complexes containing a tagged subunit was also competent to bind to microtubules and oligomerize into rings or spirals (Figure 1). That the MBP tag does not interfere with oligomerization is likely due to the rather long, 22–amino acid unstructured linker region between the MBP and the start of each fused protein.
FIGURE 1:
Binding of MBP-labeled Dam1 complexes to microtubules and assembly into rings and spirals visualized by negative-stain EM. The MBP-tagged subunit is indicated. All MBP-tagged Dam1 complexes were competent for binding to microtubules and formed rings or spirals around them.
Although the length of the linker avoided the problem of steric hindrance and allowed successful complex reconstitution, its flexibility, in fact, proved a disadvantage in some instances in our efforts to create a detailed structural map of subunit location. Flexibility in the linker prevented the MBP tag from assuming a unique position, blurring out the MBP density in our class averages. Thus we were able to locate only four of the MBP tags, which were N-terminally attached to the four largest subunits within the Dam1 complex. In addition, we previously located the position of the C-terminal end of Dam1p using an alternative approach. Our previous structural analysis of a Dam1 complex containing a truncated form of Dam1p lacking its C-terminus revealed that this region is located in the protrusion of the unassembled complex (Wang et al., 2007). Including results from this additional localization study indicated that the total mass of the labeled proteins accounts for almost three-fourths of the complex. These experiments all rely on the fact that the readout in an EM image is a map of protein density. If mass is added, through the nonintrusive MBP protein tagging, or removed, through a domain deletion analysis, the location of the mass that appeared or disappeared in the resulting class-average images compared with those from wild-type Dam1 complexes is a direct readout of the location of that tagged or truncated protein subunit.
The MBP-tagged complexes were prepared for negative-stain EM as previously described for the wild-type complex (Wang et al., 2007) with small modifications as outlined in Materials and Methods. Particles (images of protein complexes) were then manually selected from the EM micrographs. Reference-free 2D classification and alignment were used to generate class averages with good signal-to-noise ratios. Class averages corresponding to dimers of Dam1 complexes were further analyzed because, in such cases, appearance of extra density corresponding to the MBP label should be present twice and in a similar location for each complex within the dimer. Observing extra densities in two equivalent locations within each dimer class average provided an internal control that the changes seen were in fact due to the added MBP tag.
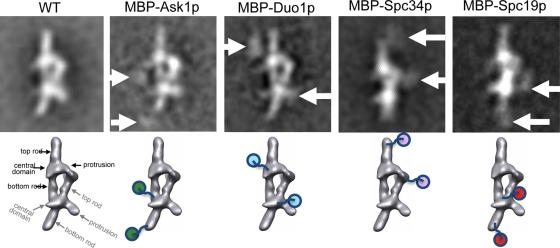
Figure 2 shows five representative class averages corresponding to a view of the Dam1 dimer complex along its main axis. The long, flexible linker between the labeled subunit and the MBP tag gave rise to diffuse density for the MBP. However, in four cases, the MBP tags on the two complexes within a dimer were clearly visible, and the position of these two instances of extra density could be explained in terms of a single location in each of the complexes, as proposed at the bottom in the figure (the leftmost structure shows the domain nomenclature used). For the MBP-Ask1–containing complex the extra densities (marked by arrows in Figure 2) are compatible with labeling at the end of the bottom rod for each Dam1 complex (indicated by ball-and-chain schematics below). For the MBP–Duo1 complex the extra densities are most compatible with an MBP location off the central domain. For the MBP–Spc34 complex the extra densities are located at the top rod, whereas for the MBP–Spc19 complex the extra densities are more consistent with the MBP located at the end of the bottom rod (near the location of Ask1).
FIGURE 2:
Mapping the locations of MBPs in labeled Dam1 complexes. Top, class averages of four different MBP-labeled complexes, with the corresponding class average from wild-type complexes shown for comparison. Arrows point to extra densities attributed to the MBP in the labeled complexes. All of the class averages correspond to dimers, and thus two extra densities are typically seen for each particle. Bottom, the interpretation of MBP densities mapped onto the Dam1 dimer structure. The four main features in each monomer (top rod, central domain, protrusion, and bottom rod) are indicated for the wild type (black for top monomer complex, gray for the bottom one).
