Abstract
Polymers have emerged as powerful biological tools; however, their ability to gain access to the intracellular environment is limited. To expand the biological utility of polymer scaffolds, we have synthesized an internalization domain using the ring-opening metathesis polymerization (ROMP). A polymer functionalized with guanidinium groups is effectively internalized by cells and localized in both vesicles and the cytoplasm. Because the synthesis of such materials is modular, we anticipate that compounds of this type can be fashioned that facilitate the delivery of cargo via end-cap derivatization or block copolymer synthesis.
New synthetic methods are fueling the use of polymers in biology.1 Indeed, polymers can function as therapeutics,1a,c biomaterials,1b imaging agents,2 and multivalent biological probes.3 The growing applications of polymers stem from the ability to tailor their properties. For example, the molecular masses of polymers can be controlled, thereby modulating their serum half-life, cell or tissue targeting, and binding avidity. In addition, because polymers are modular, a wide range of different functionality can be introduced to endow them with tailored bulk properties and the ability to engage in specific biological recognition events. One disadvantage of polymers, however, is that their ability to gain access to cytoplasmic and nuclear targets is limited. Because of their high molecular weights and functionalities, most polymers are cell impermeable. We reasoned that the utility of bioactive polymers could be expanded by devising a general method for promoting their cellular internalization. To this end, we envisioned generating polymers that contain an artificial translocation domain, or ATD.
The design of our polymer ATD was guided by the features of known internalization agents. Small, highly cationic peptides, including TAT and oligoarginine, can promote cellular uptake.4 These internalization agents, often referred to as protein transduction domains (PTDs) or cell-penetrating peptides, can facilitate the intracellular delivery of compounds ranging from small molecules to proteins to nanoparticles. In addition, PTD mimics, such as β-peptides and peptoids, have been described.5 The structural diversity of internalization agents indicates that an optimal number of guanidinium groups, not backbone composition, is a primary requirement for uptake. The model for uptake involves association of the guanidinium groups with the sulfate groups of cell-surface glycosaminoglycans, such as heparan sulfate.6 This association is followed by internalization via an endocytic pathway. Because this pathway depends upon Coulombic interactions with ubiquitous proteoglycans and not the presence of a specific receptor, compounds that exploit this route can be taken up by a variety of cell types. Thus, to develop a general route to ATDs with these properties, we envisioned appending guanidinium groups to a polymer backbone that can be synthesized in one step. The optimal number of guanidinium groups for internalization has been found to be between 8 and 16;7 therefore, for this approach to be effective, the length of the polymer must be tightly controlled.
The ring-opening metathesis polymerization (ROMP) is a powerful method for the synthesis of defined bioactive polymers.8 ROMP can be living, and initiation rates can exceed propagation rates; therefore, polymeric structures of low polydispersity can be synthesized in a controlled manner. We envisioned that ROMP could be used to assemble a short, general backbone that could be functionalized with guanidinium groups to impart cell-permeability. This strategy is modular,9 allowing for further polymerization or functionalization. For example, to visualize internalization of the compounds using microscopy, a known end-capping reagent can be used to append a ketone group to react with a functionalized fluorophore.10
To assemble the ROMP-derived ATD, ruthenium carbene 211 was added to succinimidyl ester-substituted norbornene 1 to afford a polymeric precursor.9 To ensure that the polymer length of the final product was appropriate for internalization, we employed a monomer to initiator ratio of 10:1 (Scheme 1A). The polymerization was terminated with enol ether 10-methoxydec-9-ene-2-one12 (3), to afford the succinimidyl ester-containing polymer 4 with a single ketone group available for conjugation. Integration of the polymer backbone alkene and terminal phenyl proton signals in the 1H NMR spectrum indicate that the degree of polymerization (DP) of the polymer is 10. Analysis by gel permeation chromatography (GPC) gives a similar DP (11) and a polydispersity index (PDI) of 1.20. The average length (ca. 10) and the low PDI of our polymers indicate that a majority of the individual molecules should be capable of internalization. Guanidinium groups were introduced on the polymer via amide bond formation with amine 5 to yield water-soluble polymer 6 (Scheme 1).
Scheme 1.

Synthesis of the cationic internalization domain
To study extracellular receptor clustering and signaling we used an acyl hydrazone to append a fluorophore to a polymer.13 This linkage can undergo hydrolysis upon cellular uptake.14 Though this lability could be valuable for cargo delivery, it complicates monitoring polymer uptake and trafficking. Thus, we synthesized an alkoxylamine-functionalized rhodamine B derivative (7) that would allow for installation of a fluorophore via an oxime linkage (see supporting information). In the cellular environment, oxime groups are more stable than are acyl hydrazones;15 therefore, the former can be used to append a label to assess polymer localization within the cell. The fluorophore not only serves as a reporter but also as prototype cargo. We therefore modified 6 to generate labeled, guanidinium-substituted polymer 8.
