This study shows that the eIF2 kinase PERK is required not only for translational control but also for activation of ATF6 and its target genes in the unfolded protein response. The PERK pathway facilitates both the synthesis of ATF6 and trafficking of ATF6 from the endoplasmic reticulum to the Golgi for intramembrane proteolysis and activation of ATF6.
Abstract
Disruptions of the endoplasmic reticulum (ER) that perturb protein folding cause ER stress and elicit an unfolded protein response (UPR) that involves translational and transcriptional changes in gene expression aimed at expanding the ER processing capacity and alleviating cellular injury. Three ER stress sensors (PERK, ATF6, and IRE1) implement the UPR. PERK phosphorylation of the α subunit of eIF2 during ER stress represses protein synthesis, which prevents further influx of ER client proteins. Phosphorylation of eIF2α (eIF2α∼P) also induces preferential translation of ATF4, a transcription activator of the integrated stress response. In this study we show that the PERK/eIF2α∼P/ATF4 pathway is required not only for translational control, but also for activation of ATF6 and its target genes. The PERK pathway facilitates both the synthesis of ATF6 and trafficking of ATF6 from the ER to the Golgi for intramembrane proteolysis and activation of ATF6. As a consequence, liver-specific depletion of PERK significantly reduces both the translational and transcriptional phases of the UPR, leading to reduced protein chaperone expression, disruptions of lipid metabolism, and enhanced apoptosis. These findings show that the regulatory networks of the UPR are fully integrated and help explain the diverse biological defects associated with loss of PERK.
INTRODUCTION
The endoplasmic reticulum (ER) is a site for calcium storage, lipid biosynthesis, and the folding and assembly of proteins destined for the secretory pathway. Calcium dysregulation, oxidative damage, and perturbations in posttranslational modification of proteins can lead to accumulation of misfolded protein that can cause ER stress. Accumulation of misfolded protein and the ensuing ER stress is referred to as proteotoxicity, which can contribute to the etiology of diseases including diabetes, cancer, and neurodegenerative disorders (Schroder and Kaufman, 2005; Marciniak and Ron, 2006; Ron and Walter, 2007; Wek and Cavener, 2007; Hotamisligil, 2010). ER stress activates three transmembrane proteins—ATF6, IRE1 (ERN1), and PERK (PEK/EIF2AK3)—which together induce the unfolded protein response (UPR), involving both transcriptional and translational regulation of genes that serve to expand the processing capacity of the ER and return this organelle to homeostasis (Ron and Walter, 2007; Wek and Cavener, 2007).
ATF6 is a membrane-bound transcription factor that is localized to the ER and serves as both a sensor of ER stress and a transcriptional activator of UPR target genes. There are two ER stress-responsive isoforms of ATF6 (α/β) (Haze et al., 2001; Thuerauf et al., 2004, 2007), with the predominant activator of UPR target genes, ATF6α (ATF6), being the primary focus of this study. During ER stress, ATF6 transits from the ER to the Golgi, where regulated intramembrane proteolysis by site-1-protease (S1P) and site-2-protease (S2P) liberates the N-terminal portion of ATF6 (DeBose-Boyd et al., 1999; Shen and Prywes, 2004; Ron and Walter, 2007). The resulting ATF6(N) then enters the nucleus and binds to ER stress response elements and unfolded protein response elements (ERSEs and UPREs) located in the promoters of UPR targeted genes (Yamamoto et al., 2004, 2007; Ron and Walter, 2007; Wu et al., 2007; Adachi et al., 2008). Activation of ATF6 leads to increased transcription of a network of genes, including those encoding ER chaperones, such as BiP/GRP78, and components of the ER-associated degradation (ERAD) pathway (Ron and Walter, 2007; Wu et al., 2007; Yamamoto et al., 2007; Adachi et al., 2008). Deletion of ATF6 in mice does not lead to an overt developmental phenotype; however, ATF6-null mice subjected to pharmacological induction of ER stress showed dysregulation of chaperone and metabolic genes associated with lipid homeostasis leading to hepatic steatosis not found in the similarly treated wild-type (WT) animals (Wu et al., 2007; Yamamoto et al., 2007, 2010; Rutkowski et al., 2008). Central to the UPR dysregulation in the ATF6−/− mice is a persistent activation of IRE1, an endoribonuclease that facilitates the cytoplasmic splicing of XBP1 mRNA, leading to the synthesis of an active form of the XBP1 transcription factor (Yoshida et al., 2001; Calfon et al., 2002; Schroder and Kaufman, 2005). Activated XBP1 enhances the transcription of UPR genes involved in protein quality control, disulfide linkage, ERAD, and lipid synthesis (Ron and Walter, 2007; Lee et al., 2008; Glimcher, 2010). The endoribonuclease function of IRE1 also facilitates the degradation of many mRNAs during ER stress, further contributing to changes of the transcriptome (Hollien and Weissman, 2006; Hollien et al., 2009).
The translational control arm of the UPR is directed by PERK phosphorylation of eIF2, a translation initiator factor that combines with guanosine triphosphate (GTP) and delivers the initiator Met-tRNAiMet to the translational machinery. Phosphorylation of the α subunit of eIF2 at Ser-51 blocks the activity of eIF2B required for the exchange of eIF2-GDP (guanosine diphosphate) to the active version eIF2-GTP (Wek and Cavener, 2007; Sonenberg and Hinnebusch, 2009). The subsequent reduction in protein synthesis prevents further influx of nascent polypeptides into the ER and provides the cell ample time to implement the UPR reprogramming of the transcriptome. Accompanying this repression of global protein synthesis, eIF2α phosphorylation (eIF2α∼P) also enhances the translation of select mRNAs, such as that encoding ATF4, a transcriptional activator of genes involved in metabolism, cellular redox status, and regulation of apoptosis (Harding et al., 2000a, 2003; Lu et al., 2004; Vattem and Wek, 2004; Ron and Walter, 2007; Wek and Cavener, 2007). Among the ATF4 target genes are additional transcription factors, such as CHOP (GADD153/DDIT3), which serve to amplify the restructuring of the transcriptome to manage stress or direct cell fate toward apoptosis (Fawcett et al., 1999; Harding et al., 2000a; Marciniak et al., 2004; Marciniak and Ron, 2006).
In addition to ER stress, preferential translation of ATF4 occurs in response to diverse stress conditions that regulate other eIF2α kinases, including GCN2 (EIF2AK4) activated by nutrient deprivation, HRI (EIF2AK1) induced by heme deficiency and oxidative stress, and PKR (EIF2AK2), which participates in the cellular defense against viral infection (Wek and Cavener, 2007; Sonenberg and Hinnebusch, 2009). Because ATF4 is a common downstream target of each of the eIF2α kinases and their corresponding activating stress signals, this eIF2α∼P/ATF4 pathway has been referred to as the integrated stress response (ISR; Harding et al., 2000a, 2003). The ISR network is critical for ameliorating stress conditions, such as those afflicting the ER organelle, as genetic perturbations of the eIF2α kinases have significant medical consequences. For example, PERK disruption leads to Wolcott-Rallison syndrome, which is characterized by neonatal diabetes, atrophy of the exocrine pancreas, skeletal dysplasia, growth retardation, and hepatic complications resulting in morbidity (Delepine et al., 2000; Senee et al., 2004; Julier and Nicolino, 2010). These pathologies were recapitulated in PERK-deficient mice (Harding et al., 2001; Zhang et al., 2002), in which the molecular and cellular mechanisms have been investigated (Li et al., 2003; Zhang et al., 2006b; Iida et al., 2007; Gupta et al., 2010).
Activation of ATF6, IRE1, and PERK in response to ER stress is thought to occur in parallel, but the timing or duration of each may differ. Although the mechanistic details are not yet resolved, it has been proposed that the ER luminal portions of each of these sensory proteins can be bound and repressed by BiP (Shen et al., 2002; Ron and Walter, 2007). Accumulation of unfolded proteins in the ER lumen is suggested to compete with the sensory proteins for BiP binding, enhancing release of this ER chaperone from each ER sensor and thus leading to their activation (Bertolotti et al., 2000; Ma et al., 2002; Shen et al., 2002; Ron and Walter, 2007; Pincus et al., 2010). Alternatively, it has been proposed that unfolded proteins can directly interact with the ER sensor proteins, such as IRE1, which then facilitates oligomerization and activation (Credle et al., 2005; Kimata et al., 2007; Ron and Walter, 2007; Cui et al., 2011). These models posit that activation of the sensory proteins in response to ER stress occurs largely by independent rather than by interconnected processes.
We wished to determine whether the translational control arm of the UPR is coupled to the regulation of the transcriptional components of this stress response, insuring coordinate control of the transcription and translation phases of the UPR. Our studies show that the PERK/eIF2α∼P/ATF4 pathway is required to facilitate activation of ATF6 in response to ER stress both in vivo and in cultured cells. As a consequence, liver-specific deletion of PERK leads to enhanced apoptosis in response to ER stress, along with hepatic dysfunctions, features that are similar to those described for ATF6-null mice. These findings are important because development of specific therapies targeting the UPR will depend on understanding the integration of this signaling network. In this model, loss of translational control of the UPR also significantly blocks the transcriptional arms, resulting in dysregulation of the UPR network that would ultimately contribute to disease.