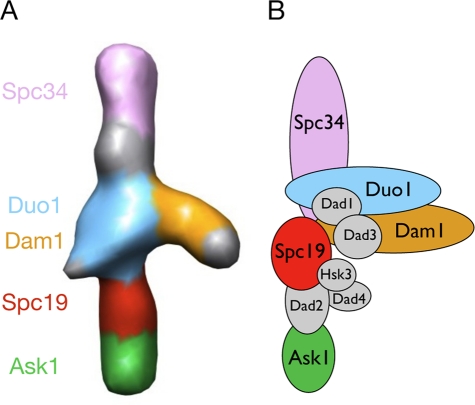
Figure 3A shows the proposed location of 5 of the 10 Dam1 complex subunits, based on the position of the four MBP labels just described and the previously reported position of the C-terminus of Dam1p (Wang et al., 2007). Each of the four largest proteins in the complex—Dam1p, Duo1p, Ask1p, and Spc34p—appears to occupy one of the three major domains—the top or bottom rod or the central domain—and would account for most of the mass density seen in each of their assigned structural regions. With half of the proteins and the great majority of the mass of the Dam1 complex now localized within the wild-type structure through EM imaging, it is possible to generate a model of the structural organization of the entire complex (Figure 3B) by adding constraints from known biochemical interactions (Shang et al., 2003; Miranda et al., 2007; Ikeuchi et al., 2010).
FIGURE 3:
(A) Structure of the Dam1 monomer complex with the locations inferred from our labeling experiments mapped onto it (left). (B) Schematic of the proposed subunit distribution within the complex using the information from our labeling studies (colored subunits localized by MBP labels; Dam1p position based on previous studies using a truncated form of Dam1p), as well as additional biochemical studies.
The Dam1 complex fits into the ring structure without a major conformational change
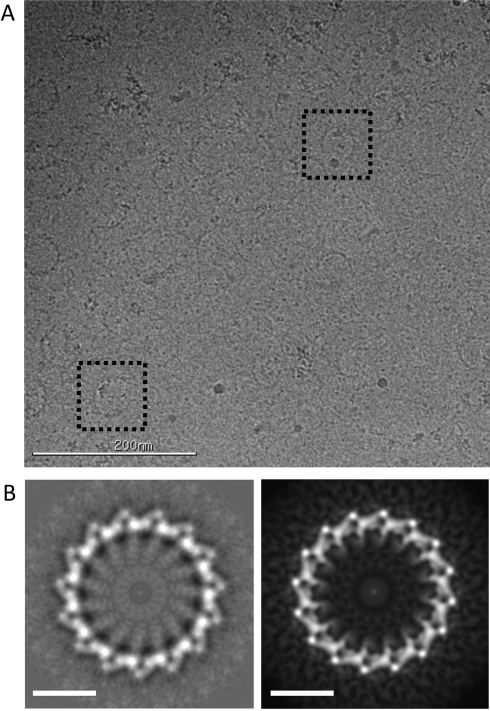
All previously reported EM imaging of the Dam1 complex in the absence of microtubules was carried out using rotary-shadowed (Miranda et al., 2005) or negatively stained samples (Westermann et al., 2005; Wang et al., 2007). The choice of these preparation methods was mostly due to the small size of the complex (∼200 kDa), which makes visualization difficult in the absence of staining agents. At the concentrations of the complex used for negative-stain EM (approximately subnanomolar) and physiological salt concentrations, the complex has a tendency to oligomerize into irregular arches of different sizes. At higher salt concentration the majority of complexes form dimers, with single complexes and trimers also present (Wang et al., 2007). We previously reported the 3D reconstruction of the Dam1 complex dimer using negative-stain and single-particle image analysis (Wang et al., 2007). In addition, cryo-EM was used to obtain the structure of the Dam1 complex assembled as double spirals around microtubules (Wang et al., 2007; Ramey et al., 2011). When we attempted to dock the structure of a single Dam1 complex (obtained by segmentation of the two monomers in the negatively stained dimer, followed by averaging) into the structure of the Dam1 spirals, an obvious fit was not clear. Each Dam1 complex is rod shaped and ∼100 Å in length, with a central domain from which a protrusion emerges. A similar “protruding” density was visible in the spirals, reaching toward the microtubule surface and referred to as the bridge (Wang et al., 2007). Therefore we assumed that the protruding, bridge densities in the spiral corresponded to the protrusion in a single Dam1 complex (as indicated in Figure 2) but repeated in the series of monomers in the spiral. To reach this conclusion, we needed to invoke a large conformational change in a single complex upon its binding to the microtubule in order to fit the cryo-EM density of the Dam1 double spiral (Wang et al., 2007). Later we were able to obtain the cryo-EM structure of the Dam1 ring, likely a more physiologically relevant assembly form of the complex (Ramey et al., 2011). Although the overall shape of the complex in the ring is not too different from the one in the double spirals, the change in shape was sufficient to make the previous docking of the single complex unsatisfactory and thus call into question the validity of our previous assumption of the necessity for a conformational change in the monomer as it assembles into a ring structure. In fact, the repeating shape in the ring was reminiscent of what we observed in the Dam1 dimer structure. To shed light on this issue, we decided to use cryo-EM to visualize the Dam1 complex in the absence of microtubules. These studies required much higher concentrations of the complex. When complex concentrations as high as 500 μM (1000-fold higher than used for negative stain) were used under physiological salt conditions (150 mM NaCl), we made the surprising observation that Dam1 rings clearly formed in the absence of microtubules (Figure 4A). This finding thus suggested that, in contrast to what we previously proposed (Wang et al., 2007), Dam1 does not need to undergo a large, tubulin-induced conformational change upon binding the microtubule in order to self-assemble into closed rings.
