We developed systems to rapidly express any yeast gene or to specifically degrade any protein, each with minimal untargeted disturbance of cell physiology. We illustrate applications of these new tools for elucidating the architecture and dynamics of genetic regulatory networks.
Abstract
We describe the development and characterization of a system that allows the rapid and specific induction of individual genes in the yeast Saccharomyces cerevisiae without changes in nutrients or temperature. The system is based on the chimeric transcriptional activator Gal4dbd.ER.VP16 (GEV). Upon addition of the hormone β-estradiol, cytoplasmic GEV localizes to the nucleus and binds to promoters containing Gal4p consensus binding sequences to activate transcription. With galactokinase Gal1p and transcriptional activator Gal4p absent, the system is fast-acting, resulting in readily detectable transcription within 5 min after addition of the inducer. β-Estradiol is nearly a gratuitous inducer, as indicated by genome-wide profiling that shows unintended induction (by GEV) of only a few dozen genes. Response to inducer is graded: intermediate concentrations of inducer result in production of intermediate levels of product protein in all cells. We present data illustrating several applications of this system, including a modification of the regulated degron method, which allows rapid and specific degradation of a specific protein upon addition of β-estradiol. These gene induction and protein degradation systems provide important tools for studying the dynamics and functional relationships of genes and their respective regulatory networks.
INTRODUCTION
Significant effort has been placed into achieving tightly controlled gene expression systems across a spectrum of organisms, guided by the transformative discoveries of gratuitous inducers of the Escherichia coli lactose (lac) operon genes. The significant desirable features of a useful induction system include 1) a complete lack of target gene activation in the absence of inducer, 2) rapid induction of a target gene in the presence of inducer, and 3) the availability of a gratuitous inducer (i.e., one that interacts with no unintended regulatory system and thus should be inert with respect to physiology). The gratuitous inducers isopropyl β-d-1 thiogalactopyranoside (IPTG) and thiomethyl β-d-galactoside, combined with the extraordinary specificity of lac gene regulation by lac repressor (lacI) and its variants, still most closely approximate the ideal.
Two common approaches in mammalian cells are direct adaptations of bacterial systems: IPTG-inducible lacI-based and tetracycline-inducible tetR-based systems (Labow et al., 1990; Gossen and Bujard, 1992). By expressing a chimeric transcription factor with a eukaryotic activation domain (e.g., VP16) fused to either lacI or tetR, transcription of a target gene downstream of a minimally active promoter with the appropriate operator sites can be maintained at high levels in the absence of inducer. The tetR-VP16 protein, also called tTA (tetracycline transactivator), unbinds DNA in the presence of tetracycline, thereby repressing transcription. This configuration is referred to as tet-off. A mutant of tetR-VP16, named rtTA (reverse tetracycline transactivator), contains a mutated tetR moiety that can bind tet operator sites only in the presence of drug, turning the tet-off system into a tet-on system (Gossen et al., 1995). An advantage of tetR-based systems over lacI-based systems is that tetR binds tetracycline much more tightly than lacI binds IPTG (ka ∼ 109 M−1 for tetR vs. ka ∼ 106 M−1 for lacI), allowing experiments to be performed with lower tetracycline concentrations (Pfahl, 1981; Takahashi et al., 1986).
Tet-based systems have become widely used in Saccharomyces cerevisiae. Tet-off is generally preferred because with tet-on there is considerable transcription in the absence of the inducer, a condition referred to as “leakiness” (Belli et al., 1998). A dual expression system, which uses factors that respond reciprocally to tetracycline addition, has significantly reduced the leakiness of tet-on, however (Belli et al., 1998). Using gene expression microarrays, it was shown that doxycycline (a commonly used tetracycline analogue) does not have a significant impact on the abundance of S. cerevisiae mRNA transcripts (Wishart et al., 2005). A major limitation of the tet-off system is that it relies on protein dilution and degradation to reduce target protein levels, both of which can be quite slow. In addition, a recent implementation of the tet-on system takes nearly 15 min to reach near-maximal levels of target transcription and by 5 min is only weakly induced (Alexander et al., 2010).
The most widely used induction system in S. cerevisiae uses Gal4p-mediated induction of targets, which are placed downstream of promoters (such as the GAL1, GAL7, or GAL10 promoters) containing UASGAL 17-mer sites, CGG-N11-CCG (Giniger et al., 1985). This motif is recognized by a Gal4p homodimer (Marmorstein et al., 1992; Hong et al., 2008). Deletion of the galactokinase GAL1, which is required for galactose utilization, allows galactose to induce GAL genes in a nearly gratuitous manner (Hovland et al., 1989). Because GAL genes are catabolite repressed (Adams, 1972), rapid activation requires that cells be grown in relatively poor, nonglucose carbon sources before the galactose pulse. This system was used by Chua et al. to investigate the transcriptional response of 50+ transcription factors that cause growth inhibition upon overexpression (Chua et al., 2006).
Fusion of the Gal4p DNA binding domain (Gal4dbd) to the electronegative portion of herpes simplex virus protein VP16 results in a strong transcriptional activator (Sadowski et al., 1988). Fusion of the hormone binding domain of the human estrogen receptor to Gal4-VP16 results in a strong transcriptional activator that is only active in the presence of β-estradiol (Louvion et al., 1993), as illustrated in Figure 1. By placing GEV expression under the control of constitutive, low-to-moderate strength promoters, errant expression of target genes is reduced to near-background levels in the absence of β-estradiol (Gao and Pinkham, 2000; Quintero et al., 2007; Veatch et al., 2009). Even though GEV relies on the Gal4dbd to bind DNA, it is not catabolite repressed, providing a significant advantage over the more standard Gal4-based gene induction approach.
FIGURE 1:
Schematic of the GEV overexpression system. GEV is constitutively expressed from an ACT1 promoter. Before activation by β-estradiol, GEV is inactive in the cytoplasm, associating with the Hsp90 chaperone complex. β-Estradiol diffuses through the cell membrane and binds to the GEV estrogen receptor domain, resulting in release of GEV from the Hsp90 chaperone complex. GEV then localizes to the nucleus, binds to the UASGAL consensus sequences, and strongly activates transcription through its VP16 domain.
We were motivated to adapt this system further and to characterize it thoroughly because we wanted to carry out a genome-scale screen of yeast transcription factor activities under circumstances that allow us to follow dynamically the consequences of induction with minimal confounding changes. To this end we eliminated any need for plasmids (as in Gao and Pinkham, 2000 and Quintero et al., 2007) and enabled the use of the system in a prototrophic background with an intact HAP1 gene. Supplemental Table 1 summarizes in more detail the relative merits of alternative expression systems.
Our adaptation of GEV provides an efficient switch that induces target genes to near maximal levels within minutes following hormone addition to the culture. We show that use of intermediate concentrations of inducer results in a graded response: all the cells in the population produce intermediate amounts of protein. Using a single-cell visualization method, we verify that GEV activation occurs through its rapid nuclear localization in the presence of β-estradiol. We show that GEV induction, even to high levels, results in relatively few gene expression changes, making it a useful tool for overexpression studies in S. cerevisiae.