RESULTS
Loss of PERK disrupts the UPR and renders the liver susceptible to ER stress
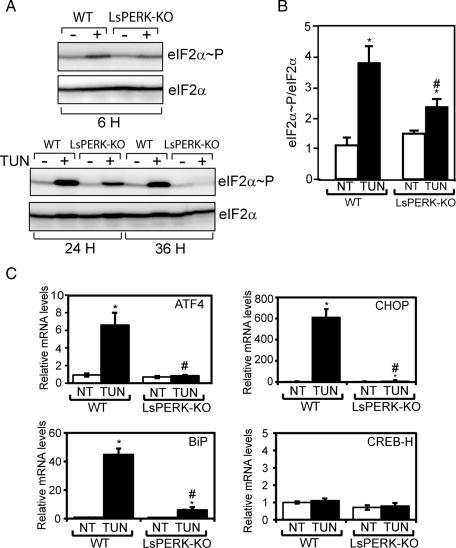
To address the role of PERK in the UPR in liver, we bred mice with a liver-specific knockout of PERK (LsPERK-KO). LsPERK-KO mice were produced by deletion of floxed PERKfl/fl using cre expression driven by the liver-specific albumin promoter, as described previously (Zhang et al., 2002; Bunpo et al., 2009). Consistent with PERK depletion from the liver, there was significantly reduced eIF2α∼P following 6 h after injection with tunicamycin (1 mg/kg), a potent ER stress agent that blocks N-glycosylation of proteins (Figure 1, A and B). Disruption of eIF2α∼P in the LsPERK-KO livers was also observed following 24- and 36-h treatments with tunicamycin. In addition to translational expression of ATF4, induced eIF2α∼P enhances the transcription of ATF4 and its target gene CHOP, and the levels of both mRNAs were sharply reduced in the LsPERK-KO mice during ER stress (Figure 1C). Expression of CREB-H, which has been previously linked to the UPR (Zhang et al., 2006a), was unchanged during the treatment with tunicamycin.
FIGURE 1:
Liver-specific knockout of PERK reduces eIF2α∼P and the ISR in response to tunicamycin treatment. (A) WT and LsPERK-KO mice received intraperitoneal injections with tunicamycin (TUN), and following 6, 24, or 36 h of exposure to this ER stress agent, the levels of eIF2α∼P and total eIF2α in livers were measured by immunoblot analyses. The + indicates treatment with tunicamycin, and the – indicates injection with only excipient. Results shown in each panel are representative of at least three independent experiments. (B) Quantification of the levels of total and phosphorylated eIF2α in the WT and LsPERK-KO livers following 6 h of treatment with tunicamycin (TUN) or with excipient (NT, no treatment). (C) The levels of ATF4, CHOP, BiP, and CREB-H mRNAs in the WT and LsPERK-KO livers exposed to tunicamycin for 6 h, or to no treatment, were measured by qPCR. * indicates statistically significant differences (p < 0.05) between the levels of tunicamycin treated (TUN) and no treatment (NT) samples; # indicates statistically significant differences (p < 0.05) between WT and LsPERK-KO mice. The error bar represents the SEM.
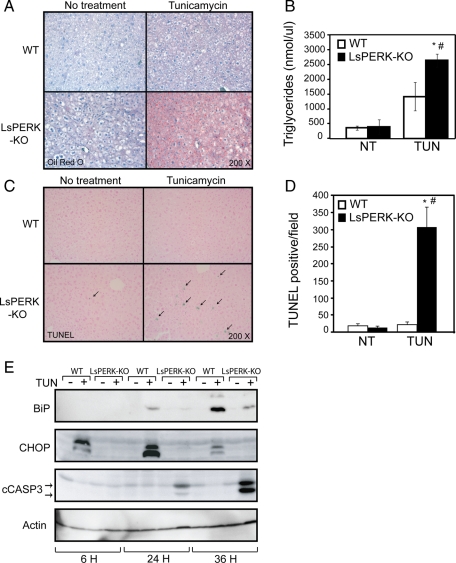
Accompanying this block in PERK activity and downstream gene expression, livers for the LsPERK-KO mice displayed substantial Oil Red O staining, indicating the presence of fat deposits in response to 12-h tunicamycin treatment (Figure 2A). This staining was significantly reduced in the WT mice. Triglyceride levels were also increased in livers of the LsPERK-KO mice treated with tunicamycin as compared with the WT (Figure 2B). After 24 h of the ER stress, increased terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining of hepatocytes in PERK-deficient mice indicated enhanced apoptosis in the liver (Figure 2, C and D). The TUNEL staining was accompanied by elevated cleavage of caspase-3 in the LsPERK-KO livers (Figure 2E). As expected, induction of CHOP protein in response to ER stress was substantially diminished in the LsPERK-KO livers, consistent with the idea that activation of the ISR was blocked as illustrated by the lowered levels of eIF2α∼P. Interestingly, the mRNA and protein levels of the ER chaperone BiP were also substantially reduced with loss of PERK function, with only modest levels of BiP protein in the LsPERK-KO liver samples following 36-h tunicamycin treatment (Figures 1C and 2E). These results suggest that PERK deficiency blocks induction of eIF2α∼P and the translational control arm of the UPR, which leads to disruption of liver homeostasis and increased apoptosis during ER stress.
FIGURE 2:
Loss of PERK enhances liver apoptosis in response to ER stress. (A) WT and LsPERK-KO mice received intraperitoneal injections with tunicamycin or excipient (NT, no treatment). Following 12 h of exposure to the ER stress agent, a histological analysis of the livers was carried out using Oil Red O stain to visualize triglycerides, and liver sections were counterstained with hematoxylin. (B) Quantification of triglyceride levels in WT and LsPERK-KO liver samples following treatment with tunicamycin (TUN) or vehicle control (NT). Dysregulation of metabolic genes was also noted and can be found in Supplemental Figure 2. (C) WT and LsPERK-KO mice were treated with tunicamycin for 24 h, and a TUNEL analysis was carried out on liver histological sections, with the arrows indicating TUNEL-positive cells in a representative field. (D) Quantification of the TUNEL-positive levels in WT and LsPERK-KO livers following treatment with tunicamycin (TUN) or with no treatment (NT). The error bar represents the SEM, and the * indicates statistically significant differences (p < 0.05) between the tunicamycin treated and no treatment samples; # indicates statistically significant differences (p < 0.05) between WT and LsPERK-KO mice. (E) WT and LsPERK-KO mice were treated with tunicamycin (+) or vehicle only (-) for 6, 24, or 36 h, and the levels of BiP, CHOP, and cleaved caspase 3 in liver lysates were measured by immunoblot analysis.
Loss of PERK in liver blocks activation of the UPR transcriptome
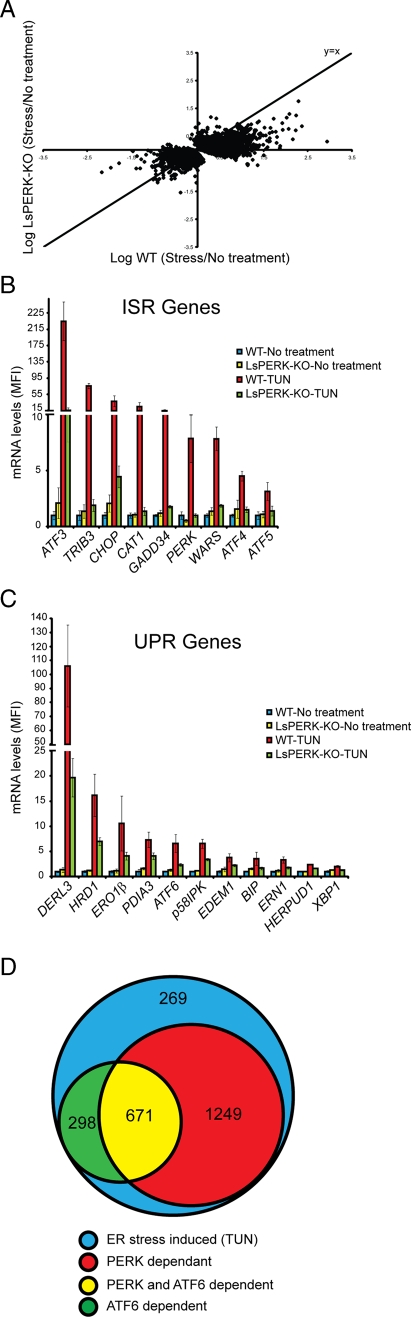
An analysis of gene expression changes in the LsPERK-KO mice indicated that there was significantly diminished induction of BiP expression in response to ER stress. Lowered BiP levels suggested that, in the liver, PERK normally enhances the expression of key ER chaperones, a function previously ascribed to ATF6 and IRE1/XBP1 (Lee et al., 2002; Yamamoto et al., 2004; Schroder and Kaufman, 2005; Ron and Walter, 2007). To investigate changes in UPR genes that require PERK, total RNA was isolated from WT and LsPERK-KO livers and analyzed by Affymetrix microarrays. In WT livers, 9645 probe sets were significantly changed by tunicamycin treatment, with nearly 50% of these being induced by twofold or greater (Table 1; p ≤ 0.05, FDR ≤ 0.15). A comparison between the induction ratios (stress/control) for the ER stress-induced genes in the WT versus the PERK-deficient livers indicated that 63% (4599/7324) displayed a significantly lower induction ratio in the LsPERK-KO mice. Furthermore, among those genes enhanced at least twofold by tunicamycin treatment, 74% (3523/4737) showed a requirement for PERK for full induction in response to ER stress (Table 1). These finding are illustrated in Figure 3A, which features a comparative analysis between the stress-induced transcriptome changes in WT (x-axis) versus LsPERK-KO (y-axis). The PERK requirement for changes in the UPR transcriptome is supported by the large number of probe sets that were increased in the WT livers upon ER stress, but lie below the diagonal where y = x.
TABLE 1:
PERK facilitates expression of a large number of UPR target genes.
| Fold change (treated/untreated)a | |||||
|---|---|---|---|---|---|
| Dependence | Total | >1.0 | ≥2.0 | <1.0 | ≤0.5 |
| WT | 9645b | 7324 | 4737 | 2321 | 797 |
| PERK-dependent | 4948c | 4599 | 3523 | 349 | 190 |
aExpression profile summary of gene transcripts, as defined by probe set, that were significantly changed after treatment with tunicamycin (1 mg/kg). The number of induced genes are shown for both the >1.0-fold and ≥2.0-fold thresholds. The repressed genes are also shown for the <1.0- and <0.5-fold thresholds.
bThe total number of transcripts significantly changed (p ≤ 0.05) by ER stress.
cThe number of PERK-dependent transcripts that were significantly changed by ER stress.