FIGURE 4:
The Dam1 complex forms rings and curved oligomers at high concentration in the absence of microtubules. (A) Cryo-EM micrograph of wild-type Dam1 complex over open holes at a concentration of 0.5 mg/ml. Two rings are highlighted by dashed boxes. (B) Averaging and symmetrization of 52 rings produced a representative image of rings in the absence of microtubules (left). Comparison with a reprojection of the structure of the Dam1 ring assembled around microtubules (right) shows that no major conformational change is induced by the microtubule and that two rings are very similar. Scale bars in B, 20 nm.
To further pursue the idea of ring assembly by Dam1 complexes without major conformational rearrangements, we carried out 2D classification and analysis of 52 ring images. Our analysis shows that these rings are composed of 15 repeats (Dam1 complex copies), in contrast with the 16 observed in rings around microtubules (Westermann et al., 2006; Ramey et al., 2011). Density averaging of the rings and imposition of this symmetry resulted in detailed class averages (Figure 4B, left). When compared with the reprojection of the structure of the Dam1 ring assembled around microtubules (Figure 4B, right), it is clear that the repeating subunit has a very similar structure in both and that no major conformational change is induced by the microtubule. One small but significant difference between these two images in Figure 4B, which helps to explain how interpretation of previous results was erroneous, is that the bridge density facing the microtubules is not as clear in the absence of the tubulin polymer (Figure 4B, left), suggesting that it corresponds to a flexible region of the complex that is averaged out in studies carried out in the absence of microtubules but becomes at least partially ordered when involved in microtubule interactions. Indeed, the idea that the connections between the Dam1 complex and the microtubule involve flexible “arms” has been proposed based on biochemical data (Miranda et al., 2007), which would explain the absence of these domains in the averaged, unassembled structure: they only become stabilized and therefore visible after averaging when bound to microtubules.
Without side views of the Dam1 rings in the absence of microtubules, we were prevented from obtaining a 3D reconstruction of these assemblies. We therefore used the present 2D analysis of the frozen-hydrated Dam1 rings as a guide to pursue the docking of the previous negative-stain Dam1 dimer structure (EMDB 1372; Wang et al., 2007) into the density of the full ring around microtubules (EMDB 5254; Ramey et al., 2011). Of importance, if the bridge density in the cryo-EM reconstruction of the ring is not considered, eight negative-stain dimer structures can be directly placed around one ring, repeating the contacts between each complex within the dimer around the ring (Figure 5). Thus the ring assembly on a microtubule is just a continuation of the self-association that Dam1 complexes experience on their own, likely facilitated by concentrating the complexes onto the microtubule lattice via interactions mediated by flexible structural elements. This conclusion is in agreement with our previously published observation that some Dam1 rings form in the absence of microtubules when the complex is bound to a lipid monolayer containing Ni-bound nitriloacetic acid lipids (which interacted with the His tag in the complex) or negatively charged phospholipids (Westermann et al., 2005). The availability of this new docking model for the full ring around the microtubule allows us to place our Dam1 subunit localization findings within this larger context.
