Abstract
Mycobacterium bovis is the causative agent of bovine tuberculosis, with a wide host range. Fifty human M. bovis isolates were typed using spoligotyping and variable number tandem repeats (VNTR). Fifteen of these spoligotypes have not yet been recorded in cattle. The predominant spoligotype in humans and cattle was subdivided by VNTR.
Mycobacterium bovis has a wide host range, infecting many domestic and wild animals. Although occurring relatively rarely, M. bovis can also infect humans. In the United Kingdom, only about 1% of clinically diagnosed cases of tuberculosis (TB) that are subsequently proven bacteriologically are attributed to M. bovis, but in the developing world, M. bovis is still a cause for concern (6). The resurgence of bovine TB in cattle in the United Kingdom is raising concerns that transmission from cattle to humans might be a serious public health issue. It is therefore important to be able to quickly identify where rates of M. bovis in cattle are high and pose a potential risk of transmission to humans. M. bovis was once a major source of TB in humans in the United Kingdom but was almost eradicated after the introduction of control measures to reduce bovine tuberculosis in cattle together with the pasteurization of milk for human consumption. The majority of bovine TB cases in the 1980s and early 1990s presented either in the elderly or in those who had been infected abroad and returned or migrated to the United Kingdom (13). Many animals, such as badgers, foxes, ferrets, and deer (1, 3, 9), are believed to act as vectors for transmission to livestock, and some have also been associated with transmission to humans (8, 16, 18). Enhanced surveillance of M. bovis infections in humans was initiated in 1998. However, in 2001 a revised system which allows more timely collection of data was introduced (4, 5). Advances in molecular typing have provided tools to enhance our knowledge of M. bovis dissemination. Restriction fragment length polymorphism using the insertion sequence IS6110 is considered to provide the best discrimination of M. tuberculosis isolates. However, M. bovis isolates from cattle usually have a single copy of IS6110 (7); therefore, alternative techniques such as spacer oligonucleotide typing (spoligotyping) and variable number tandem repeats (VNTR) have been used successfully in discriminating between strains of M. bovis (1, 7, 11, 12, 15, 17).
This study examines the molecular epidemiology of M. bovis cases within the United Kingdom using two molecular typing techniques and compares the typing patterns obtained to those prevalent in United Kingdom cattle today.
All available viable M. bovis isolates (50 isolates) from humans diagnosed in the United Kingdom between 1997 and 2000 were identified; 40 were recovered at the Mycobacterium Reference Unit, London, and 10 were recovered at the Scottish Mycobacteria Reference Laboratory, Edinburgh. DNA was extracted by using a quick extraction method (19). Briefly, one colony was removed using a 1-μl loop and placed in 150 ml of water. An equal volume (150 ml) of chloroform was added, and the mixture was vortexed and then boiled at 80°C for 20 min to kill the cultures.
Spoligotyping was performed using the method described by Kamerbeek et al. (15), and VNTR was performed using the method described by Frothingham and Meeker-O'Connell (11). The size of each exact tandem repeat at each locus (A to E) was determined by running the PCR product on an agarose gel containing size markers (100-bp ladder; Promega, Southampton, United Kingdom) (20-bp ladder; Sigma-Aldrich, Dorset, United Kingdom). Deletion typing was carried out on a strain with a spoligotype not typical of M. bovis, using the method described by Brosch et al. (2). Seven regions of difference (RD) were examined: RD 4, 7, 8, 9, 10, 12, and 13. The Hunter-Gaston index (HGI), which is based on the probability of two unrelated strains from a test population being placed into different typing groups, was calculated to determine the discriminatory power of each typing method alone and in combination (14).
Epidemiological information was obtained from internal laboratory records at the Mycobacterium Reference Unit and Scottish Mycobacteria Reference Laboratory and from existing surveillance data held at the Health Protection Agency Communicable Disease Surveillance Centre.
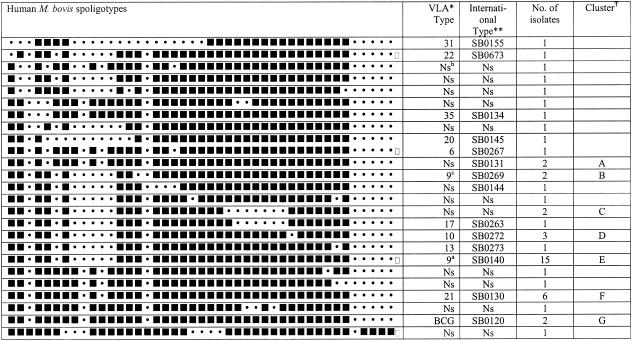
Spoligotyping of the 50 human M. bovis isolates produced 25 individual spoligotypes (Fig. 1) and had an HGI of 0.90. Thirty-two isolates were divided into seven clusters, A to G (Fig. 1). The spoligotypes were compared with M. bovis spoligotypes from a bank of over 15,000 cattle isolates collected from all over the United Kingdom held at the Veterinary Laboratory Agency (VLA) and dating between 1987 and 2002. The largest cluster of human M. bovis isolates (15 isolates, 30%) had been seen in cattle before and was sequentially numbered type 9 (international type SB140; http://www.Mbovis.org) at the VLA. Type 9 is the most frequently seen spoligotype of M. bovis (over 30% of all isolates have this spoligotype) isolated from cattle and has a wide geographical range in the United Kingdom (10).