We also developed a modified version of the inducible protein depletion method of Taxis et al. (2009) that allows rapid, specific target protein degradation simply by making the system inducible by β-estradiol using our GEV system. We use GEV to induce the tobacco etch virus (TEV) protease enabling N-end rule degradation (Bachmair et al., 1986) of an appropriately modified target gene driven by its native promoter. We show that the kinetics of degradation are also rapid, resulting in substantial degradation within 15 min after induction of the system. Finally, we show examples illustrating the power of these systems to perturb regulatory networks specifically by introducing (and/or overexpressing) a single regulatory protein by induction with β-estradiol.
RESULTS
The GEV system
We constructed two isogenic strains of opposite mating type (DBY12020, DBY12021) to use for studies of gene induction/overexpression in S. cerevisiae. These strains (see Table 1) contain an integrated copy of PACT1-GEV, so that GEV is produced constitutively and at relatively high levels. To study dynamically the consequences of induction of a gene, its promoter is replaced by the GAL1 promoter (PGAL1) in either DBY12020 or DBY12021. If one wants a fluorescent reporter to monitor induction, one can use DBY12039. To study the consequences of the degradation of a protein, a peptide sequence recognized by TEV (i.e., N-degron sequence) can be appended to the N-terminus of the gene in DBY12132, as described in Materials and Methods.
TABLE 1:
Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| DBY11389 | MATa, ura3Δ0 | This study |
| DBY11408 | MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, gal10Δ::KanMX4 | Open Biosystems |
| DBY11415 | MATa/MATα, (PGAL10+gal1)Δ::loxP/GAL1, leu2Δ0::PACT1-GEV-GFP-KanMX/LEU2, gal4Δ::LEU2/GAL4, HTB2-mCherry-CaUra3/HTB2, his3/HIS3, leu2/LEU2, hap1–/HAP1+ | This study |
| DBY12020 | MATa, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, HAP1+ | This study |
| DBY12021 | MATα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, HAP1+ | This study |
| DBY12027 | MATα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, KanMX4-Pgal1-MET4, HAP1+ | This study |
| DBY12039 | MATα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, ybr032wΔ::PGAL1-GFP-NatMX, HAP1+ | This study |
| DBY12040 | MATα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, KanMX4-PGAL1-CBF1, HAP1+ | This study |
| DBY12055 | MATa, NDeg-MET4-13Myc-KanMX, gal4Δ::LEU2, gal1Δ::TEV-HphMX4, leu2Δ0::PACT1-GEV-NatMX, HAP1+ | This study |
| DBY12086 | MATα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, KanMX4-PGAL1-TPS2, HAP1+ | This study |
| DBY12099 | MATα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, KanMX4-PGAL1-MET4, met6Δ::HphMX4, HAP1+ | This study |
| DBY12100 | MATα, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, ura3Δ::HphMX4, HAP1+ | This study |
| DBY12132 | MATα, gal1Δ::TEV-URA3, ura3Δ0, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, HAP1+ | This study |
| DBY12168 | MATa/MATα, gal4Δ::LEU2/GAL4, (PGAL10 + gal1)Δ::loxP/GAL1-mCherry-NatMX, leu2Δ0::PACT1-GEV-GFP-KanMX/LEU2, HAP1+/HAP1+ | This study |
| DBY11440 | MATa/MATα, MET4-13Myc-KanMX/MET4, ura3Δ0/URA3, gal1Δ::TEV-URA3/GAL1, gal4Δ::LEU2/GAL4, leu2Δ0::PACT1-GEV-KanMX/LEU2, hap1–/HAP1+ | This study |
| DBY12032 | MATa, leu2Δ0, HAP1+ | This study |
| DBY12194 | MATα, NDeg-MET31, gal1Δ::TEV-URA3, fcy1Δ::CreEDB78, HAP1+ | This study |
| DBY12197 | MATa, MET31-13Myc-KanMX, can1Δ::PACT1-GEV, lys2, gal1Δ::TEV-URA3, HAP1+ | This study |
| DBY12199 | MATa, NDeg-MET31-13Myc-KanMX, gal1Δ::TEV-URA3, fcy1Δ::CreEDB78, HAP1+ | This study |
| DBY12200 | MATα, gal1Δ::TEV-URA3, can1Δ::PACT1-GEV, gal4Δ::LEU2, HAP1+ | This study |
| DBY12234 | DBY12199 x DBY12200 | This study |
| DBY12235 | DBY12197 x DBY12194 | This study |
All strains are s288c. HAP1+ indicates a repaired HAP1 allele (Hickman and Winston, 2007), and hap1– indicates the partial loss-of-function s288c allele (Gaisne et al., 1999; Hickman and Winston, 2007).
Note that a gene whose promoter has been replaced with a GAL regulated promoter should be expressed only after addition of β-estradiol to the growth medium; this has been the case with the several dozen constructs we have made. Importantly, although GEV binds DNA via the Gal4p DNA binding domain, it is not subject to inhibition or repression by glucose, making feasible induction and overexpression experiments in standard glucose-containing media simply by the addition of the inducer β-estradiol.
Quantitation of GEV localization
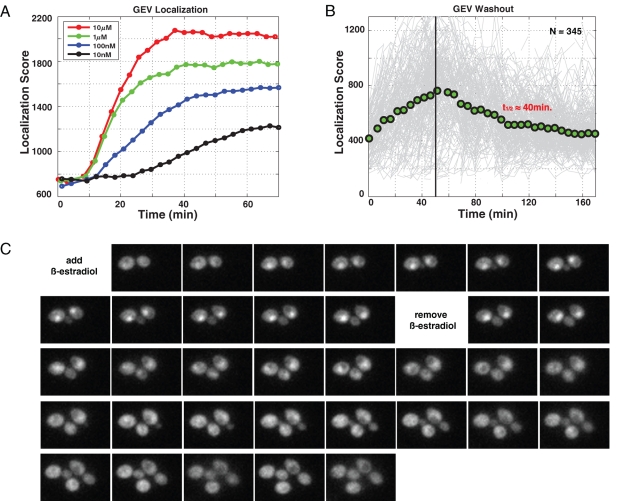
Generally, hormone receptors localize to the nucleus and activate transcription after binding their cognate ligands (Pratt and Toft, 1997). To our knowledge, this model has not been experimentally verified in the case of GEV. Therefore, to study GEV nuclear localization following hormone addition, we constructed a functional (Supplemental Figure 1), chromosomally integrated, C-terminus–tagged GEV-GFP (green fluorescent protein) reporter and measured its localization following pulses of β-estradiol (Figure 2A and Supplemental Movie 1) in single cells. We observed a level of GEV localization that is proportional to the amount of added β-estradiol for a range of concentrations. At higher doses (100 nM to 10 μM), we can see the nuclear localization signal emerging from the background 6–8 min following hormone addition. The initial rate of localization achieved its maximum at concentrations at or above 1 μM β-estradiol.