FIGURE 3:
PERK is required for full induction of a majority of the UPR-targeted genes. (A) The scatter plot is the log ratio of stressed vs. no stress for WT (x-axis) and LsPERK-KO (y-axis). This comparative analysis of gene transcripts induced or repressed by ER stress in the livers of WT mice and LsPERK-KO mice displays all significant probe sets (p < 0.05) following treatment with 2 μM tunicamycin for 6 h. (B and C) Microarray measurements of gene transcripts targeted by the ISR (B) and the UPR (C). The mean fluorescence intensity (MFI) for each transcript is shown as a histogram, along with the SD. Each of these representative genes showed a significant induction in response to ER stress that was dependent on PERK (p ≤ 0.05, FDR ≤ 0.15). (D) Comparative analysis of genes induced by tunicamycin in mice livers. The LsPERK-KO microarray analysis described here was compared with microarray measurements derived from ATF6 liver-specific knockout mice as reported by Rutkowski et al. (2008). Genes significantly induced by at least twofold were plotted as Venn diagrams. Shown here are genes induced by tunicamycin in liver that were present in both microarray data sets (blue), with ATF6-dependent genes (green), PERK-dependent genes (red), or PERK- and ATF6-dependent genes (yellow).
These results indicate that PERK is a major participant in the transcriptional phase of the UPR, with a majority of activated genes requiring PERK function for full induction. As expected, these PERK-dependent genes included canonical ISR target genes such as ATF4, CHOP, GADD34, ATF3, and TRIB3 (Figure 3B). Many UPR target genes, however, also required PERK for full induction in response to tunicamycin treatment, including those involved in protein folding and ERAD (Figure 3C). To address the apparent overlap between PERK and ATF6 transcriptional target genes, we compared a previously reported microarray characterization of livers from ATF6 knockout mice treated with tunicamycin with our LsPERK-KO data set (Rutkowski et al., 2008). A total of 2487 genes were significantly induced by ER stress in both data sets, with 39% (969/2487) of these requiring ATF6 for full induction (Figure 3D). Importantly, among these ATF6-target genes, 69% (671/969) were also dependent on PERK for full induction. We conclude that PERK facilitates the expression of not only the ISR genes, but also a majority of the UPR genes, including those targeted by ATF6.
Turnover of a large number of gene transcripts is enhanced in response to ER stress (Hollien and Weissman, 2006; Hollien et al., 2009). There were 2321 gene transcripts that were significantly reduced in WT livers upon tunicamycin treatment, with 797 being lowered by at least twofold (Table 1). PERK contributed to 24% of those genes repressed by twofold or greater (190/797). This result is illustrated in Figure 3A, with the repressed transcripts in WT livers (x-axis) lying above the y = x diagonal. Turnover of a large number of mRNAs is known to be enhanced in response to ER stress, and the endoribonuclease activity of IRE1 is thought to facilitate the decay of many of these mRNAs (Hollien and Weissman, 2006; Hollien et al., 2009). These results suggest that PERK is also a significant contributor to the repression of the transcriptome in response to ER stress.
PERK facilitates activation of ATF6 in response to ER stress
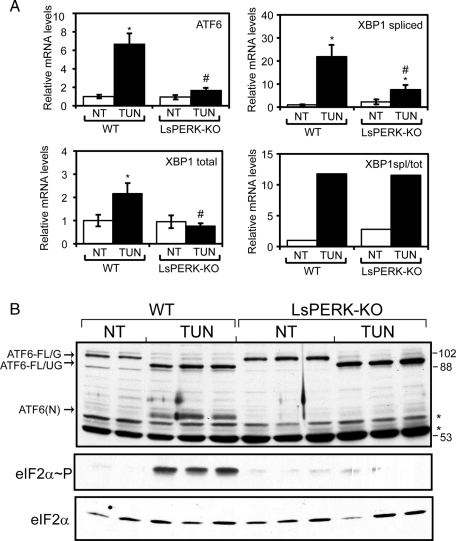
Our microarray analysis suggested that both ATF6 and XBP1 mRNA expression were substantially diminished in LsPERK-KO mice subjected to tunicamycin treatment compared with WT, results that were confirmed by quantitative PCR (qPCR) measurements (Figures 3C and 4A). These findings support the idea that PERK can facilitate activation of the UPR transcriptome by contributing to the activation of ATF6 and XBP1 during ER stress. Although loss of PERK lowered the total levels of XBP1 mRNA and, as a consequence the amount of spliced XBP1 mRNA in response to ER stress, the ratio of spliced/total XBP1 transcript was unchanged between the LsPERK-KO and WT mice (Figure 4A). This finding suggests that, within this time frame of ER stress, IRE1 endoribonuclease activity for XBP1 mRNA cleavage is not significantly altered by loss of PERK function.
FIGURE 4:
PERK facilitates activation of ATF6 in livers following tunicamycin treatment. (A) WT and LsPERK-KO mice were treated with tunicamycin (TUN) or excipient (NT, no treatment) for 6 h, and the levels of ATF6, XBP1, and spliced XBP1 mRNA were measured by qPCR. The ratio of spliced XBP1 mRNA vs. total XBP1 transcripts was also determined (XBP1spl/XBP1tot). The error bar represents the SD, and the * indicates statistically significant differences (p < 0.05) between tunicamycin treatment and non–ER stress. The # symbol indicates significant differences (p < 0.05) between the WT and LsPERK-KO livers. (B) Activation of ATF6 was measured by immunoblot analysis using whole-cell lysates prepared from WT and LsPERK-KO livers following tunicamycin treatment for 6 h or no treatment (NT) control. The arrows indicate full-length glycosylated ATF6 (ATF6-FL/G), full-length unglycosylated ATF6 (ATF6-FL/UG), and the cleaved ATF6(N) proteins. Two or three different liver samples are shown for each treatment group in the immunoblot panels. The position of the molecular weight standards (kDa) in the SDS–PAGE are shown to the right of the panel, and * designations indicate proteins that cross-react with the antibody prepared against recombinant ATF6 protein. Additionally, the levels of eIF2α∼P and total eIF2α were measured by immunoblot analyses using antibodies specific to each version of this translation initiation factor.
Given the larger than expected role for PERK in UPR-directed gene expression, we next addressed whether loss of this eIF2α kinase could adversely affect the activation of ATF6 in response to ER stress. Consistent with our previous results, PERK deficiency significantly reduced eIF2α∼P following 6-h tunicamycin treatment (Figure 4B). Immunoblot analyses comparing the levels of the full-length (ATF6-FL/G) and processed ATF6(N) protein in WT and LsPERK-KO mice indicated that activation of ATF6 was dramatically reduced with loss of PERK function (Figure 4B). Following treatment of the WT mice with tunicamycin, there was accumulation of unglycosylated full-length ATF6 (ATF6-FL/UG), along with the proteolytically cleaved and activated ATF6(N). Cleavage of ATF6 was dependent on PERK following 6 h of ER stress. We note that the size of the ATF6(N) protein is consistent with that determined using an ATF6 expression plasmid encoding residues 1–373 (HA-ATF6-1-373) transfected into cultured mouse embryonic fibroblasts (MEF; Supplemental Figure 1). Furthermore, ATF6 was reported to be glycosylated at multiple sites (Hong et al., 2004), and, as described later in this article, the ATF6-FL/UG was detected in MEF cells treated with tunicamycin, but not detected in response to thapsigargin, another well-characterized ER stress agent that does not directly inhibit protein glycosylation (Hong et al., 2004; Schroder and Kaufman, 2005). These results indicate that PERK facilitates activation of ATF6 in response to ER stress, providing an important rationale for why loss of PERK has such a broad impact on the UPR transcriptome.
We next addressed whether PERK also facilitates activation of ATF6 in cultured MEF cells exposed to ER stress agents. WT MEF cells were treated with either tunicamycin or thapsigargin for up to 6 h, and ATF6 levels were measured by immunoblot analyses. We found that, consistent with our liver analysis, tunicamycin triggered accumulation of the cleaved ATF6(N), initiating at 1 h of treatment and continuing through the 6-h regimen (Figure 5A). Accumulation of ATF6(N) coincided with the induction of ATF4 expression, which was readily detected following 2 h of the ER stress. During the time course of tunicamycin exposure, there was also a gradual decrease of ATF6-FL/G coincident with increasing ATF6-FL/UG. The ATF6-FL/UG is suggested to represent ATF6 that is newly synthesized during ER stress, as the addition of cycloheximide, a potent inhibitor of protein synthesis, along with tunicamycin resulted in minimal detection of ATF6-FL/UG (Figure 5B). Thapsigargin treatment also triggered accumulation of ATF6(N); however, in this case there was no detectable ATF6-FL/UG (Figure 5A). As a consequence, we did not detect measurable reduction of ATF6-FL/G during the time course of thapsigargin treatment, as the synthesis of ATF6 is sufficient to replace that being processed into the activated ATF6(N).
FIGURE 5:
PERK is required for full activation of ATF6 in MEF cells in response to different ER stress conditions. (A) WT MEF cells were treated with 2 μM tunicamycin (TUN) or 1 μM thapsigargin (TG) for between 1 and 6 h, as indicated, or with no stress treatment (0). Equal amounts of the protein lysates prepared from the cultured cells were separated by SDS–PAGE, and the levels of ATF6, ATF4, and actin were measured by immunoblot analysis. The arrows indicate the glycosylated (ATF6-FL/G) and unglycosylated (ATF6-FL/UG) versions of full-length ATF6, and the cleaved ATF6(N) proteins. (B) WT MEF cells were treated with 2 μM tunicamycin (TUN), tunicamycin and 50 μg/ml cycloheximide (TUN/CHX), or cycloheximide alone (CHX) for up to 6 h as indicated. The levels of ATF6-FL/G, ATF6-FL/UG, and actin were measured by immunoblot analysis. (C and D) WT and PERK−/− MEF cells were treated with tunicamycin (TUN) or thapsigargin (TG) for up to 6 h, and the levels of ATF6, eIF2α∼P, and total eIF2α were measured by immunoblot. The position of the molecular weight standards (kDa) are shown to the right of the ATF6 panel.