FIG. 1.
Spoligotyping patterns for human M. bovis isolates in the United Kingdom. Symbols: *, comparison of human M. bovis spoligotypes with a bank of M. bovis spoligotypes seen in United Kingdom cattle; **, international spoligotype website is http://www.Mbovis.org; _T arbitrarily labeled clusters. Superscript numbers: a, most predominant spoligotype (type 9); b, spoligotype not seen in United Kingdom cattle before; c, identical to type 9 except for absence of spacer 15.
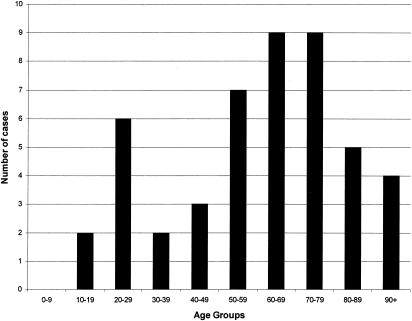
Human type 9 isolates were seen across the United Kingdom, suggesting that transmission between cattle and humans might occur. Interestingly, 15 of the human M. bovis spoligotypes had not been seen at the VLA in isolates from cattle. When these 15 types were compared to the international spoligotype database, only 2 were recognized. The first was isolated in Argentina, the second was isolated in Australia, and the remaining 13 spoligotypes were all unique to the United Kingdom. In the majority of these cases, it is likely that disease was due to reactivation of a past infection that had been acquired prior to milk pasteurization rather than to primary infection, because 72.3% of the patients were over the age of 50. (Fig. 2). Therefore, these 13 unique spoligotypes may reflect M. bovis strains circulating in the United Kingdom over 50 years ago.
FIG. 2.
Number of M. bovis cases by age group.
M. bovis spoligotypes do not usually contain spacers 39 to 43; however, one spoligotype from the panel contained spacers 40 to 43, which are more commonly seen in M. tuberculosis. Phenotypic and biochemical tests demonstrated that this isolate had typical M. bovis characteristics; it was microaerophilic, TCH (thiophen-2-carboxylic acid hydrazide) negative, and pyrazinamide resistant, and it grew better on pyruvate than glycerol Lowenstein-Jensen slopes.
Deletion analysis was performed to ascertain the identity of this strain. The strain contained RD 4, 12, and 13 but lacked RD 7, 8, 9, and 10, indicating that this strain is actually M. Africanum and not M. bovis.
VNTR typing alone produced 18 different patterns and had an HGI of 0.85. Combining spoligotyping with VNTR vastly improved the level of discrimination, producing 34 different types and a very high HGI of 0.96. Furthermore, VNTR was very useful in subdividing type 9 spoligotypes, separating the group into six subtypes (Table 1).
TABLE 1.
Subdivision of spoligotyping clusters by VNTR
| Spoligo clustera (no. of isolates) | No. of VNTR subtypes | VNTR profiles |
|---|---|---|
| A (2) | 2 | 55543 75543 |
| B (2) | 2 | 55543 75543 |
| C (2) | 1 | 75553 |
| D (3) | 2 | 63543 75543 |
| E (15) | 6 | 56543 63543 65542 |
| 65543 75543 75553 | ||
| F (6) | 2 | 65543 66543 |
| G (2) | 2 | 54544 55343 |
Arbitarily labeled clusters.
Epidemiological information showed that the study population was widely distributed across the United Kingdom, had an average age of 58.7 years, and had approximately equal proportions of males and females (21:17). Where ethnicity was known (a total of 15 patients), 14 patients were white and 1 was of black-African origin and was originally from Nigeria but had lived in the United Kingdom since 1996. This person had a unique spoligotype; therefore, it is possible that she was infected in Nigeria before arriving in the United Kingdom. Of interest, 59% (13 of 22) of cases had some contact with a farm, ranging from having a Saturday job milking cows, to living on a dairy farm as a child, to being a farmer (now retired). One spoligotype cluster represents an outbreak on a farm in Gloucester. Two siblings (a 20-year-old male and a 17-year-old female) living on their parents' farm became infected with M. bovis. The brother occasionally helped his father on the farm by restraining the cattle and would often be sprayed with nasal mucus. Cattle infected with M. bovis of the same spoligotype had been detected on the farm in previous years. Transmission from cattle to human is thought to have occurred by the inhalation of infected aerosols from cattle. The brother is thought to have subsequently infected his sister, as she had no contact with the cattle but was also diabetic and pregnant, i.e., immuno-compromised. This is thought to be the first case of human-to-human transmission since 1990 (R. M. M. Smith, F. Drobniewsky, A. L. Gibson, J. D. E. Montague, M. N. Logan, D. Hunt, R. G. Hewinson, R. L. Salmon, and B. O’Neill, unpublished data).