FIGURE 2:
Single-cell analyses of a GEV-GFP fusion protein. (A) Measuring nuclear localization of GEV-GFP in the presence of different amounts of β-estradiol in DBY11415. (B) DBY11415 was grown in the presence of 1 μM β-estradiol for 50 min. The input medium was then switched to medium lacking β-estradiol, and the kinetics of delocalization were quantified. (C) Images of single cells from the washout experiment in (B). For each cell, the localization score is defined as the mean of the top 5% of pixel intensities minus the mean pixel intensity over the entire cell.
To determine the nuclear residence time of activated GEV after removal of β-estradiol, cells were grown in a flow chamber. For the first 50 min, medium containing 1 μM β-estradiol was flowed over the cells. Then the medium inflow was switched (Figure 2B, black line) to a medium lacking β-estradiol, resulting in an approximately exponential reduction in nuclear fluorescence with time. In this way we determined that the nuclear half-life of GEV is ∼40 min (Figure 2, B and C, and Supplemental Movie 2).
Kinetics of GEV-mediated gene induction
To investigate the kinetics of target gene induction by GEV, we inserted PGAL1 in front of various reporter genes (Liko et al., 2006). First, we constructed a strain in which PGAL1 is driving transcription of the CBF1 gene at its native chromosomal locus. The level of CBF1 transcript, as determined by microarray analysis, reaches ∼70% its maximum by 5 min following addition of 1 μM β-estradiol (Figure 3A). Using fluorescence in situ hybridization (FISH) to monitor transcription in single cells, we observed both the cytoplasmic transcripts and nuclear transcription sites of CBF1, as shown in the image from Figure 3B, which was taken 8 min following addition of 1 μM β-estradiol to the culture. We used a PGAL1-GFP reporter to find the point at which GEV becomes saturated by inducer. The percentage of induced cells is determined (see Materials and Methods) over time, and we find that GEV is saturated between 100 nM and 1 μM β-estradiol, as the induction kinetics are similar between these two concentrations (Figure 3C).
FIGURE 3:
The kinetics of GEV-mediated genetic switch. (A) We followed transcription of PGAL1-CBF1 induced by GEV by microarray (strain = DBY12040), with DBY12001 RNA as a reference. Values are zero-normalized. (B) Maximal projection image of CBF1 transcripts from DBY12040 8 min after β-estradiol addition to the medium (FISH). Active transcription sites of PGAL1-CBF1 can be seen in single cells. (C) GEV activation of a PGAL1-GFP reporter in DBY12039 as measured by flow cytometry.
GEV-mediated gene induction shows a graded response
The generally accepted model for the mechanism of nuclear receptors predicts that subsaturating concentrations of inducer result in a response that is proportional to the concentration of inducer (i.e., exhibit a graded as opposed to switch-like response). Indeed this behavior was reported for GEV by Takahashi and Pryciak (2008). To verify a graded response for our system, we performed experiments in which the population of cells containing PGAL1-GFP was induced by β-estradiol for 12 h and then quantified by flow cytometry.
At concentrations less than 1 nM β-estradiol, the system is ineffective: the average intensity of the reporter is at nearly the background level (Figure 4A). At 10 nM β-estradiol, GFP intensity is increased but is well below what is achieved at higher inducer concentrations (Figure 4B); the significant point is that all the cells exposed to 10 nM β-estradiol exhibit significant fluorescence, as shown by the shift of the entire distribution to the right (Figure 4A). At 100 nM, the average induction is near maximum and the distribution of cells closely resembles that found for higher, completely saturating concentrations of β-estradiol.
FIGURE 4:
GEV activity shows a graded response in response to β-estradiol. (A) Flow cytometry data at different doses of β-estradiol. Cells were grown to mid-log phase and incubated with the indicated amount of β-estradiol for 12 h. (B) The mean GFP intensity from histograms in (A) as a function of β-estradiol dose. The Hill coefficient (nH) is 0.95 (95% CI, nH = 0.72 to 1.18). The strain used in this experiment is DBY12039.
A trade-off between GEV activation and cell growth
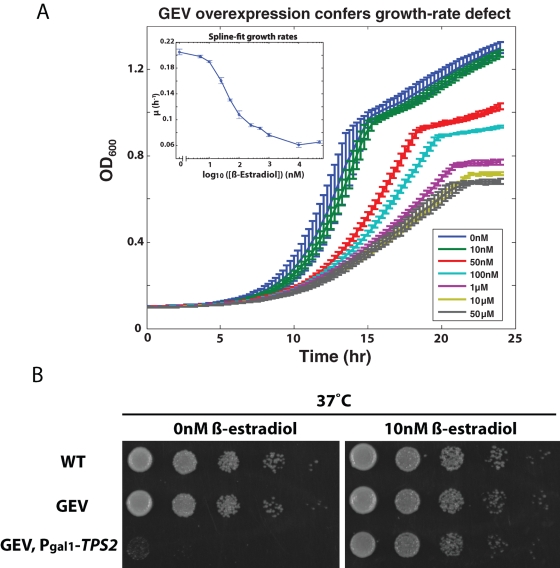
We find that strong GEV activation achieved at high concentrations of β-estradiol results in a slowing of growth (Figure 5A). This is not due to β-estradiol toxicity because the slow growth does not occur in a strain lacking the GEV system. We speculate that this effect could be due to “squelching” (Gill and Ptashne, 1988), whereby strong transcriptional activators repress off-target genes through titration of the RNA polymerase machinery, and thus reduce the cell's capacity for growth. Lang et al. (2009) have provided evidence in yeast that there is indeed a fitness cost exacted by synthesis of proteins not required for asexual proliferation.
FIGURE 5:
GEV is a gratuitous inducer at 10 nM β-estradiol without a growth defect. (A) DBY12021 grown in YPD liquid in the presence of β-estradiol. A600 was monitored over time, and growth rates were spline-fit (inset). Error bars represent ±1 SD of three replicates. (B) The heat sensitivity of tps2Δ is repaired by GEV-mediated induction of TPS2 with 10 nM β-estradiol. (WT = DBY12001; GEV = DBY12021; GEV + PGAL1-TPS2 = DBY12086).
At lower concentrations of β-estradiol (10 nM), the GEV-containing cells no longer show the growth defect (Figure 5A), but are able to induce the PGAL1-GFP reporter essentially in all the cells nonetheless (Figure 4A). Inducing GEV at 1 μM β-estradiol results in a greater than twofold reduction in growth rate (Figure 5A, inset).
To determine whether physiologically significant expression of target genes can be induced at intermediate (10 nM) levels of induction, where no growth consequence is observed, we designed an experiment in which a lethal defect might be complemented in a GEV-dependent manner. TPS2 encodes an enzyme that converts trehalose-6-phosphate to trehalose. Deletion of TPS2 causes a heat-sensitive growth phenotype, likely due to the buildup of trehalose-6-phosphate (Devirgilio et al., 1993). By placing TPS2 downstream of PGAL1, cells become heat sensitive in the absence of β-estradiol (Figure 5B). Growth is restored to wild-type levels in the presence of 10 nM β-estradiol, and the resulting growth is indistinguishable from wild type (Figure 5B).