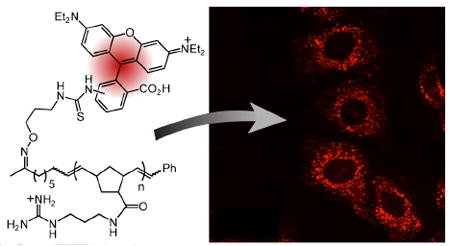
To evaluate the cell-permeability of the polymer, HeLa cells were incubated for with various concentrations (1, 2, or 5 μM) of 8 for 1 h at 37 °C (Fig. 1A). Often cells are visualized after fixation, but this process causes artifactual distribution of PTDs within the cytoplasm and nucleus.16 Accordingly, we analyzed the localization of the polymer using live cell microscopy. We observed punctate fluorescence consistent with vesicular localization. Diffuse fluorescence was also apparent, indicating that the polymer is present throughout the cytoplasm. Cytotoxicity assays demonstrate that cells exposed to polymer 6 (10 μM) for 4 h are >95% viable. To evaluate the influence of polymer length on uptake and cytoxicity, we generated longer polymers (DP=25). These materials are internalized less efficiently and are more cytoxic (5-fold, see supporting information).
Figure 1.

A) HeLa cells were incubated with 8 (1, 2, or 5 μM) for 1 h and viewed by confocal microscopy. The ATD is seen in endocytic vesicles (punctate fluorescence) as well as in the cytoplasm (diffuse fluorescence). B) Cells were treated with 8 (5 μM) for varying amounts of time. The ATD is present faintly in vesicles and the cytoplasm after incubation times as brief as 5 min; fluorescence increases in both locations over time. Bar = 25 μm.
Although the mechanism of intracellular delivery remains under debate, both the TAT peptide and oligoarginine peptides appear to use an energy-dependent endocytic pathway.4b To determine whether the uptake pathway of the guanidinium-substituted polymer is similar, cells were incubated with 8 (5 μM) for 1 h at 4 °C. No internalization was observed (data not shown). These results indicate that uptake of the ATD is energy-dependent. To determine the timescale for internalization and escape from the endosomes, the cells were treated with 8 (5 μM) for various time periods (5 min → 1 h) (Fig. 1B). After only 5 minutes, polymer was visible in both vesicles and in the cytoplasm. The confocal microscopy data demonstrate that the polymer is not only internalized but also enters the cytoplasm rapidly. Thus, the ROMP-derived polymer is an effective ATD.
We have used ROMP to develop a cell-permeable polymer that is not only physiologically stable but also modular in design. Our polymeric ATD strategy allows for either reversible or irreversible attachment of molecules via the end cap, facilitating delivery of cargo into the cell. Because the polymerization is living, block copolymers can be generated. We envision an ATD block could enable the internalization of multivalent ligands displaying different epitopes. We anticipate that this strategy will facilitate the study of intracellular multivalency and provide a means to recruit proteins and promote protein assemblies.
Supplementary Material
Acknowledgments
This research was supported by the NIH (GM49975). We thank K. Dickson, C. Chao, and R. T. Raines for helpful discussions. Confocal microscopy was performed at the W.M. Keck Laboratory for Biological Imaging at the University of Wisconsin–Madison. EMK acknowledges the NIH Chemistry–Biology Interface Training Program for a predoctoral fellowship (GM008505). The NMR facility is supported in part by the NSF (CHE-0342998 and CHE-9629688) and the NIH (1 S10 RR13866-01).