FIGURE 5:
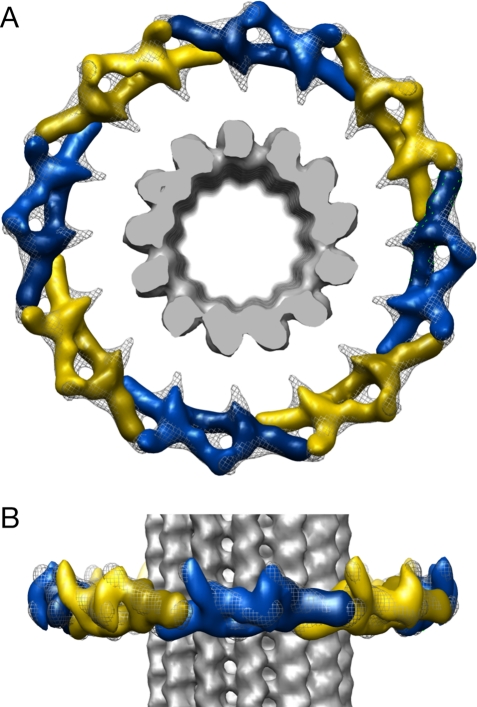
Docking of dimer structures into the assembled ring. (A) Top view and (B) side view. Each dimer is shown in either yellow or blue, with varying hues to identify monomer subunits. The ring structure is shown as a gray mesh, and the microtubule is shown as solid gray.
As shown in Figures 5 and 6, a Dam1 dimer clearly fits into two consecutive repeats of the ring, with no observable conformational rearrangement necessary, if the protrusion in the structure of the complex is allowed to point outward instead of toward the microtubule, in opposition to what was previously proposed (Wang et al., 2007). This much more parsimonious docking perfectly agrees with the idea that ring assembly and closure are attained by repeating the contacts we previously described in the context of a Dam1-complex dimer. The bridge density in the ring that becomes more ordered upon binding to the microtubule appears to emanate from the central domain of the complex, from which density can be seen at a lower threshold both in the 3D reconstruction (Wang et al., 2007) and in 2D class averages (like those shown in Figure 2) of the negatively stained samples in the absence of microtubules.
FIGURE 6:
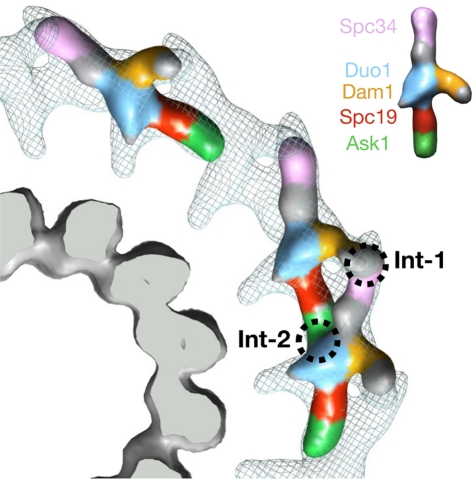
Docking of the subunit-“painted” monomer and dimer structures into the Dam1 ring structure. The microtubule (bottom left) is rendered as solid gray. One-fourth of the Dam1 ring is shown as a gray mesh. The excellent agreement of the densities for both dimer and monomer indicates that no large conformational changes occur during ring assembly and that the two interaction sites (Int1 and Int2) between monomers within the dimer are used for growth and closure of the ring. The color code for the subunits is indicated in the Dam1 monomer shown in the top right corner.