This result suggests that maximal induction is not likely to be necessary to achieve physiological levels of protein in all experiments. Intermediate levels of inducer suffice to avoid the growth-inhibiting side effects and complement the deletion. In practice, the system is fast-acting at high (>100 nM β-estradiol) concentrations, but has increased growth rate inhibition. Even at the highest inducer concentrations, few unintended genes are expressed at early times after induction. Thus the user has a trade-off: if the goal of experiments is just to see which genes are induced first, high concentrations will yield this result; if the goal is longer-term specificity, where speed is not of the essence, low concentrations of inducer can be used. It is not clear that this problem is avoidable: using a less strong promoter than that of ACT1 to drive GEV produces a system with the mitigated growth effect but at the price of less speedy induction.
GEV induction in chemostats is suitable for dynamically studying the physiological consequences of gene action
Because yeast are extremely sensitive to slight changes in the extracellular environment, the ideal growth setting for induction experiments is in the chemostat. Briefly, a chemostat works by flowing fresh medium into a growth vessel, and culture medium is diluted from the vessel at a rate determined by the experimenter. With this experimental setup, we can grow to steady state a culture of cells containing the GEV construct and PGAL1 fused to any gene of interest. In the case in which a gene's native promoter is replaced with PGAL1 (in a haploid), the cells are effectively deletion mutants of that gene in the absence of β-estradiol due to the tight repression of PGAL1 in the presence of 2% glucose. By adding β-estradiol to such cultures, we can rapidly and strongly induce the gene of interest and follow the physiological consequences. For the particular case of genes encoding transcription factors, the consequence to be followed is the genome-wide transcriptional response of the cells. The goal is to infer a transcription factor's immediate targets and quantify the strength of their response. For those target genes that respond most quickly, the inference of causation is strengthened.
GEV induction in a steady-state culture is nearly gratuitous
For this approach to be effective, GEV needs to be nearly gratuitous, meaning that it does not have a large effect on the transcriptional landscape of the cell. Although this definition is slightly ambiguous, we can define a large effect as one that results in strong increases/decreases in many genes such that it becomes difficult to separate changes due to overexpressing the target gene from those due to background. By growing a strain (DBY12021) that contains GEV in a chemostat, we find that GEV can be nearly gratuitous (Figure 6A). In DBY12021, the GAL1 and GAL4 genes are deleted, as is the GAL10 promoter. Two hours following addition of saturating (1 μM) β-estradiol, 105 genes decreased twofold (Supplemental data set 1) and 94 genes increased twofold (Supplemental data set 2; Supplemental Figure 7). This finding accounts for ∼3% of all S. cerevisiae genes. Although this can be seen as modest compared with all yeast genes, it is nevertheless more than one sees in many deletion mutants, including transcription factors (Hughes et al., 2000; Hu et al., 2007). By using the gene ontology (GO) term finder to find enriched processes within these groups, we found that repressed genes are most enriched for glucose catabolic processes (corrected p-value = 8.49 × 10−9) and activated genes are most enriched for oxidation-reduction processes (corrected p-value = 1.73 × 10−7). Note that at 18 min following addition of 1 μM β-estradiol, a total of 57 genes have changed twofold (Supplemental Figure 7).
FIGURE 6:
(A) Hierarchical clustering of gene expression of DBY12021 grown to steady state in a phosphate-limited chemostat with a doubling time of 4.3 h, and at t = 0 min, pulsed with 1 μM β-estradiol. (B) The transcriptional response of the GAL genes from (A). (C) Transcriptional response of YPL066W and YPL067C in (A). (D) The 10 most strongly repressed genes in (A). Clusters in (B) and (D) were hierarchically clustered.
GEV induction results in strong activation of GAL2 and GAL7 and moderate induction of GAL3, GAL5, and GAL80 (Figure 6B). Surprisingly, despite the removal of the GAL10 promoter by loxP in DBY12021, we observe slight induction of GAL10 over the course of the experiment.
We found two genes (YPL067C and YPL066W) that are divergently transcribed in response to GEV induction (Figure 6C). These genes are reminiscent of GAL1 and GAL10, which are divergently transcribed in response to Gal4p activation. YPL066W and YPL067C are separated by a 385-nucleotide region. This region contains a single UASGAL sequence, which is conserved upstream of the YPL066W and YPL067C orthologues in Saccharomyces mikatae and Saccharomyces paradoxus. Despite the conservation of the Gal4p binding site, these genes have no known role in galactose metabolism and are of unknown function.
The most strongly repressed gene by 2 h following GEV activation is HSP30 (17.4-fold). The 10 genes most strongly repressed at 2 h following GEV activation are shown in Figure 6D. We could not find any significant correlation with annotated functions that these genes might hold in common.
Using GEV overexpression to probe kinetics of transcriptional regulatory networks
Large-scale ChIP-chip studies have defined extensive transcription factor–DNA binding maps, yielding a highly informative, although static, view of genetic regulation. Using the GEV system to induce rapid and high-level expression of a single transcription factor, we can look at the dynamics of target gene activation or repression.
As an example and proof of principle, we induced the MET4 gene, which encodes a well-studied, strong transcriptional activator of sulfur metabolic genes in S. cerevisiae. A strain containing PGAL1-MET4 was grown to steady state in a phosphate-limited chemostat containing high levels of extracellular methionine, a condition in which Met4p is supposed to be less active. By hierarchically clustering the transcriptional profile of the sulfur metabolic genes as a time series up to 90 min following MET4 induction, we were able to observe that the kinetics of MET4-target induction can vary from gene to gene (Figure 7).
FIGURE 7:
Quantifying the kinetics of MET4's downstream targets. DBY12027 (GEV, PGAL1-MET4) was grown to steady state in a phosphate-limited chemostat with excess methionine (200 mg/l) with a doubling time of 4.3 h. At t = 0 min, cells were pulsed with 1 μM β-estradiol. Left, hierarchical clustering of the sulfur metabolic genes with three clearly identifiable categories based on kinetics. Right, the mean expression of genes in each category is plotted (color) along with individual traces (gray). Error bars represent ±1 SD of the mean.
We divide the targets into three categories based on their kinetic profiles: strongly induced (group 1), weakly induced (group 2), and uninduced (group 3) (G1, G2, and G3, respectively, in Figure 7). The MUP1 and MUP3 genes, which encode the high- and low-affinity methionine permeases, respectively, and SAM1 and SAM2, which encode enzymes that catalyze the synthesis of S-adenosylmethionine (AdoMet), are strongly induced group 1 genes. AdoMet is the primary source of methyl donor groups in the cell.
The MET31 and MET32 genes encode homologous zinc-finger proteins (46% identical), which aid in the recruitment of Met4p to many sulfur metabolic gene promoters. MET32 is weakly induced (group 2), and we do not detect any induction of MET31 (group 3). Whereas met31Δ and met32Δ mutant cells are able to produce their own methionine when grown on minimal medium, met31Δmet32Δ double-mutants are methionine auxotrophs. Despite the sequence and functional similarity of these proteins, the observation of differential regulation is suggestive of distinct roles for MET31 and MET32, at least under particular conditions, resulting in retention of both copies during evolution.