Similar ER stress treatments of PERK−/− MEF cells revealed a substantial reduction of ATF6(N) after 6 h of tunicamycin or thapsigargin exposure, although there was some cleaved ATF6(N) product at the earlier time points (Figure 5, C and D). A/A MEF cells expressing eIF2α containing alanine substituted for the phosphorylation residue, Ser-51, also showed reduced ATF6(N) levels following tunicamycin or thapsigargin treatments (Figure 6, A and B). Lowered ATF6(N) levels were most apparent after 6 h of ER stress, with minimal detectable ATF6(N) in the mutant cells devoid of eIF2α∼P. It is noted that some ATF6(N) was detected in the A/A cells at earlier exposure times to tunicamycin or thapsigargin. As expected, induced eIF2α∼P levels were significantly reduced in the PERK−/− cells in response to these ER stress conditions, and eIF2α∼P was absent in the A/A cells (Figures 5, C and D, and 6, A and B). These results indicate that eIF2α∼P by PERK is required for full activation of ATF6 in response to different ER stress conditions both in vivo and in cultured cells.
FIGURE 6:
Phosphorylation of eIF2α and ATF4 facilitates activation of ATF6 in response to ER stress. (A and B) WT and A/A MEF cells were treated with 2 μM tunicamycin (TUN) or 1 μM thapsigargin (TG) for between 1 and 6 h, as indicated, or with no stress treatment (0). Equal amounts of the protein lysates prepared from the cultured cells were analyzed by immunoblot using antibodies specific to ATF6, eIF2α∼P, and total eIF2α. The arrows indicate full-length ATF6-FL/G, ATF6-FL/UG, and the cleaved ATF6(N) proteins. Molecular weight standards in kilodaltons are shown to the right of the panels. (C) To measure ATF6 transcriptional activity, a firefly luciferase reporter with a promoter containing 5X ATF6-binding elements was cotransfected with a Renilla normalization plasmid into PERK−/−, ATF4−/−, A/A, and WT MEF cells. The transfected cells were treated with thapsigargin for 6 h or no stress agent, and the ATF6 transcriptional activity was measured by using a dual luciferase reporter assay. The values represent the fold change in RLU after treatment with ER stress for each transfected cell type. The error bar represents the SD, and the * indicates a significant (p < 0.05) difference when compared with WT cells. (D and E) WT and ATF4−/− MEF cells were treated with thapsigargin (TG) or tunicamycin (TUN) for up to 6 or 8 h, respectively, and the levels of ATF6, ATF4, and actin were measured by immunoblot. Panels are representative of three independent experiments. The bottom panel of (E) shows levels of the ATF6-FL/G and ATF6-FL/UG proteins from the immunoblot analysis as measured by densitometry. Measurements of ATF6-FL/G (diamonds) and ATF6-FL/UG (squares) are shown as a percentage of the ATF6-FL/G band from the WT untreated lane. (F) WT and PERK−/− MEF cells were treated with tunicamycin for up 6 h or with no ER stress (0). Where indicated, Sal003 was added to the PERK−/− cells 1 h before addition of tunicamycin. Levels of ATF6, ATF4, and actin were measured by immunoblot.
To determine whether reduced ATF6(N) protein in PERK−/− and A/A MEF cells also resulted in lowered ATF6 transcriptional activity, we used an ATF6-sensitive luciferase reporter expressed from a promoter containing five optimized UPREs (Wang et al., 2000). This reporter system is potently induced by ATF6, although it is noted that XBP1 has also been shown to contribute to activation of the UPRE-based promoters (Yamamoto et al., 2004). WT cells transfected with this reporter plasmid showed an approximately 12-fold increase in ATF6 transcriptional activity after 6-h tunicamycin treatment (Figure 6C). By comparison, ATF6 activity was lowered by more than 50% in the PERK−/− and A/A mutant cells during ER stress. Collectively, these results indicate that disruption of PERK phosphorylation of eIF2α reduces both ATF6 proteolysis and transcriptional activity.
ATF4 is required for activation of ATF6
Because eIF2α∼P by PERK directs the preferential translation of ATF4, we next sought to determine whether ATF4 was required for ATF6 activation in response to ER stress. In response to thapsigargin, there was a significant reduction of ATF6(N) in the ATF4−/− MEF cells, and ATF6-FL/G was substantially reduced by 6 h of treatment (Figure 6D). We also addressed the role of ATF4 in the activation of ATF6 in response to up to 8 h of tunicamycin treatment (Figure 6E). In the absence of stress (0 h), there was a greater than 40% reduction of ATF6-FL/G in ATF4−/− MEF cells when compared with the WT control. Thus, in both thapsigargin and tunicamycin treatments, there was minimal accumulation of ATF6(N) in ATF4−/− MEF cells. Following the time course, we also found that in WT MEF cells there was a greater rate of reduction of ATF6-FL/G and, as a consequence, more accumulation of the active ATF6(N), when compared with ATF4−/− MEF cells (Figure 6E). Furthermore, the accumulation of the newly synthesized ATF6-FL/UG required to replenish ATF6 protein levels was delayed in ATF4−/− cells during the exposure to tunicamycin (Figure 6E). Finally, MEF cells obtained from an independently derived ATF4−/− knockout animal (Hettmann et al., 2000) confirmed that loss of ATF4 led to reduced ATF6(N) levels upon ER stress (Supplemental Figure 3).
We next addressed whether preconditioning of the ISR and induction of ATF4 could restore ATF6 activation in PERK−/− MEF cells treated with ER stress (Figure 6F). For this experiment, induction of ATF4 was achieved by treating PERK−/− MEF cells with a derivative of salubrinal (Sal003), which is an inhibitor of the PP1c regulatory subunits, CReP and GADD34-PP1c, that are required for eIF2α∼P dephosphorylation (Costa-Mattioli et al., 2007). Sal003 treatment for 1 h before adding the ER stress agent tunicamycin resulted in ATF4 protein levels that were sustained in PERK−/− cells during the ER stress time course. Induction of ATF4 expression in PERK−/− cells with this treatment regimen also rescued the processing of ATF6, supporting the idea that eIF2α∼P and ATF4 are required for ATF6 activation (Figure 6F). These results indicate that the ATF4 transcription factor is also central to the activation of ATF6 in response to different ER stress conditions. Further supporting this idea, loss of ATF4 resulted in significantly reduced ATF6 transcriptional activity upon ER stress, as measured by the luciferase reporter plasmid containing the optimized ATF6-binding sites (Figure 6C).
We considered three models to explain the underlying mechanisms by which the PERK/eIF2α∼P/ATF4 pathway facilitates ATF6 activation, and it is noted that these ideas are not mutually exclusive. The first is the Stability model, in which the PERK pathway facilitates stabilization of ATF6(N), thus contributing to enhanced levels of this activated form of ATF6. The second is the Synthesis model, which posits that ATF4 enhances ATF6 expression, therefore increasing the amount of full-length ATF6 entering the secretory pathway for processing in the Golgi. Last, the Trafficking model focuses on ER-to-Golgi transport and suggests that the PERK/eIF2α∼P/ATF4 pathway is important for key features of ATF6 anterograde transport to the Golgi for cleavage by S1P and S2P. Each of these models—Stability, Synthesis, and Trafficking—will be experimentally addressed next.
Stability of ATF6(N) is independent of PERK and ATF4
The first model to be addressed concerns the role of the PERK pathway in the stabilization of ATF6. ATF6(N) is suggested to have a short half-life (t1/2 ∼ 1 h) by a mechanism involving degradation through the ubiquitin/proteasome pathway (Thuerauf et al., 2002). Basic zipper transcription factors, such as ATF6 and ATF4, are often labile and have been shown to require partner proteins for stability (Lassot et al., 2001; Thuerauf et al., 2007; Chiribau et al., 2010). We sought to determine whether induction of the PERK/eIF2α∼P/ATF4 pathway stabilized ATF6(N). First, WT and ATF4−/− MEF cells were treated with tunicamycin in the presence or absence of MG132, a potent proteasome inhibitor (Lee and Goldberg, 1998). Enhanced ATF6(N) levels were detected in both WT and ATF4−/− cells, although the total accumulation of ATF6(N) was significantly greater in WT cells (Figure 7A). This finding is consistent with an earlier report suggesting that ATF6 is subject to degradation by the proteasome pathway. It is also noted that, in these MEF cells, 6-h treatment with MG132 alone does not induce ER stress, as judged by accumulation of ATF6(N) and splicing of XBP1 mRNA (Jiang and Wek, 2005, and unpublished data). As a control, other ISR inducible transcription factors, ATF4 and CHOP, which are also short-lived (Lassot et al., 2001; Chiribau et al., 2010), showed similar increases when MG132 was combined with the tunicamycin treatment in WT cells. In the ATF4−/− cells, there was no detectable ATF4 transcription factor, and its target CHOP showed some enhanced accumulation in the combined tunicamycin and MG132 treatments, although significantly less than that measured in WT cells. Consistent with our earlier experiments, there were also minimal detectable ATF6-FL/UG levels in the ATF4−/− cells in response to tunicamycin treatment in the presence or absence of MG132 (Figure 7A). This finding suggests that ATF4 contributes to the synthesis of ATF6 in response to ER stress, an idea that will be explored more fully later in this article.
FIGURE 7:
ATF6(N) protein has a short half-life that is independent of ATF4. (A) WT and ATF4−/− MEF cells were treated with tunicamycin (TUN) or cotreated with tunicamycin and MG132 (TUN/MG132) for up to 6 h, as indicated. The levels of ATF6-FL/G, ATF6-FL/UG, ATF6(N), ATF4, CHOP, and actin were measured by immunoblot analysis. Each of the panels was derived from a single immunoblot and is representative of three independent experiments. (B and C) WT and ATF4−/− cells were subject to the same treatment regimen. The half-life of ATF6(N) was measured in these cells by introducing a plasmid expressing an HA-tagged version of ATF6 encoding residues 1-373 (HA-ATF6-1-373). Cells were pretreated with 2 μM tunicamycin for 3 h, or exposed to no ER stress, followed by the addition of 50 μg/ml cycloheximide. Cells were then cultured for up to 8 h as indicated, and the HA-ATF6-1-373 (HA), CHOP, or actin proteins were measured by immunoblot analysis.