It is important to monitor bovine tuberculosis in humans, especially in those who are at high risk of primary infection, such as agricultural and abattoir workers, and to identify any transmission between animals and humans. A combination of spoligotyping and VNTR is an efficient discriminatory tool for the molecular surveillance of M. bovis and also addresses the problem of analyzing isolates with single copies of IS6110. The combined VNTR and spoligotyping approach is of value in typing M. tuberculosis isolates. Further improvements in these techniques might produce a combined system capable of high discrimination for all M. tuberculosis complex isolates in humans or other mammals.
Acknowledgments
This work was funded by the Department for Environment, Food, and Rural Affairs and supported by the Health Protection Agency, formerly the Public Health Laboratory.
REFERENCES
- 1.Aranaz, A., E. Liebana, A. Mateos, L. Dominguez, D. Vidal, M. Domingo, O. Gonzolez, E. F. Rodriguez-Ferri, A. E. Bunschoten, J. D. van Embden, and D. Cousins. 1996. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J. Clin. Microbiol. 34:2734-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clifton-Hadley, R. S., J. W. Wilesmith, M. S. Richards, P. Upton, and S. Johnston. 1995. The occurrence of Mycobacterium bovis infection in cattle in and around an area subject to extensive badger (Meles meles) control. Epidemiol. Infect. 114:179-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Communicable Disease Report Weekly. 1998. Enhanced surveillance of Mycobacterium bovis in humans. Commun. Dis. Rep. Wkly. 8:281, 284. [PubMed] [Google Scholar]
- 5.Communicable Disease Report Weekly. 2001. Enhanced surveillance of Mycobacterium bovis disease in humans in England and Wales from January 2001. Commun. Dis. Rep. Wkly. 11:2-3. [PubMed] [Google Scholar]
- 6.Cosivi, O., J. M. Grange, C. J. Daborn, M. C. Raviglione, T. Fujikura, D. Cousins, R. A. Robinson, H. F. A. K. Huchzermeyer, I. de Kantor, and F. X. Meslin. 1998. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 4:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cousins, D. V., R. A. Skuce, R. R. Kazwala, and J. D. van Embden. 1998. Towards a standardized approach to DNA fingerprinting of Mycobacterium bovis. International Union Against Tuberculosis and Lung Disease, Tuberculosis in Animals Subsection. Int. J. Tuberc. Lung Dis. 2:471-478. [PubMed] [Google Scholar]
- 8.Dalovisio, J. R., M. Setter, and S. Mikota-Wells. 1992. Rhinoceros' rhinorrhea: cause of an outbreak of infection due to airborne Mycobacterium bovis in zookeepers. Clin. Infect. Dis. 15:598-600. [DOI] [PubMed] [Google Scholar]
- 9.Delahay, R. J., C. L. Cheeseman, and R. S. Clifton-Hadley. 2001. Wildlife disease reservoirs: the epidemiology of Mycobacterium bovis infection in the European badger (Meles meles) and other British mammals. Tuberculosis 81:43-49. [DOI] [PubMed] [Google Scholar]
- 10.Durr, P. A., R. S. Clifton-Hadley, and R. G. Hewinson. 2000. Molecular epidemiology of bovine tuberculosis. II. Applications of genotyping. Rev. Sci. Tech. 19:689-701. [DOI] [PubMed] [Google Scholar]
- 11.Frothingham, F., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 12.Haddad, N., A. Ostyn, C. Karoui, M. Masselot, M. F. Thorel, S. L. Hughes, J. Inwald, R. G. Hewinson, and B. Durand. 2001. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J. Clin. Microbiol. 39:3623-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardie, R. M., and J. M. Watson. 1992. Mycobacterium bovis in England and Wales: past, present and future. Epidemiol. Infect. 109:23-33. [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalak, K., C. Austin, S. Diesel, M. J. Bacon, P. Zimmerman, and J. N. Maslow. 1998. Mycobacterium tuberculosis infection as a zoonotic disease: transmission between humans and elephants. Emerg. Infect. Dis. 4:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Soolingen, D., P. E. de Haas, J. Haagsma, T. Eger, P. W. Hermans, V. Ritacco, A. Alito, and J. D. van Embden. 1994. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J. Clin. Microbiol. 32:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson, S. M., R. McNerney, P. M. Nye, P. D. Godfrey-Faussett, N. G. Stoker, and A. Voller. 1993. Progress toward a simplified polymerase chain reaction and its application to diagnosis of tuberculosis. J. Clin. Microbiol. 31:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]