GEV-mediated induction of TEV results in rapid degradation of N-degron–tagged proteins
We chose the MET4 and MET31 genes to use in a proof-of-principle experiment to test our version of the Taxis et al. (2009) protein depletion method (Figure 8). We constructed Ndeg-modified MET4 and MET31 alleles in separate strains containing PACT1-GEV and PGAL1-TEV (see Materials and Methods). Cells were grown in rich medium (YPD) and treated with 1 μM β-estradiol at ∼2 × 107 cells/ml. Western blots were performed on proteins extracted before the addition of β-estradiol, and at several time points after induction (Figure 9). Treatment of cultures with 1 μM β-estradiol had no effect on levels of Met4p-13Myc or Met31p-13Myc lacking NDeg (Figure 9, A and B). However, 1 μM β-estradiol treatment of NDeg-MET4 or NDeg-MET31 cells showed rapid reduction of the NDeg-tagged proteins (Figure 9, A and B). By performing a best fit of the data from 10-, 20-, and 40-min time points to a power law, we find that the half-life of NDeg-Met4p-13Myc is 16 min and that of NDeg-Met31p-13Myc is 13.75 min (Figure 9C).
FIGURE 8:
(A) GEV is expressed under the regulation of the constitutive ACT1 promoter. Treatment with β-estradiol induces nuclear entry of GEV and subsequent induction of PGAL1-driven TEV. (B) The target gene of interest (YFG) is regulated by its native promoter. TEV protease binding to its cleavage site in NDeg-modified YFG, facilitated by p14-SF3b binding, results in cleavage, exposing the destabilizing amino acid phenylalanine at the N-terminus. Proteasome-mediated digestion of YFG is the result of N-end rule degradation pathway.
FIGURE 9:
(A) Western blot of DBY12055 (NDeg-Met4p-13Myc) and DBY11440 (Met4p-13Myc) before (0′) and after (10′, 20′, and 40′) 1 μM β-estradiol addition to the culture. (B) Western blot of DBY12234 (NDeg-Met31p-13Myc) and DBY12235 (Met31p-13Myc) before (0′) and after (10′, 20′, and 40′) 1 μM β-estradiol addition to the culture. (C) Normalized protein levels from experiments in (A) and (B) with DBY12055 and DBY12199. Protein levels were quantified in ImageJ and then fitted to a power law for the 10′, 20′, and 40′ time points.
A met4 null allele has a growth defect and produces small colonies on YPD plates (Hickman et al., 2011). We therefore started with a diploid cell for construction of NDeg-MET4. Sporulation of this diploid followed by dissection of the resulting tetrads showed that the spore colonies had the same wild-type size, confirming that the modified gene was functional.
Met4p is known to be ubiquitinated (Rouillon et al., 2000) and is, as a result, unstable. We observed three clear bands in our Met4p Western blots, consistent with the presence of nonubiquitinated Met4p in addition to heavier ubiquitinated forms (Figure 9A). When MET4 is tagged with NDeg, both the ubiquitinated and nonmodified Met4p proteins are targeted for degradation by TEV (Figure 9A). This finding demonstrates that even heavily modified forms of a protein can be cleaved by TEV.
Finally, we assayed the phenotypic effect of β-estradiol treatment of NDeg-MET4–containing cells. As expected, only cells treated with β-estradiol displayed a methionine auxotrophy (met –) phenotype (Supplemental Figure 8), as Met4p is required for methionine synthesis (Masselot and Robichon-Szulmajster, 1975).
DISCUSSION
The data presented show that the GEV system, as we configured it, satisfies the requirements of an effective and nearly gratuitous induction system. It can be used to rapidly induce a target gene in every cell in a culture while conferring minimal induction of off-target genes. GEV provides efficient induction. Transcription rates in S. cerevisiae measured in the literature range from 0.7 to 2 kb/min (Mason and Struhl, 2005; Zenklusen et al., 2008), the upper bound of which has been measured from a PGAL1-driven gene (Mason and Struhl, 2005). If a gene is 2 kb in length and is downstream of PGAL1, as in the case of PGAL1-MET4, generating a full transcript should take 1 min. By 2.5 min, we observed detectable amounts of PGAL1-driven transcript (Supplemental Figure 2). By 2.5 min, therefore, the gene has gone through several rounds of transcription. By 5 min, the amount of induction is near-maximal.
In our localization experiments, we detect GEV-GFP in the nucleus at 6–8 min following addition of β-estradiol to the media. If nuclear localization takes 6–8 min, how do we measure full-length transcripts at 2.5 min following β-estradiol to the medium? There are at least two potential explanations. First, the GEV-GFP fusion may have slightly slower kinetics of nuclear localization than GEV. Second, the kinetics of GEV-GFP reflects that of GEV without the fluorescent tag, but measuring localization is too insensitive to detect the first few GEV-GFP molecules that become localized to the nucleus and activate transcription.
Opportunities for dynamic studies of regulation
One of the most significant advantages of the system is the potential for following the consequences of the induction of a specific protein dynamically. This advantage is clearest in the case of a transcriptional regulator like Met4p, which we discuss further below. Not only can our understanding of the induction kinetics allow us to distinguish primary from secondary targets, but the cascade of consequences can be followed as time series that can shed light on the sequence of events under a variety of physiological conditions.
Through overexpression of MET4, we have shown that we can distinguish the kinetics of induction of the sulfur metabolic genes (Figure 7). As we will present in a future study, the kinetics of target induction can depend strongly on the choice of nutrient limitation (i.e., phosphate vs. methionine limitation). ChIP-chip studies can show the presence or absence of transcription factor–DNA interactions. They fail, however, to capture the strength and kinetics of induction of a particular gene in response to a single factor, something that we are able to readily measure via GEV-mediated overexpression of particular transcription factors. Because of these advantages afforded by the GEV system, we are currently in the process of constructing a GEV-based, prototrophic collection of transcription factor overexpression strains.
On removing β-estradiol from the media, GEV-GFP begins to delocalize within minutes (Figure 2B). How does delocalization occur so quickly? In the presence of β-estradiol, we can say that cytoplasmic GEV binds β-estradiol and enters the nucleus at the rate kin. Nucleus-localized GEV is either bound to or unbound from DNA. Unbound GEV diffuses throughout the nucleus and exits the nucleus at the rate kout. Because kin approaches 0 in the absence of β-estradiol, we can immediately measure unbound GEV leaving the nucleus the rate kout. The true rate of nuclear delocalization of GEV, however, includes the unbinding of GEV from DNA, which can be much slower.
Given that the nuclear half-life of GEV is ∼40 min and that protein half-lives can be many hours, removing β-estradiol from the media is not an effective way to quickly deplete the levels of a target protein. Similarly, methods that rely on blocking transcription upon addition of inducer to cells are not effective for rapidly depleting target proteins. These facts motivated us to drive the expression of TEV by GEV, which allows us to rapidly turn off target proteins containing an N-degron sequence. This sequence can readily be added to genes via homologous recombination. Therefore, we now have a method for rapidly shutting off the levels of single genes expressed from their native promoters. Upon β-estradiol addition and subsequent depletion of the target gene by TEV, we can study the cell's transient physiological response (such as the transcriptional profile) to losing both essential and nonessential genes. Importantly, these types of studies could help us to rigorously define the transcriptional regulatory network of essential genes in S. cerevisiae.