Organization of Dam1 complex subunits within the Dam1 ring around microtubules
The updated docking now allows us to fit the assembled structure of Figure 3, showing the mapped subunits of the complex, and generate a model for the interaction sites in the assembled Dam1 ring around microtubules (Figure 6). The Dam1 ring is assembled via two points of interaction between adjacent subunits. The first point (Int1) is distal from the microtubule: the protrusion extending from the central domain interacts with the top rod of the consecutive Dam1 complex, which was labeled in this work by MBP–Spc34. The central region of the protrusion, shown in gold in Figure 6, is reduced in the unassembled structure of ΔC-Dam1 mutant complex (Wang et al., 2007), indicating that it is formed in part by the C-terminus of Dam1p.
Ask1p appears to form the bulk of the bottom rod of the Dam1 complex monomer (Figures 3 and 6). The end of this domain joins with the central domain of the next complex along a ring, forming the second oligomerization interface, Int2. The N-terminal domain of Ask1p localized by our MBP tag was previously shown to be the region that interacts with the rest of the complex (Ikeuchi et al., 2010). It is noticeable that in that study the vast majority of Ask1p does not appear to participate in binding interactions with the rest of the complex. Its large C-terminal region contains two Cdc28 phosphorylation sites. In our structural model this region would likely correspond to the tip of the bottom rod, which touches the central domain of the adjacent monomer. If this interface did contain the Cdc28 phosphorylation sites, it would constitute an additional mechanism for the regulation of Dam1 assembly. One important question concerning the interpretation of the interaction study by Ikeuchi et al. (2010) is whether the pairwise interactions represent intracomplex or intercomplex interactions.
To independently verify the subunit arrangement proposed in Figure 6, we obtained cryo-EM reconstructions for two of the MBP-tagged complexes. Unfortunately, the reconstruction of the double-Dam1 spiral formed using the MBP–Spc34p complex did not render a clear density difference map when compared with the assembly of the wild-type complex (data not shown). However, the double spiral formed by the MBP–Ask1 Dam1 complex does show extra density on the inner face of the spiral, in agreement with the position of Ask1 in our model (Supplemental Figure S3).
Of interest, the C-terminal domains of both Dam1p and Ask1p were shown not to participate in intracomplex interactions and to be posttranslationally modified by mitotic regulatory proteins (Cheeseman et al., 2002; Li and Elledge, 2003; Zhang et al., 2005; Shimogawa et al., 2006). These regions are both, however, positioned at interfaces between complexes in the ring, suggesting that these C-terminal domains may be important interfaces for complex oligomerization and that their posttranslational modification may regulate their function in vivo by regulating their assembly state. Phosphomimetic mutants of both Dam1p and Spc34p were shown to specifically decrease binding between those two proteins in what we are proposing is an intercomplex interface (Shang et al., 2003). Indeed, our previous EM analysis of phosphomimetic Dam1p complexes showed decreased dimer and ring formation but no obvious destabilization of the decameric complex, suggesting that the Spc34p–Dam1p interaction modulated by phosphorylation is between complexes (Wang et al., 2007). According to our structural model, phosphorylation of Dam1p, Spc34p, or both would weaken this outer interface and therefore reduce oligomerization.
Dam1p is a major target of the spindle checkpoint kinase Ipl1p, and phosphorylation of the Dam1 complex promotes detachment of kinetochores, thereby “resetting” the kinetochore for another try at correct bioriented spindle attachment (Cheeseman et al., 2002). Of interest, a phosphomimetic mutant of the Ipl1p sites in Dam1p (three of four of which are at the Dam1p C-terminus) shows little effect on the binding of the Dam1 complex to the microtubule but causes a reduction in ring assembly (Westermann et al., 2005). This finding agrees with a distant location of this region from the microtubule and its close location to the protrusion tip involved in Int1. Thus our proposed docking would suggest a model in which phosphorylation of Dam1p by Ipl1p weakens this distal interface and disrupts oligomerization, causing ring breakdown while leaving the flexible domain that is responsible for microtubule binding unchanged. Our model suggests that one role of Ipl1p might be to regulate the formation of the Dam1 ring and cause the kinetochore to lose this processive attachment to the spindle.