We next directly measured the half-life of ATF6(N) in WT and ATF4−/− cells. Because the steady-state levels of ATF6(N) are a consequence of both the processing of full-length ATF6 and the turnover of ATF6(N), a HA-tagged version of ATF6(N) containing amino acid residues 1-373 (HA-ATF6-1-373) was expressed in both WT and ATF4−/− MEF cells. The synthesis of the HA-ATF6-1-373 would therefore occur independently of intramembrane proteolysis, and the HA tag would allow for the expressed ATF6 protein to be distinguishable from endogenous ATF6. MEF cells expressing HA-ATF6-1-373 were either untreated or exposed to tunicamycin for 3 h. Induction of CHOP was used as a control, as it is robustly induced after 3 h of ER stress by a mechanism requiring ATF4. These cells were then subjected to treatment with cycloheximide, and the rate of turnover of the tagged ATF6(N) protein was monitored. There was no appreciable difference in the rate of decay of the HA-ATF6-1-373 protein in the WT MEF cells in the presence or absence of ER stress (Figure 7B). In both cases, the activated version of ATF6 displayed a half-life of ∼3 h. Furthermore, loss of ATF4 did not alter the rate of turnover of the HA-ATF6-1-373 protein (Figure 7C). On the basis of these findings, we conclude that the PERK/ATF4 pathway is not a significant contributor to the stability of the activated ATF6(N).
The PERK/eIF2α∼P/ATF4 pathway facilitates increased ATF6 expression during the UPR
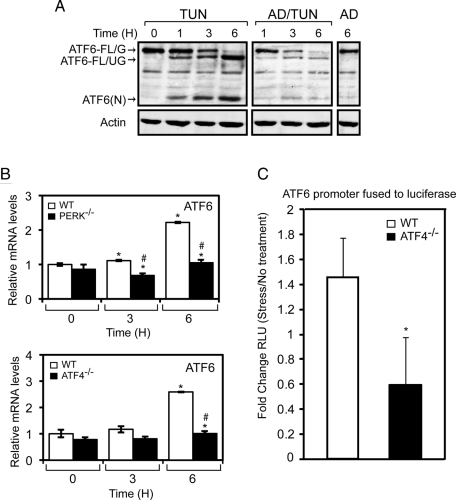
An important part of the activation of ATF6 during ER stress is its induced synthesis, which is required to replenish the full-length ATF6 that is cleaved to generate ATF6(N). The Synthesis model suggests that disruption of the PERK/eIF2α∼P/ATF4 pathway lowers the levels of ATF6(N) by blocking the induced synthesis of full-length ATF6. Synthesis of the full-length protein is required for continued availability of this substrate for intramembrane proteolysis during ER stress. We initially addressed the role of ATF4 in ATF6 synthesis by determining the requirement for global transcription in the accumulation of ATF6(N). Increasing amounts of ATF6(N) were detected between 1 and 6 h of tunicamycin treatment (Figure 8A). This increase was accompanied by a depletion of ATF6-FL/G that was nearly absent by 6 h of exposure to tunicamycin and by accumulation of ATF6-FL/UG, which represented the ATF6 protein newly synthesized during the ER stress condition. By comparison, a 30-min pretreatment with actinomycin D, a potent inhibitor of RNA polymerase II, led to a more rapid depletion of ATF6-FL/G, and reduced levels of ATF6-FL/UG and ATF6(N), during the time course of tunicamycin treatment. These results suggest that transcription is an important contributor to accumulation of ATF6(N) during ER stress. This result is consistent with the idea that synthesis of ATF6 replenishes the processing pathway of ATF6, ensuring a continued source of full-length ATF6 for cleavage into the labile ATF6(N) transcription factor.
FIGURE 8:
PERK and ATF4 facilitate increased expression of the ATF6 gene in response to ER stress. (A) WT MEF cells were treated with tunicamycin (TUN), 10 μM actinomycin D and tunicamycin (AD/TUN), or actinomycin D (AD) alone for up to 6 h, as indicated. The levels of ATF6-FL/G, ATF6-FL/UG, and actin were measured by immunoblot analysis. (B) PERK−/− and ATF4−/− MEF cells, and their WT counterparts, were treated with tunicamycin for up to 6 h, and the levels of ATF6 mRNA were measured by qPCR. (C) A 1.5-kb segment (−1500 to −1) of the ATF6 promoter was fused to a firefly luciferase reporter and assayed for expression in MEF cells treated with either tunicamycin for 6 h or no stress agent. The fold change in ATF6 promoter activity in response to ER stress was determined by the dual luciferase assay. In panels B and C, the error bar represents the SD, and the * indicates significance between untreated and treated samples; the # indicates significance between cell types, with p < 0.05.
Induced ATF6 synthesis is suggested to involve increased transcription of ATF6, as there was a two- to threefold increase in ATF6 mRNA during tunicamycin treatment (Figure 8B). As described earlier regarding the LsPERK-KO mice (Figure 4A), loss of PERK or ATF4 in the MEF cells blocked the increase in ATF6 mRNA in response to ER stress (Figure 8B). Also consistent with the earlier LsPERK-KO experiments, there was significantly lowered induction of total and spliced XBP1 mRNA levels during tunicamycin treatment in PERK−/− or ATF4−/− cells (Supplemental Figures 4 and 5). To further support the idea that ATF4 facilitates the transcription of ATF6, we measured the activity of the ATF6 promoter fused to a firefly luciferase reporter that was transfected into WT or ATF4−/− MEFs. Although there was an ∼1.5-fold increase in the ATF6 promoter activity in WT cells upon treatment with tunicamycin, loss of ATF4 blocked induction of the ATF6 promoter (Figure 8C). These results indicate that ATF4 enhances the transcription of the ATF6 gene.
ATF4 facilitates trafficking of ATF6 from the ER to the Golgi
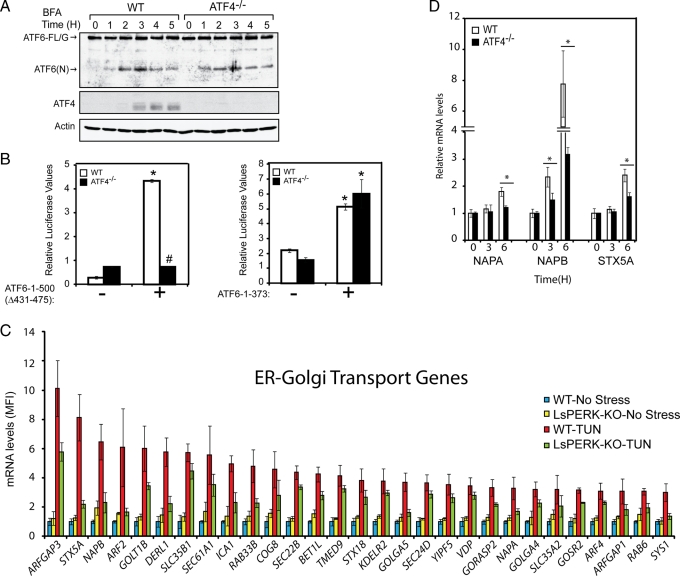
A final model to explain the role of the PERK/eIF2α∼P/ATF4 pathway in the activation of ATF6 suggests that this pathway facilitates the cellular trafficking of ATF6 during ER stress. In support of this model, our earlier observations showed that the rate of decrease of ATF6-FL/G in response to ER stress was slower in the ATF4−/− cells than in the WT, and was accompanied by reduced levels of cleaved ATF6(N) (Figure 6E). We used two experimental approaches to address whether ATF4 facilitates ATF6 trafficking. First, we used brefeldin A, a chemical inhibitor of anterograde ER-Golgi vesicle trafficking that causes S1P and S2P relocalization from the Golgi compartments to the ER (DeBose-Boyd et al., 1999; Shen and Prywes, 2004, 2005). We reasoned that brefeldin A would trigger a redistribution of Golgi proteins, including the S1P and S2P proteases, to the ER causing cleavage of ATF6 that was independent of trafficking. Consistent with an earlier study (Shen and Prywes, 2004), there was an accumulation of ATF6(N) in WT cells within 2 h of brefeldin A exposure (Figure 9A). This accumulation was accompanied by expression of ATF4 after 3 h of treatment, suggesting that at later time points brefeldin A can stress the ER organelle. Importantly, this pattern of ATF6 activation by brefeldin A was not changed in the absence of ATF4, with similar levels of ATF6(N) accumulation at each of the time points (Figure 9A). This finding demonstrates that the S1P and S2P proteases are active in the ATF4−/− cells, and supports the idea that ATF4 is required to facilitate passage of ATF6 from the ER to the Golgi.
FIGURE 9:
ATF4 is dispensable for activation of ATF6 in response to brefeldin A treatment. (A) WT and ATF4−/− MEF cells were treated with 5 μg/ml brefeldin A (BFA) for up to 5 h, and the levels of ATF6-FL/G, ATF6(N), ATF4, and actin were measured by immunoblot analysis. (B) Plasmids encoding ATF6-1-500Δ431-475 or ATF6-1-373 (indicated by +), along with a firefly luciferase reporter containing a promoter with 5X ATF6-binding elements, were introduced into WT and ATF4−/− MEF cells. Luciferase values normalized to a Renilla internal control are shown, * represents significance (p < 0.05) with respect to untransfected control, and # shows significance between cell types. Error bars represent the SD. (C) Graphical representation of the expression of genes involved in ER-Golgi trafficking that are PERK dependent. The mean fluorescence intensity (MFI) for each transcript is shown as a histogram, along with the SD. (D) WT and ATF4−/− MEF cells were treated with tunicamycin for up to 6 h, as indicated. The mRNA levels of NAPA, NAPB, and STX5A were measured by qPCR. The * indicates that a significant difference was detected between the cell types.