Opportunities for enhancement of the system
There are still a few dozen genes that are induced or repressed significantly when GEV is induced with β-estradiol. One way to improve the system will be to replace GEV's Gal4dbd with a DNA binding domain from either a non-yeast transcription factor (such as LexA) or from custom-designed zinc fingers that recognize DNA sequences not found in S. cerevisiae. Ideally, we can choose a DNA binding domain with high target DNA specificity and low enough affinity for binding DNA that the chimeric protein binds only to a single sequence of interest.
Another area for extension of the system is the adaptation of other nuclear receptors that work by the same mechanism as that of GEV, so that independent induction of two target genes can be accomplished. With two independent inducing systems one could also induce synthesis of a target protein by adding one inducer, and then destroy, at will, the protein by inducing the TEV protease with the other inducer.
Conclusion
To conclude, we believe that the GEV and TEV induction and protein degradation systems will allow new kinds of regulatory control of cells in vivo. Mutant studies generally require growing a defective strain up to study; with GEV one can avoid this by making expression of the gene dependent on gratuitous induction with β-estradiol, as we showed in the case of TPS2 (Figure 5). This feature was put to use in studying the growth effects of met4 deletions, as was the opportunity to study the effect of loss of Met4p dynamically using the TEV system described here (Hickman et al., 2011). For accurate assessment of the roles of genes and proteins in networks, dynamic perturbation of steady states still seems the most attractive approach, which we imagine will be facilitated by the technology introduced here.
MATERIALS AND METHODS
Strains
We constructed prototrophic HAP1+ yeast strains that can be readily modified to either 1) induce expression of a single gene or 2) rapidly degrade a single target protein. These strains contain an integrated copy of GEV driven by an ACT1 promoter and are gal4Δ gal1Δ.
For induction/overexpression studies, we constructed GEV-containing strains of either mating type: DBY12020 (MATa) and DBY12021 (MATα). These strains can be transformed with a KanMX4-PGAL1 linear fragment (Liko et al., 2006) in front of any non-essential gene to make that gene inducible by β-estradiol.
For protein degradation studies, we constructed DBY12132 and DBY12200, which contain both GEV and the TEV protease. In DBY12132 and DBY12200, PACT1-GEV is at the LEU2 or CAN1 locus, respectively. By modifying a target gene in these strains to contain a degron sequence recognized by TEV at its N-terminus, a gene can be expressed under its native promoter and degraded in the presence of β-estradiol.
Details of the strain construction can be found in the Supplementary Information. Relevant primers are provided in Supplemental Table 2.
Media and growth conditions
For microscopy and flow cytometry experiments, cells were grown in low-fluorescence medium (LFM). LFM is chemostat minimal medium (Brauer et al., 2008) lacking riboflavin and folic acid, and contains, in addition to glucose (20 g/l), 5 g/l (NH4)2SO4, 1 g/l KH2PO4, 0.5 g/l MgSO4, 0.1 g/l NaCl, 0.1 g/l Ca2Cl, 0.5 mg/l H3BO4, 0.04 mg/l CuSO4, 0.1 mg/l KI, 0.2 mg/l FeCl3, 0.4 mg/l MnSO4, 0.2 mg/l Na2MoO4, 0.4 mg/l ZnSO4, 2 μg/l biotin, 0.4 mg/l calcium pantothenate, 2 mg/l inositol, 0.4 mg/l niacin, 0.2 mg/l para-aminobenzoic acid, 0.4 mg/l pyridoxine HCl, and 0.4 mg/l thiamine.
For induction experiments, yeast were cultured in 500 ml chemostats (Sixfors, Infors AG, Bottmingen, Switzerland) under phosphate limitation (20 mg/l potassium phosphate) and pulsed with 1 μM β-estradiol (Tocris Biosciences, Ellisville, MO) diluted from a 2.5 mM stock in ethanol. For the MET4 induction experiment, phosphate-limited medium was supplemented with 200 mg/l methionine. Culture volume was maintained at 300 ml. Approximately 18 h of batch phase growth before pump turn-on was initiated from a 1:60 dilution of cells grown overnight in the same medium. Cultures were grown at 30°C, stirred at 400 rpm, and aerated at ∼6 l/min with filtered humidified air.
For all other experiments, cells were grown on rich medium (YPD; 2% peptone, 1% yeast extract, 2% glucose).
Microscopy
For GEV localization studies, fluorescence was measured with an epifluorescence Nikon Eclipse-TI inverted microscope using a Nikon 40×/0.95 NA DIC Plan Apo objective. The microscope is equipped with a PerfectFocus system (Nikon) for maintaining the correct focal plane, a Clara CCD Camera (Andor DR328G; South Windsor, CT) for recording fluorescence emission, and a TI-S-ER motorized stage with encoders to ensure near-perfect return to marked stage positions (Nikon MEC56100). GEV-GFP emission was visualized at 525 nm (50-nm bandwidth) upon excitation at 470 nm (40-nm bandwidth; Chroma 49002_Nikon ETGFP filter cube). HTB2-mCherry emission was visualized at 620 nm (60-nm bandwidth) upon excitation at 545 nm (30-nm bandwidth; Chroma 96364_Nikon ET-DSRed filter cube). Image acquisition was automatically controlled using NIS Elements software (Nikon Instruments, Melville, NY).
For FISH, images were acquired with an Olympus IX81 inverted fluorescence microscope as described in Silverman et al. (2010).
GEV-GFP localization experiment
Cultures of DBY11415 were grown in LFM overnight to saturation. Overnight cultures were diluted 1:50 in fresh LFM to a Klett of ∼10 (Klett-Summerson Colorimeter). The Klett was monitored, and at Klett 40 cells were loaded into 96-well glass-bottom optical plates for microscopy (Nunc 265300; Thermo Fisher Scientific, Rochester, NY). Each well in the 96-well plate was incubated with 30 μl of concanavalin A solution (2 mg/ml concanavalin A [MP Biomedicals 150710] dissolved in 5 mM MnCl2 5 mM CaCl2, pH 6.5) to fix cells to the bottom of the plate. After a 5 min incubation, excess concanavalin A was aspirated off, and 4 μl of cell culture + 91 μl of fresh LFM + 1 μl of 100% ethanol were added to each well. Cells were allowed to settle to the bottom of each well while stage positions were marked on the microscope using ambient room light.
Cells were imaged immediately before and immediately after addition of 4 μl of β-estradiol to each well. Each experiment was done in triplicate, and two images were taken per well of the 96-well plate, resulting in six image frames per concentration of β-estradiol. Images were acquired every 170 s during induction of GEV-GFP localization.