As mentioned earlier, the second interface, labeled Int2 in Figure 6, is formed by the interaction between the Ask1–MBP lobe and the large, central domain of the next Dam1 complex. This central domain likely contains several subunits. From previous interaction maps and our labeling studies, this domain is likely to include a significant part of Duo1p and Dam1p, the small proteins Dad1p and Dad3p, and possibly elements of Spc34p and Spc19p. Therefore it is unclear exactly which subunit(s) are involved in this second interface. As with Int1, Int2 may contain modified residues that could regulate assembly. Both Dam1p and Ask1p are posttranslationally modified. Dam1p is methylated in vivo by Set1 at lysine 233 (Zhang et al., 2005) and phosphorylated by Ipl1 on serines S20, S257, S265, and S292 (Cheeseman et al., 2002) and Mps1 on serines S13, S49, S217, S218, S221, and S232 (Shimogawa et al., 2006). Mutating these regulatory sites causes spindle defects in vivo (Cheeseman et al., 2002; Zhang et al., 2005). Ask1p is phosphorylated at two sites on its C-terminus by Cdc28. Abolishing phosphorylation at these sites in vivo sickens temperature-sensitive ask1 mutant yeast (Li and Elledge, 2003).
The present model locates the largest subunits of the complex, Dam1p and Spc34p, most outwardly in the ring and thus points at them as major potential sites of interaction with other kinetochore components. In agreement with this proposal, two-hybrid and binding assays identified these two Dam1-complex subunits as interacting with the Ndc80p subunit of the Ndc80 complex (Shang et al., 2003). Of importance, this interaction was shown to be regulated by phosphorylation. A functional, direct interaction between these two complexes was indeed recently demonstrated in vitro (Lampert et al., 2010; Tien et al., 2010).
On the other hand, Duo1p is the most proximal subunit to the microtubule surface based on our studies. Both Dam1p and Duo1p were reported to bind to microtubules (Hofmann et al., 1998; Cheeseman et al., 2001). Now that we have mapped the C-terminus of Dam1p within the outer arms of the ring, it seems likely that the N-terminal region of Dam1p and some or all of Duo1p are contributing to the flexible domains seen extending inward toward the microtubule (Figure 6). Limited proteolysis experiments showed Dam1p, Ask1p, and Duo1p to be digested after a 2-h treatment with elastase, both with and without microtubules (Miranda et al., 2007). Of interest, the N-terminal portion of Duo1p seemed to be cleaved specifically. This suggests that the C-terminal domain of Duo1p and the N-terminal domain of Dam1p make up the microtubule-binding interface. Ask1p may be a flexible attachment to other elements in the kinetochore or the microtubule, although no microtubule-binding activity has been attributed to it.
In summary, through labeling experiments, using both mutants and MBP fusion constructs, we have now localized five of the 10 proteins subunits within the Dam1 complex. This information, combined with protein–protein interaction studies, has led us to propose a model for the layout of the complex and to suggest the involvement of different subunits in self-assembly contacts and in interactions with microtubules and other kinetochore components. The N-termini of Ask1p and Spc34p and the C-terminal domain of Dam1p are all at or near interfaces between Dam1 complexes within the ring. Each of these subunits is posttranslationally modified during mitosis by regulatory kinases and/or methyltransferases, suggesting that these interactions may be modulated by the spindle checkpoint in order to ensure correct chromosome segregation.
Previous structural data led us to conclude that the Dam1 complex underwent a large conformational change upon binding the microtubule (Wang et al., 2007). Our image analysis of Dam1 rings formed in the absence of microtubules and the cryo-EM structure of the ring form of Dam1 around microtubules now allow us to propose a more convincing and parsimonious docking of the monomer complex within the ring. These studies strongly indicate that in fact there is not a large conformational change on the Dam1 complex upon binding to the microtubule and that the intercomplex interfaces seen in the Dam1 complex dimer are indeed preserved within the ring around microtubules.
With the complex defined at the resolution of 3 nm in several different forms, much could be learned from an atomic structure of either the full complex or any of the subcomplexes that are stable. Unfortunately, no Dam1 component structures have been reported. The labeling data and unique shape of the complex would allow for an unambiguous fit of an atomic structure into both the assembled and unassembled structures. This could lead to a mechanistic understanding of Dam1 function and would undoubtedly suggest many further experiments.