Trafficking and activation of ATF6 is suggested to involve multiple steps, including the reduction of the disulfide bonds that facilitates deoligomerization of ATF6 into ATF6 monomers, the release of inhibitor BiP (Shen et al., 2002; Nadanaka et al., 2007), and the loading of ATF6 into COPII vesicles for transport from the ER to the Golgi (Schindler and Schekman, 2009). Deletion of the BiP binding domain and the cysteine residues from the luminal portion of ATF6 (1-500 Δ431-475) were reported to lead to a constitutively active transcription factor that is transported to the Golgi for cleavage independent of ER stress (Shen et al., 2002). Consistent with this idea, we found that expression of ATF6-1-500 Δ431-475 in nonstressed WT MEF cells led to a significant increase in ATF6 activity as measured by luciferase reporter expressed from a promoter with optimized ATF6-binding elements (Figure 9B). The ATF6 activity was blocked, however, when the ATF6-1-500 Δ431-475 was expressed in the ATF4−/− cells. In contrast, when WT and ATF4−/− MEF cells were transfected with the ATF6(N) (residues 1–373) construct that is not dependent on Golgi processing, transcriptional activity was similarly induced in both WT and ATF4−/− cells. These results suggest that even when ATF6 is readily released from the ER, loss of ATF4 function interferes with the passage of ATF6 to the Golgi for processing by S1P to S2P proteases.
ATF4 facilitates gene expression important for trafficking to the Golgi
Given that ATF4 facilitates trafficking of ATF6 from the ER to the Golgi, we reasoned that ATF4 may induce the expression of genes involved in ER-to-Golgi transport. In our microarray analysis, a number of the genes that showed a PERK-dependent induction in response to tunicamycin are known to participate in this trafficking process (Figure 9C). These genes include COPII vesicle coat proteins, GTPase activating proteins, and genes associated with vesicle formation and fusion such as STX5a (Rowe et al., 1998; Geng et al., 2008) and α/β-SNAP required for recycling of SNARE complexes at target membranes (Ungar and Hughson, 2003). Further supporting this idea, measurements of STX5a and α/β-SNAP mRNAs by qPCR confirmed that ATF4 contributes to increased expression of these ER-to-Golgi trafficking genes in MEF cells during ER stress (Figure 9D). These results suggest that the PERK/eIF2α∼P/ATF4 pathway directs increased expression of genes involved in protein trafficking, and highlights the importance of the ISR in facilitating ATF6 transport from the ER to the Golgi for cleavage by S1P and S2P.
DISCUSSION
In this study we showed that PERK-mediated eIF2α∼P not only triggers translational control in response to ER stress, but also facilitates the activation of ATF6 and the transcriptional phase of the UPR. Among the UPR genes induced by twofold, 74% showed a significant reduction in gene expression in the LsPERK-KO livers (Table 1 and Figure 3A). Furthermore, activation of ATF6 in response to ER stress was significantly diminished with loss of PERK (Figure 4B). These central findings were observed not only in liver tissues, but also in MEF cells containing mutations that blocked each of the steps in the PERK/eIF2α∼P/ATF4 pathway (Figures 5 and 6). ATF4 facilitates induction of ATF6 during ER stress by at least two mechanisms. First, ATF4 enhances the transcription of the ATF6 gene, ensuring that there are sufficient amounts of newly synthesized ATF6 available for continued processing into activated ATF6(N) (Figures 4A and 8B). Second, ATF4 contributes to the trafficking of ATF6 from the ER to the Golgi for subsequent proteolysis and activation by S1P and S2P. Supporting this idea is the finding that mutations that adversely affect each step of the PERK/eIF2α∼P/ATF4 pathway significantly diminish the levels of activated ATF6(N) in response to either tunicamycin or thapsigargin treatments (Figures 4B, 5, and 6). By comparison, ATF4 is dispensable for the processing of ATF6(N) in MEF cells upon exposure with brefeldin A, which triggers redistribution of S1P and S2P to the ER (Figure 9B). This result suggests that although S1P and S2P are functional in ATF4−/− cells, the passage of ATF6 to the Golgi is impeded with loss of ATF4. ATF4 contributes to the expression of many genes involved in ER-to-Golgi transport (Figure 9, C and D), and diminished levels of key proteins that are important for ATF6 trafficking is suggested to be an important underlying reason for the reduced activation of ATF6(N) in ATF4-deficient cells. These findings provide a mechanistic understanding of earlier reports that suggested that PERK may affect the expression of ER chaperones and the efficiency of secretory processes (Adachi et al., 2008; Rutkowski et al., 2008; Gupta et al., 2010). In this regard, this study suggests that ATF4 plays a role in the expression of genes contributing to the assembly and processing of secretory proteins, in addition to its earlier described role in regulating genes involved in metabolism, resistance to oxidative stress, and regulation of apoptosis (Harding et al., 2003).
The PERK/eIF2α∼P/ATF4 pathway alleviates ER stress by multiple mechanisms
Phosphorylation of eIF2α by PERK directs essential adaptive functions that alleviate cellular injury during ER stress. This idea is illustrated by our finding that the LsPERK-KO livers showed enhanced activation of caspase-3 and apoptosis during ER stress (Figure 2). One underlying mechanism by which PERK can alleviate stress damage is by reducing translation, which would lower the influx of nascent proteins into the ER organelle that is inundated with misfolded proteins (Harding et al., 2000b). Our study suggests that the PERK/eIF2α∼P/ATF4 pathway is also central for activation of ATF6 during ER stress and, as a consequence, is critical for full implementation of the transcription of UPR genes, including those involved in protein folding, ERAD, and trafficking in the secretory system. This reconfiguration of the transcriptome would contribute to the expansion of the processing capacity of the secretory pathway, which would be essential for returning the stressed ER to homeostasis. Consistent with these ideas, the livers of ATF6−/− mice also showed increased apoptosis when challenged with tunicamycin (Wu et al., 2007). It was suggested that lowered protein chaperone expression in the ATF6-deficient mice compromised the ability of the ER to cope with stresses that disrupt protein folding and assembly. Livers from the ATF6−/− mice treated with tunicamycin also exhibited significantly more intracellular triglycerides compared with WT, accompanied by evidence of microvesicular steatosis (Rutkowski et al., 2008). Together, these results suggest that perturbations in the hepatic lipid homeostasis in the ER-stressed LsPERK-KO mice may result at least in part from the failure to appropriately activate ATF6 and its targeted gene expression (Figures 2A, 3, and 4B).
PERK is also suggested to contribute to cell death during prolonged and severe ER stress. Although the underlying mechanisms by which eIF2α∼P can contribute to apoptosis is not well understood, it is thought that chronic ER stress can induce CHOP expression to a certain threshold that induces the expression of genes critical for triggering cell death (Marciniak and Ron, 2006; Ron and Walter, 2007). In this sense, PERK has a dynamic and balanced range of cellular responsiveness that is optimal for cellular adaption to ER stress (Wek and Anthony, 2009). The eIF2α∼P by PERK would lower protein synthesis and facilitate activation of the UPR, which would serve to expand the secretory pathway. Perturbations in the ISR that lead to levels of eIF2α∼P outside this adaptive zone would trigger extremes in translational control that would be detrimental to cells. For example, hypophosphorylation of eIF2α as can be seen in Wolcott-Rallison syndrome can prevent proper implementation of UPR transcription, along with heightened protein synthesis, which can exacerbate protein trafficking deficiencies during ER stress. This UPR dysfunction can lead to cell death by mechanisms independent of CHOP induction (Harding et al., 2000a). Hyperphosphorylation of eIF2α during chronic ER stress can heighten CHOP expression, which potentiates signaling pathways that can culminate in apoptosis.
The range of eIF2α∼P levels is important to consider when examining the biological effects of eIF2α∼P and the UPR during different stress conditions. Chemically induced ER stress, such as that elicited by tunicamycin used in this study, can produce an acute and terminal stress response, whereas physiological stresses that the liver can encounter may be more transient and within the adaptive zone (Rutkowski and Kaufman, 2007; Rutkowski and Hegde, 2010). For example, a high-fat diet can elicit eIF2α∼P in the liver, and it was reported that attenuated eIF2α∼P in transgenic animals expressing elevated levels of GADD34 can diminish liver triglycerides and hepatosteatosis (Oyadomari et al., 2008). In this case, dephosphorylation of eIF2α would be viewed as protective in the liver, and our LsPERK-KO study would suggest an important role for the ISR for cellular survival. The mechanistic rationale for these differences is not well understood. Clearly the expression of important regulators of intermediary metabolism, including those involved in lipogenesis and lipid oxidation, are affected by the PERK pathway (Supplemental Figure 2), and the expression of these genes may be influenced by the diversity, intensity, and duration of the critical stress signals triggered by physiological changes in the liver.
Earlier studies have suggested divergent effects of PERK and IRE1 on cell viability (Lin et al., 2007, 2009). Sustained PERK signaling was proposed to be maladaptive to cells, and similar durations of IRE1 activation were suggested to be restricted to remedying ER stress. These ideas are complicated by the findings that PERK is required for full activation of ATF6 and implementation of the UPR transcriptome. Furthermore, the PERK/eIF2α∼P/ATF4 pathway reduced XBP1 expression, although it did not appear to alter IRE1 activity as measured by splicing of XBP1 mRNA (Figure 4A and Supplemental Figures 4 and 5). Therefore, each of the branches of the UPR are fully integrated, and perturbations of PERK can reduce the critical signaling functions required for cell survival during ER stress.