GEV-GFP washout experiment
DBY11415 cells were maintained in a flow cell (Bioptechs FCS2; Butler, PA) during microscopy. Cells were adhered to the bottom of a 40-mm round coverslip by first incubating the coverslip with concanavalin A, and then the coverslip was assembled into the FCS2 chamber. Cells were maintained at room temperature during the course of the experiment (25°C). The Klett of the culture was 110 when cells were loaded into the flow cell. Two peristaltic pumps (Instech P720; Plymouth Meeting, PA) were alternately used to perfuse medium to cells during microscopy. One pump perfused LFM, and the other perfused LFM with 1 μM β-estradiol. The pump flow rates were automatically controlled by computer (Bioptechs Perfusion Controller), and by switching between pumps we were able to exchange media within the cell chamber in ∼2 min. GEV nuclear localization was first induced with LFM + 1 μM β-estradiol for 50 min. The pump flow rates were then switched, and images were acquired every 300 s to monitor delocalization of GEV-GFP. Conditions for imaging GFP and mCherry were identical to the induction experiment except that a 1.5X optovar was inserted into the optical path to magnify images from 40X to 60X.
GEV-GFP image processing
Image processing was performed by preprocessing the images in ImageJ and then using a custom-written Matlab script to extract the intensities in the nuclei relative to the cytoplasm. Code is available upon request. Briefly, the image processing was done as follows: Fluorescence images were background subtracted using the rolling ball background subtraction command in ImageJ using a ball radius of 50 pixels. The method has been previously described (Sternberg, 1983). Images were examined by eye in ImageJ, and out-of-focus images were removed before further analysis.
These preprocessed images were then analyzed using a custom-written Matlab script. In brief, the code first identifies cells using various thresholding commands on the GFP images to create a “mask” of the image (an image in which pixels inside of cells are identified as 1 and pixels outside of cells are identified as 0). The user is then given an opportunity to correct any errors in this mask by hand. The code then uses similar thresholding commands on the mCherry image to identify cell nuclei and create a mask for the nuclei. Using these two masks the script then extracts intensity information from the GFP image for each cell identified in the cell mask, using the nucleus mask to differentiate between the cytoplasm and the nucleus. A particle-tracking routine (track.m from http://physics.georgetown.edu/matlab/index.html) is then used to align values for cells and nuclei between images taken at different time points. Finally, the various parameters are exported as cell-by-time matrices in which each row represents a single cell identified throughout the time course. The Localization Score in all figures is determined for each cell by first averaging the top 5% of pixel intensities and then subtracting from this number the average pixel intensity over the entire cell (Cai et al., 2008).
FISH
FISH probe design and experiments were performed as described by Silverman et al. (2010). DBY12040 was grown to early-log phase and pulsed with 1 μM β-estradiol. Cells were fixed before β-estradiol addition to the culture, and at 2 and 8 min (Figure 3B) following β-estradiol addition. The Cy3-labeled DNA probes used for FISH are listed in Supplemental Table 3.
Flow cytometry
Cell fluorescence was measured with a BD LSRII Multi-Laser Analyzer with HTS (BD Biosciences, Sparks, MD). High fluorescence in single cells was determined from the distribution of fluorescent intensities in cells lacking GFP (the distribution was determined from the fluorescence values of 100,000 cells). The cutoff for high fluorescence was set at the top 0.2% of the distribution (GFP signal = 190 au).
Growth curves
Cells and media (100 μl) were grown on flat-bottom, 96-well plates (Costar 3631). A600 was measured every 15 min in a Synergy HT (Bio Tek, Winooski, VT) microplate reader. Cells were grown at 30°C with medium shaking. Growth rates were computed with spline-fits using the grofit R package (Kahm et al., 2010).
Microarrays and functional annotation
RNA from cells was extracted, labeled, and hybridized to Agilent (Santa Clara, CA) expression microarrays as described by Brauer et al. (2008) with slight modifications. Briefly, RNA was extracted from frozen cells with a standard acid–phenol method and then cleaned with RNeasy (Qiagen, Valencia, CA). Cleaned mRNA was then converted to cDNA and subsequently converted to labeled cRNA with the Agilent Quick-Amp Labeling Kit (Part No. 5190-0424). Hybridization to 8 x 15k Agilent microarrays (Agilent, Santa Clara, CA) was performed for 17 h at 65°C while rotating on a rotisserie at 20 rpm. Microarrays were washed and then scanned with Agilent Feature Extractor Software version 9.5. Flagged features marked as unreliable were marked N/A. Raw green- and red-channel intensities were floored to a value of 350 (i.e., intensities < 350 were set equal to 350). If the numerator (red-channel, sample RNA) for a given measurement was floored, the denominator (green-channel, reference RNA) for that measurement was set to the average green channel intensity for that gene. The purpose of this smoothing process is to make genes with negligible red-channel signal intensity less sensitive to small fluctuations in the reference intensity. After flooring of the data, the log2 ratios were computed.
For time series experiments, 5 ml of cells was taken per culture per time point. Reference RNA was taken from DBY12001 grown in a phosphate-limited chemostat at a dilution rate of 0.18h−1. Enrichment for Gene Ontology (GO) terms was determined using the Generic Gene Ontology Term Finder (Boyle et al., 2004).
Construction of the degron system
The degron system consists of two parts, each of which ends up inserted into the chromosome. One of these is the protease cassette, consisting of the GAL1 promoter, a modified TEV protease-coding region, the spliceosome subunit p14 peptide, a CYC1 terminator, and a selectable (hygromycin) marker, all integrated to replace the genomic GAL1 coding sequence. The details of this construction are described in the Supplemental Material. The second part is a simple way to insert the degron/marker cassette at any promoter of interest. To this end, we constructed a generic template for PCR (plasmid pRB3326) that enables the placement of our modified degron/marker cassette at the initiator methionine codon of any open reading frame (details are also found in the Supplemental Material).
Modification of target genes to make them TEV-sensitive
Because native proteins are not sensitive to the TEV protease, a generic system was adapted from Taxis et al. (2009) to make the required modifications of any gene routine. The overall idea is to generate a native-promoter driven, degron-modified gene. A particular new feature of our method is the inclusion of a removable (CRE-LOX) segment that results in loss of the selectable marker. The details of this construction, and its application to the specific cases of MET4 and MET31, are given in the Supplemental Material.
Construction of can1Δ::PACT1-GEV strains
The PACT1-GEV construct was amplified from the pAGL plasmid (Veatch et al., 2009) using oligonucleotides containing 56-59 bases of sequence identity to the flanking region of the CAN1 gene (ACT1-GEV_to_CAN1 FOR and ACT1-GEV_to_CAN1 REV). This PCR product was then transformed into either MATa or MATα wild-type prototrophic yeast cells by using a standard protocol selecting for canavanine resistance.
Western blots
Total protein was extracted from ∼10 ml aliquots of cultures at ∼2 × 107 cells/ml by using the YPX extraction kit (Protein Discovery, Knoxville, TN) with the addition of protease inhibitors (Roche tablets, cat#11836170001; Basel, Switzerland). Proteins were separated using Invitrogen NuPage precast 10% Bis-Tris gels and transferred to Invitrolon polyvinylidene fluoride (Invitrogen cat# LC2005; Carlsbad, CA) according to the manufacturer's protocols. Primary antibody for the 13-Myc epitope was mouse anti–human c-Myc from BD PharMingen (cat# 51-1485GR; San Diego, CA). Secondary antibody was goat anti–mouse-horseradish peroxidase (HRP; cat# 115-035-003) from Jackson ImmunoResearch (West Grove, PA). Primary antibody dilution was typically 1:1000, and secondary antibodies were used at a 1:10,000 dilution. Immunoreactive peptides were detected by incubation with ECL+ detection reagent (Amersham/GE RPN2132; Little Chalfont, UK) and imaging on HyBlot CL autoradiography film (Denville Scientific, Metuchen, NJ). As a loading control, we probed blots with mouse monoclonal anti–β-actin antibody (cat# ab8224; Abcam, Cambridge, MA). Secondary antibody was as above, goat anti–mouse-HRP. Protein levels were quantified with the “Gel Analysis” feature in ImageJ and normalized to the t = 0 levels in each time course.