MATERIALS AND METHODS
Purification of MBP-tagged recombinant Dam1 complexes
Tagged Dam1 complexes were produced by the MacroLab user facility (QB3, University of California, Berkeley) using the following strategy. The 6xHis tag present on Hsk3p in a previously described PolyCistronic vector containing all 10 wild-type proteins (Westermann et al., 2005) was removed. Each of the 10 wild-type genes was cloned into an MBP fusion plasmid (6xHis-MBP-[22 amino acid linker]-protein). Generation of the tagged complexes involved coexpression of two plasmids at a time, the PolyCistronic vector for the 10 wild-type proteins lacking a tag, and one of the MBP-protein plasmids containing also a 6xHis tag. Each complex was then purified through the 6xHis tag using nickel bead chromatography followed by gel filtration.
Electron microscopy and image analysis
The tagged complexes were prepared for negative-stain EM as previously described for the wild-type complex, with small modifications (Wang et al., 2007), except that the samples were prepared without an extra “sandwich” layer of carbon. Each complex was imaged on a Tecnai 12 microscope with either film or a 4k × 4k digital CMOS Tietz camera (Tietz Video and Image Processing Systems, www.tvips.com). Data from both sources were treated using the same computational approaches. Images were assessed visually for good stain, and particles were then manually selected without regard for whether they appeared to be monomeric or dimeric versions of the complex. Reference-free class averages were then generated using an iterative alignment and classification script using the IMAGIC (van Heel et al., 1996) and CAN (Ogura et al., 2003; Ramey et al., 2009) software packages. The docking of the negatively stained Dam1 complex dimer density into the cryo-EM ring reconstruction was performed using the symmetrical fit command in the UCSF Chimera package (Pettersen et al., 2004), which allowed us to account for the symmetry of the ring and the multiple dimer copies in an automated and objective way.
Supplementary Material
Acknowledgments
We are grateful to Patricia Grob and Tom Houweling for technical support and to David Drubin for his comments on the manuscript. Figures 5 and 6 and Supplemental Figure 3 were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR001081). This work was funded by National Institute of General Medical Sciences grants to E.N. (2PO1GM51487C) and G.B. (R01GM47842). E.N. is a Howard Hughes Medical Institute Investigator.
Abbreviations used:
- EM
electron microscopy
- MBP
maltose-binding protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0659) on September 30, 2011.
REFERENCES
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park SS, Garcia G 3rd, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR 3rd, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2008;105:10326–10331. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi A, Nakano H, Kamiya T, Yamane T, Kawarasaki Y. A method for reverse interactome analysis: high-resolution mapping of interdomain interaction network in Dam1 complex and its specific disorganization based on the interaction domain expression. Biotechnol Prog. 2010;26:945–953. doi: 10.1002/btpr.403. [DOI] [PubMed] [Google Scholar]
- King RW. When 2 + 2 = 5: the origins and fates of aneuploid and tetraploid cells. Biochim Biophys Acta. 2008;1786:4–14. doi: 10.1016/j.bbcan.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Elledge SJ. The DASH complex component Ask1 is a cell cycle-regulated Cdk substrate in Saccharomyces cerevisiae. Cell Cycle. 2003;2:143–148. [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Miranda JJ, King DS, Harrison SC. Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol Biol Cell. 2007;18:2503–2510. doi: 10.1091/mbc.E07-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Ramey VH. Structure-function insights into the yeast Dam1 kinetochore complex. J Cell Sci. 2009;122:3831–3836. doi: 10.1242/jcs.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol. 2003;143:185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ramey VH, Wang HW, Nakajima Y, Wong A, Liu J, Drubin D, Barnes G, Nogales E. The Dam1 ring binds to the E-hook of tubulin and diffuses along the microtubule. Mol Biol Cell. 2011;22:457–466. doi: 10.1091/mbc.E10-10-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey VH, Wang HW, Nogales E. Ab initio reconstruction of helical samples with heterogeneity, disorder and coexisting symmetries. J Struct Biol. 2009;167:97–105. doi: 10.1016/j.jsb.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa MM, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by Aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Wang HW, Ramey VH, Westermann S, Leschziner AE, Welburn JP, Nakajima Y, Drubin DG, Barnes G, Nogales E. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. The Set1 methyltransferase opposes Ipl1 Aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.