IRE1 is also suggested to be a contributor to apoptosis during certain ER stress arrangements. IRE1 can bind the scaffolding protein TRAF2, which serves to activate JNK, a potent inducer of apoptosis (Urano et al., 2000; Ron and Walter, 2007). This underlying explanation may be important for the enhanced apoptosis of LsPERK-KO livers during ER stress (Figure 2). IRE1-dependent JNK signaling is suggested to be significantly enhanced with reductions in XBP1 expression (Glimcher, 2010), and the levels of XBP1 mRNA were significantly reduced in LsPERK-KO livers, despite IRE1 endonuclease activity being fully retained (Figure 4A).
Role of ATF4 in the implementation of the UPR
ATF4 is suggested to be required for full activation of ATF6 in response to ER stress by enhancing ATF6 transcription and by facilitating the expression of genes that contribute to ATF6 trafficking from the ER to the Golgi. ATF4 may carry out these functions by directly binding to the promoters of UPR genes, through additional transcription factors targeted by ATF4, or by critical cellular changes resulting from ATF4-directed transcription. ATF4-deficient cells are suggested to be defective for the expression of genes involved in nutrient import and metabolism that predispose cells to oxidative stress (Harding et al., 2003). This stress may interfere with cellular processes such as the release of ATF6 from the ER, which requires the reduction of disulfide linkages within ATF6 to promote deoligomerization required for loading and passage into COPII vesicles for anterograde transport to the Golgi (Nadanaka et al., 2007).
MATERIALS AND METHODS
Animals
The animal study protocol was approved by the Institutional Care and Use Committee at the Indiana University School of Medicine–Evansville. Mice homozygous for the LoxP allele of PERK (PERKfl/fl) were mated with transgenic mice heterozygous for Cre recombinase expressed from the albumin gene promoter (AlbCre) to create a liver-specific deletion of PERK, LsPERK-KO (AlbCre+•PERKfl/fl). This mating also produced littermates that are Cre negative that express a WT version of PERK (AlbCre−•PERKfl/fl; Bobrovnikova-Marjon et al., 2008; Bunpo et al., 2009). The lsPERK-KO mice were derived from a cross between C57BL/6 and 129SvE mice. Genotyping that showed efficient PERK gene deletion in the LsPERK-KO livers was reported (Bunpo et al., 2009), and is consistent with lowered eIF2α∼P levels in response to tunicamycin treatment (Figures 1, A and B, and 4B). Male and female adult mice (n = 5–8 per treatment group), ages 3-6 mo, were individually housed in wire-bottomed cages, maintained on a 12-h light/dark cycle, and provided unrestricted access to food (7017 NIH-31 Open Formula Mouse/Rat Sterilizable Diet; Harlan Teklad, Indianapolis, IN) and water. The key gene expression, triglyceride, and apoptotic findings that we reported were similar between the male and female mice. Among the assays in which n = 8, there were between four and seven males, and between one and three females. Analysis of ATF6 activation in Figure 4B includes independent liver samples prepared from at least one male and one female in each treatment group. Mice received intraperitoneal (i.p.) injections of tunicamycin at a dose of 1 mg/kg body weight or an equivolume of excipient (0.3% dimethyl sulfoxide in phosphate-buffered saline; Nair et al., 2007). Mice were killed by decapitation at 6, 12, 24, or 36 h after injection. Dissected livers were rinsed in ice-cold PBS, weighed, and either snap-frozen in liquid nitrogen or fixed in 4% paraformaldehyde.
Immunoblot analyses
Liver tissue samples were prepared for SDS–PAGE as previously described (Bunpo et al., 2009). Tissues were homogenized in seven volumes of a solution consisting of 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.4), 100 mM KCl, 0.2 mM EDTA, 2 mM EGTA, 1 mM dithiothreitol, 50 mM sodium fluoride, 50 mM β-glycerophosphate, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 0.5 mM sodium orthovanadate. Following electrotransfer to polyvinylidene fluoride membranes, immunoblots were incubated with primary and secondary antibodies, and then were visualized using enhanced chemiluminescence reagents (ECL; GE Biosciences, Piscataway, NJ). Cellular lysates were prepared from cultured MEF cells using RIPA-buffered solution containing 50 mM Tris-HCl (pH 7.9), 150 mM sodium chloride, 1% Nonidet P-40, 0.1% SDS, 100 mM sodium fluoride, 17.5 mM β-glycerophosphate, 0.5% sodium deoxycholate, and 10% glycerol supplemented with EDTA-free protease inhibitor cocktail tablet (Roche, Basel, Switzerland) and were subjected to SDS–PAGE. Antibodies for eIF2α∼P (Ser-51), cleaved caspase-3, and Grp78/BiP were purchased from Cell Signaling Technology (Danvers, MA), total eIF2 and CHOP were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and actin was obtained from Bethyl Laboratories (Montgomery, TX). Secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA) and BioRad (Hercules, CA). ATF6 antibody was prepared in rabbits using the mouse ATF6 N-terminal amino acids 6–307 as the antigen and was used at a concentration of 1:500 in 5% milk powder (wt/vol) in PBS. ATF4 antibody prepared against recombinant human ATF4 was affinity purified and used in immunoblot analyses. The Odyssey infrared imaging system (LI-COR, Lincoln, NE) was used in conjunction with the ECL method for antibody detection as described (Teske et al., 2011). Densitometric measurements (integrated density) were taken in Adobe Photoshop using a scanned image (600 dpi) that was inverted for histogram analysis; background measurements were also recorded and subtracted from final measurements.
To measure the half-life of ATF6(N), plasmid p913 encoding an N-terminal HA-tagged version of ATF6 residues 1–373 (HA-ATF6-1-373) was transfected into WT or ATF4−/− MEF cells. After 24 h, cells were pretreated with 2 μM tunicamycin for 3 h, or exposed to no ER stress, followed by the addition of 50 μg/ml cycloheximide. Cells were then incubated for up to 8 h. Lysates were prepared and analyzed by SDS–PAGE and immunoblot analysis using antibodies specific to the HA-tag, CHOP, or actin.
Reverse transcription and real-time PCR
Total RNA was extracted from frozen tissue using TriReagent (Molecular Research Center, Cincinnati, OH) followed by a DNase treatment (VersaGene DNase kit; Gentra Systems, Minneapolis, MN). To inactivate the reaction, the samples were heated to 70°C for 5 min. RNA samples were further purified using the RNeasy mini kit (Qiagen, Chatsworth, CA), yielding preparations with an A260/A280 ratio between 1.8 and 2.0. The levels of the indicated mRNAs were determined by qPCR. Approximately 1 μg each of the RNA solutions was used for reverse transcription, which used the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. TaqMan Gene Expression Master Mix (Roche) and TaqMan Gene Expression Assays (Applied Biosystems, Carlsbad, CA) were used for the qPCR. Amplification and detection were performed on the LightCycler 480 (Roche). For measurement of the spliced version of the XBP1 mRNA, the forward primer 5′-GAGTCCGCAGCAGGTG-3′ and reverse primer 5′-CTCTGGGAGTTCCTCCAGACT-3′ were used in the qPCR. All measurements were in triplicate and normalized to β-actin or 18S mRNA. Results were obtained by the comparative Ct method, and results were expressed as fold change with respect to the nonstressed control.
Histology
Tissues fixed in 4% paraformaldehyde were frozen and then sectioned (10 μm) using a cryostat. TUNEL assays were performed according to the manufacturer's instructions (Trevigen TACS 2 TdT-Blue Label In Situ Apoptosis Detection Kit; R&D Systems, Minneapolis, MN). TUNEL-positive cells were measured from three equal sized areas per section. Digital images of selected areas captured at 200× were imported into Scion Image for Windows (Scion Corporation, Frederick, MD), and TUNEL-positive cells were manually marked and counted using the counting tool. The TUNEL-staining showed pyknotic bodies, and the nucleus was stained darker than the cytoplasm portion in the tissue preparations. Frozen sections were also stained with Oil Red O and counterstained with hematoxylin to visualize lipid content and tissue morphology.
Triglyceride measurements
Triglycerides were measured using the Triglyceride Quantification Kit from BioVision Research Products (Mountain View, CA) following the manufacturer's instructions. Tissue samples (∼40 mg) were weighed and homogenized in 10 volumes of a 5% NP-40 solution. Samples were heated to 85°C then subjected to centrifugation at 10,000 × g for 2 min at room temperature. The resulting supernatants were diluted 1:500 (vol/vol) with sterile water, a volume of 50 μl of diluted supernatant was added to 50 μl of reaction mix, and absorbances were measured at 570 nm. Absorbances of standards were plotted, and test sample readings were applied to the standard curve to calculate the triglyceride concentration.
Microarray analysis
The microarray analysis was carried out using RNA prepared from livers obtained from WT and LsPERK-KO mice that received injections (1 mg/kg body weight) of tunicamycin or excipient for 6 h, as highlighted in the Animals section of Materials and Methods. Three mice from each control and four mice from each experimental group were analyzed, for a total of 14 preparations analyzed. Total RNA was isolated from liver tissue using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions, and a RNeasy mini kit (Qiagen) was used to further purify samples. The RNA was then labeled using the standard Affymetrix protocol for 3′-IVT arrays (Affymetrix, Santa Clara, CA). Labeled cRNA was hybridized for 17 h to the Affymetrix Mouse Genome 430 2.0 Array. The signal values and detection calls were derived using the MAS5 algorithm in Affymetrix GeneChip Operating Software. Probe sets were eliminated from further analysis if an absent call was determined in greater than 50% of the samples in both the WT treated/untreated and LsPERK-KO treated/untreated. The filtered data were imported into Partek Genomics Suite (Partek, St. Louis, MO), where hierarchical clustering and principal components analysis were used to detect any outlier arrays; none were found. Values of p were determined for treatment condition (stress/no stress), genotype (WT/LsPERK-KO), and interaction terms by two-way analysis of variance (ANOVA) using the Partek software. False discovery rates (FDR) were calculated according to Benjamini and Hochberg (1990). Probe sets that were not significantly changed during tunicamycin treatment were filtered out (p > 0.05 or FDR > 0.15), such that the resulting data set would further represent ER stress-dependent genes. From these ER stress-dependent data, probe sets that had an interaction p value (condition * genotype) of p > 0.05 were removed to enrich the subset of PERK-dependent genes. Values of p for each statistical test and FDR values for each comparison are listed in Supplemental Tables 1, 2, and 3. The following gene ontology (GO) terms were used to identify genes in the PERK-dependent data set that were linked with ER-to-Golgi transport: Primary GO term, Biological process: 0006810 (transport). Secondary GO terms, Gene Ontology Cellular Compartment: 0005783 (endoplasmic reticulum), 0005789 (endoplasmic reticulum membrane), 0000139 (Golgi membrane), 0005794 (Golgi apparatus). Microarray data have been deposited in GEO (gene expression omnibus; www.ncbi.nlm.nih.gov/geo) under the accession number GSE29929.