Supplementary Material
Acknowledgments
This research was supported by grants GM046406 (to D.B.) and by the National Institute of General Medical Sciences Center for Quantitative Biology (GM071508). R.S.M. acknowledges support from the National Science Foundation Graduate Research Fellowship. P.A.G. acknowledges support from T32 CA 009528-24. Lance Parsons and Tina DeCoste helped with FISH analysis and flow cytometry, respectively. We thank Saw Kyin of the Princeton Molecular Biology SynSeq facility for synthesizing the amino-allyl modified DNA oligonucleotides used for FISH, Ryan Briehof for technical assistance, and Michael Knop and Dan Gottschling for providing reagents. We thank Greg Lang, Xin Wang, Christopher Crutchfield, and Ned Wingreen for helpful discussions.
Abbreviations used:
- FISH
fluorescence in situ hybridization
- Gal4dbd
Gal4p DNA binding domain
- GEV
Gal4dbd.ER.VP16
- GFP
green fluorescent protein
- GO
gene ontology
- IPTG
isopropyl β-d-1 thiogalactopyranoside
- LFM
low-fluorescence medium
- NDeg
TEV cleavable peptide
- rtTA
reverse tetracycline transactivator
- TEV
tobacco etch virus
- tTA
tetracycline transactivator
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0466) on September 30, 2011.
REFERENCES
- Adams BG. Induction of galactokinase in Saccharomyces cerevisiae—kinetics of induction and glucose effects. J Bacteriol. 1972;111:308–&. doi: 10.1128/jb.111.2.308-315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, et al. RiboSys, a high-resolution, quantitative approach to measure the in vivo kinetics of pre-mRNA splicing and 3 ‘-end processing in Saccharomyces cerevisiae. RNA. 2010;16:2570–2580. doi: 10.1261/rna.2162610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast (vol 26, pg 942, 1998) Nucleic Acids Res. 1998;26:U8–U8. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng SA, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:U485–U416. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci USA. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devirgilio C, Burckert N, Bell W, Jeno P, Boller T, Wiemken A. Disruption of Tps2, the gene encoding the 100-Kda subunit of the trehalose-6-phosphate synthase phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur J Biochem. 1993;212:315–323. doi: 10.1111/j.1432-1033.1993.tb17664.x. [DOI] [PubMed] [Google Scholar]
- Gaisne M, Becam AM, Verdiere J, Herbert CJ. A “natural” mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1) Curr Genet. 1999;36:195–200. doi: 10.1007/s002940050490. [DOI] [PubMed] [Google Scholar]
- Gao CY, Pinkham JL. Tightly regulated, beta-estradiol dose-dependent expression system for yeast. Biotechniques. 2000;29:1226–1231. doi: 10.2144/00296st02. [DOI] [PubMed] [Google Scholar]
- Gill G, Ptashne M. Negative effect of the transcriptional activator Gal4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Giniger E, Varnum SM, Ptashne M. Specific DNA binding of Gal4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Hickman MJ, Petti AA, Ho-Shing O, Silverman S, McIsaac RS, Lee TA, Botstein D. Coordinated regulation of sulfur and phospholipid metabolism reflects the importance of methylation in the growth of yeast. Mol Biol Cell. 2011;22:4192–4204. doi: 10.1091/mbc.E11-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MJ, Winston F. Heme levels switch the function of hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol Cell Biol. 2007;27:7414–7424. doi: 10.1128/MCB.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MQ, Fitzgerald MX, Harper S, Luo C, Speicher DW, Marmorstein R. Structural basis for dimerization in DNA recognition by Gal4. Structure. 2008;16:1019–1026. doi: 10.1016/j.str.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland P, Flick J, Johnston M, Sclafani RA. Galactose as a gratuitous inducer of Gal gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- Hu ZZ, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Kahm M, Hasenbrink G, Lichtenberg-Frate H, Ludwig J, Kschischo M. grofit: fitting biological growth curves with R. J Stat Softw. 2010;33:1–21. [Google Scholar]
- Labow MA, Baim SB, Shenk T, Levine AJ. Conversion of the Lac repressor into an allosterically regulated transcriptional activator for mammalian cells. Mol Cell Biol. 1990;10:3343–3356. doi: 10.1128/mcb.10.7.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci USA. 2009;106:5755–5760. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liko D, Slattery MG, Phillips CL, Heideman W. Using the yeast gene deletion collection to customize gene expression. Biotechniques. 2006;40:728. doi: 10.2144/000112192. [DOI] [PubMed] [Google Scholar]
- Louvion JF, Havauxcopf B, Picard D. Fusion of Gal4-Vp16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 1993;131:129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by Gal4—structure of a protein DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Masselot M, Robichon-Szulmajster H. Methionine biosynthesis in Saccharomyces cerevisiae. Molecular and General Genetics. 1975;139:121–132. doi: 10.1007/BF00264692. [DOI] [PubMed] [Google Scholar]
- Pfahl M. Characteristics of tight-binding repressors of the Lac operon. J Mol Biol. 1981;147:1–10. doi: 10.1016/0022-2836(81)90075-9. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Quintero MJ, Maya D, Arevalo-Rodriguez M, Cebolla A, Chavez S. An improved system for estradiol-dependent regulation of gene expression in yeast. Microb Cell Fact. 2007;6:10. doi: 10.1186/1475-2859-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCFMet30 complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. Gal4-Vp16 Is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Silverman SJ, et al. Metabolic cycling in single yeast cells from unsynchronized steady-state populations limited on glucose or phosphate. Proc Natl Acad Sci USA. 2010;107:6946–6951. doi: 10.1073/pnas.1002422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SR. Biomedical image processing. Computer. 1983;16:22–34. [Google Scholar]
- Takahashi M, Altschmied L, Hillen W. Kinetic and equilibrium characterization of the Tet repressor tetracycline complex by fluorescence measurements—evidence for divalent metal-ion requirement and energy transfer. J Mol Biol. 1986;187:341–348. doi: 10.1016/0022-2836(86)90437-7. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol. 2008;18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Stier G, Spadaccini R, Knop M. Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol Syst Biol. 2009;5:267. doi: 10.1038/msb.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart JA, Hayes A, Wardleworth L, Zhang NS, Oliver SG. Doxycycline, the drug used to control the tet-regulatable promoter system, has no effect on global gene expression in Saccharomyces cerevisiae. Yeast. 2005;22:565–569. doi: 10.1002/yea.1225. [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.