Cell culture
PERK−/−, ATF4−/−, and A/A MEF cells, and their WT counterparts, have been described previously (Jiang et al., 2004). We also analyzed ATF4−/− MEF cells that were derived from an independent knockout (Hettmann et al., 2000), as presented in Supplemental Figure 3. These MEF cells were grown in DMEM (4.5 g/l glucose), supplemented with 1X MEM nonessential amino acids (HyClone SH30238.01), 1X MEM essential amino acids (HyClone SH30598.01), and 50 μM β-mercaptoethanol, as described previously (Harding et al., 2003). Cultured cells were treated with 2 μM tunicamycin, 1 μM thapsigargin, 1 μM MG132, 10 μM actinomycin D, 50 μg/ml cycloheximide, 5 μg/ml brefeldin A, and 10 μM Sal 003 as indicated. Specific experimental details regarding pretreatments or cotreatments are provided in figure legends when more than one drug was used.
Luciferase assays
Luciferase assays were carried out in six-well plates using the Dual-Luciferase reporter assay system (Promega, Madison, WI) following the manufacturer's instructions. The ATF6 expression plasmids were described by Prywes and colleagues (Wang et al., 2000). For measurements of ATF6 transcriptional activity, plasmid p912 encoding five ATF6-binding elements fused to the firefly luciferase reporter gene was cotransfected with a Renilla normalization plasmid into PERK−/−, ATF4−/−, A/A, and WT MEF cells. Measurements were determined as the relative light units (RLU) of the firefly luciferase normalized to Renilla luciferase. Plasmid p1018 expressing ATF6 encoding residues 1-500Δ431-475 (Shen et al., 2002) was cotransfected with p912 and the Renilla normalization plasmid into WT and ATF4−/− MEF cells, and the ATF6 transcriptional activity with the cotransfection of ATF6-1-500Δ431-475 was determined. The activity of the ATF6 gene promoter was measured by using a luciferase reporter assay. A DNA segment encoding −1500 to −1 base pairs relative to the transcriptional start site of the ATF6 gene was amplified using the primers F5′-GGGACGCGTGCAAACGTGCAGCCTGGTCTGTATGTGGGTCCCC-3′ and R5′-GGGCTCGAGCACCGCCCCGTGGCCTCCTGCCGCGCCCAGCCTTTCTAGG-3′, with the Mlu1 and Xho1 restriction sites underlined. The resulting PCR product was then digested, and the DNA fragment was inserted using the Mlu1 and Xho1 restriction sites of the pGL3-basic vector (Promega), resulting in p1052. The plasmid p1052 was cotransfected with the Renilla vector into ATF4−/−, and WT MEF cells and the ATF6 promoter activity was determined using the dual luciferase assay.
Statistics
Data were analyzed using two-way ANOVA to assess main and interaction effects, with drug and genotype as the independent variables (StatSoft, Tulsa, OK). When a significant overall effect was detected, differences among treatment groups were assessed with Duncan's multiple range post hoc test. The level of significance was set at p < 0.05 for all statistical tests.
Supplementary Material
Acknowledgments
We acknowledge support from the National Institutes of Health (grant GM049164 to R.C.W.), from predoctoral fellowships (T32DK064466 to B.F.T.), and from the American Heart Association (11PRE7240012 to B.F.T.). The microarray analyses were carried out in conjunction with the Center for Medical Genomics at the Indiana University School of Medicine. We thank Ronald Prywes (Columbia University) for generously providing the ATF6 reporter and expression plasmids and Douglas Cavener (Pennsylvania State University) for providing the mice engineered with the floxed PERK. We also thank Danielle Brueck for technical assistance and Thomas Baird for helpful comments during manuscript preparation.
Abbreviations used:
- eIF2α∼P
phosphorylation of eIF2α
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- GTP
guanosine triphosphate
- ISR
integrated stress response
- MEF
mouse embryonic fibroblasts
- qPCR
quantitative PCR
- S1P
site-1-protease
- S2P
site-2-protease
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- UPR
unfolded protein response
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0510) on September 14, 2011.
REFERENCES
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci USA. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. J Biol Chem. 2009;284:32742–32749. doi: 10.1074/jbc.M109.047910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chiribau CB, Gaccioli F, Huang CC, Yuan CL, Hatzoglou M. Molecular symbiosis of CHOP and C/EBP{beta} isoform LIP contributes to endoplasmic reticulum stress-induced apoptosis. Mol Cell Biol. 2010;30:3722–3731. doi: 10.1128/MCB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Li J, Ron D, Sha B. The structure of the PERK kinase domain suggests the mechanism for its activation. Acta Crystallogr D Biol Crystallogr. 2011;67:423–428. doi: 10.1107/S0907444911006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-a kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding-protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J. 1999;339:135–141. [PMC free article] [PubMed] [Google Scholar]
- Geng L, Boehmerle W, Maeda Y, Okuhara DY, Tian X, Yu Z, Choe CU, Anyatonwu GI, Ehrlich BE, Somlo S. Syntaxin 5 regulates the endoplasmic reticulum channel-release properties of polycystin-2. Proc Natl Acad Sci USA. 2008;105:15920–15925. doi: 10.1073/pnas.0805062105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher LH. XBP1: the last two decades. Ann Rheum Dis. 2010;69((suppl 1),):i67–i71. doi: 10.1136/ard.2009.119388. [DOI] [PubMed] [Google Scholar]
- Gupta S, McGrath B, Cavener DR. PERK (EIF2AK3) regulates proinsulin trafficking and quality control in the secretory pathway. Diabetes. 2010;59:1937–1947. doi: 10.2337/db09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H, Zeng H, Zhang Y, Jungreis R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in Perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000a;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000b;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, Mori K. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettmann T, Barton K, Leiden JM. Microphthalmia due to p53-mediated apoptosis of anterior lens epithelial cells in mice lacking the CREB-2 transcription factor. Dev Biol. 2000;222:110–123. doi: 10.1006/dbio.2000.9699. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hong M, Luo S, Baumeister P, Huang JM, Gogia RK, Li M, Lee AS. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J Biol Chem. 2004;279:11354–11363. doi: 10.1074/jbc.M309804200. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Li Y, McGrath BC, Frank A, Cavener DR. PERK eIF2 alpha kinase is required to regulate the viability of the exocrine pancreas in mice. BMC Cell Biol. 2007;8:38. doi: 10.1186/1471-2121-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Wek RC. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem. 2005;280:14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- Jiang HY, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, Wang X, Ron D, Cavener DR, Wek RC. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol Cell Biol. 2004;24:1365–1377. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier C, Nicolino M. Wolcott-Rallison syndrome. Orphanet J Rare Dis. 2010;5:29. doi: 10.1186/1750-1172-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassot I, Segeral E, Berlioz-Torrent C, Durand H, Groussin L, Hai T, Benarous R, Margottin-Goguet F. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(betaTrCP) ubiquitin ligase. Mol Cell Biol. 2001;21:2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. PERK eIF2alpha kinase regulates neonatal growth by controlling the expression of circulating insulin-like growth factor-I derived from the liver. Endocrinology. 2003;144:3505–3513. doi: 10.1210/en.2003-0236. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Vattem KM, Wek RC. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem. 2002;277:18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadanaka S, Okada T, Yoshida H, Mori K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol. 2007;27:1027–1043. doi: 10.1128/MCB.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, Jain MR, Liew C, Chan JY, Kong AN. Toxicogenomics of endoplasmic reticulum stress inducer tunicamycin in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Toxicol Lett. 2007;168:21–39. doi: 10.1016/j.toxlet.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rowe T, Dascher C, Bannykh S, Plutner H, Balch WE. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science. 1998;279:696–700. doi: 10.1126/science.279.5351.696. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler AJ, Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc Natl Acad Sci USA. 2009;106:17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Senee V, et al. Wolcott-Rallison syndrome: clinical, genetic, and functional study of EIF2AK3 mutations and suggestion of genetic heterogeneity. Diabetes. 2004;53:1876–1883. doi: 10.2337/diabetes.53.7.1876. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shen J, Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J Biol Chem. 2004;279:43046–43051. doi: 10.1074/jbc.M408466200. [DOI] [PubMed] [Google Scholar]
- Shen J, Prywes R. ER stress signaling by regulated proteolysis of ATF6. Methods. 2005;35:382–389. doi: 10.1016/j.ymeth.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Baird TD, Wek RC. Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 2011;490:333–356. doi: 10.1016/B978-0-12-385114-7.00019-2. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Marcinko M, Belmont PJ, Glembotski CC. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem. 2007;282:22865–22878. doi: 10.1074/jbc.M701213200. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Morrison L, Glembotski CC. Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J Biol Chem. 2004;279:21078–21084. doi: 10.1074/jbc.M400713200. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Morrison LE, Hoover H, Glembotski CC. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J Biol Chem. 2002;277:20734–20739. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream open reading frames regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Wek RC, Anthony TG. Beta testing the antioxidant function of eIF2alpha phosphorylation in diabetes prevention. Cell Metab. 2009;10:1–2. doi: 10.1016/j.cmet.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;9:2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamotot A, Okada T, Mori K. XPB1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006a;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Feng D, Li Y, Iida K, McGrath B, Cavener DR. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006b;4:491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.