Footnotes
Supporting Information Available: Experimental preparations and characterization of compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Duncan R. Nat Rev Drug Disc. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]; (b) Hawker CJ, Wooley KL. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]; (c) Langer R, Tirrell DA. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 2.(a) Kim JH, Park K, Nam HY, Lee S, Kim K, Kwon IC. Prog Polym Sci. 2007;32:1031–1053. [Google Scholar]; (b) Allen MJ, Raines RT, Kiessling LL. J Am Chem Soc. 2006;128:6534–6535. doi: 10.1021/ja061383p. [DOI] [PubMed] [Google Scholar]
- 3.(a) Kiessling LL, Gestwicki JE, Strong LE. Angew Chem Int Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kramer RH, Karpen JW. Nature. 1998;395:710–713. doi: 10.1038/27227. [DOI] [PubMed] [Google Scholar]; (c) Sil D, Lee JB, Luo D, Holowka D, Baird B. ACS Chem Biol. 2007;2:674–684. doi: 10.1021/cb7001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Snyder EL, Dowdy SF. Pharm Res. 2004;21:389–393. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]; (b) Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]; (c) Futaki S. Biopolymers. 2006;84:241–249. doi: 10.1002/bip.20421. [DOI] [PubMed] [Google Scholar]; (d) Goun EA, Pillow TH, Jones LR, Rothbard JB, Wender PA. ChemBioChem. 2006;7:1497–1515. doi: 10.1002/cbic.200600171. [DOI] [PubMed] [Google Scholar]
- 5.(a) Huang K, Voss B, Kumar D, Hamm HE, Harth E. Bioconjugate Chem. 2007;18:403–409. doi: 10.1021/bc060287a. [DOI] [PubMed] [Google Scholar]; (b) Umezawa N, Gelman MA, Haigis MC, Raines RT, Gellman SH. J Am Chem Soc. 2002;124:368–369. doi: 10.1021/ja017283v. [DOI] [PubMed] [Google Scholar]; (c) Schroder T, Schmitz K, Niemeier N, Balaban TS, Krug HF, Schepers U, Brase S. Bioconjugate Chem. 2007;18:342–354. doi: 10.1021/bc0602073. [DOI] [PubMed] [Google Scholar]; (d) Okuyama M, Laman H, Kingsbury SR, Visintin C, Leo E, Eward KL, Stoeber K, Boshoff C, Williams GH, Selwood DL. Nat Methods. 2007;4:153–159. doi: 10.1038/nmeth997. [DOI] [PubMed] [Google Scholar]; (e) Daniels DS, Schepartz A. J Am Chem Soc. 2007;129:14578–14579. doi: 10.1021/ja0772445. [DOI] [PubMed] [Google Scholar]
- 6.(a) Tyagi M, Rusnati M, Presta M, Giacca M. J Biol Chem. 2001;276:3254–3261. doi: 10.1074/jbc.M006701200. [DOI] [PubMed] [Google Scholar]; (b) Fuchs SM, Raines RT. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Melikov K, Chernomordik L. Cell Mol Life Sci. 2005;62:2739–2749. doi: 10.1007/s00018-005-5293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Poon GMK, Gariepy J. Biochem Soc Trans. 2007;35:788–793. doi: 10.1042/BST0350788. [DOI] [PubMed] [Google Scholar]; (e) Asoh S, Ohsawa I, Mori T, Katsura K, Hiraide T, Katayama Y, Kimura M, Ozaki D, Yamagata K, Ohta S. Proc Natl Acad Sci USA. 2002;99:17107–17112. doi: 10.1073/pnas.262460299. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Soane L, Fiskum G. J Neurochem. 2005;95:230–243. doi: 10.1111/j.1471-4159.2005.03359.x. [DOI] [PubMed] [Google Scholar]
- 7.(a) Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]; (b) Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y, Takehashi M, Tanaka S, Ueda K, Simpson JC, Jones AT, Sugiura Y, Futaki S. Mol Ther. 2004;10:1011–1022. doi: 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]; (c) Rothbard JB, Garlington S, Lin Q, Kirschberg T, Kreider E, McGrane PL, Wender PA, Khavari PA. Nat Med. 2000;6:1253–1257. doi: 10.1038/81359. [DOI] [PubMed] [Google Scholar]
- 8.(a) Schrock RR, Hoveyda AH. Angew Chem Int Ed. 2003;42:4592–4633. doi: 10.1002/anie.200300576. [DOI] [PubMed] [Google Scholar]; (b) Smith D, Pentzer EB, Nguyen ST. Polym Rev. 2007;47:419–459. [Google Scholar]; (c) Bielawski CW, Grubbs RH. Prog Polym Sci. 2007;32:1–29. [Google Scholar]; (d) Lee Y, Sampson NS. Curr Opin Struct Biol. 2006;16:544–550. doi: 10.1016/j.sbi.2006.05.015. [DOI] [PubMed] [Google Scholar]; (e) Smith D, Pentzer EB, Nguyen ST. Polymer Reviews. 2007;47:419–459. [Google Scholar]
- 9.Strong LE, Kiessling LL. J Am Chem Soc. 1999;121:6193–6196. [Google Scholar]
- 10.(a) Gordon EJ, Gestwicki JE, Strong LE, Kiessling LL. Chem Biol. 2000;7:9–16. doi: 10.1016/s1074-5521(00)00060-0. [DOI] [PubMed] [Google Scholar]; (b) Owen RM, Gestwicki JE, Young T, Kiessling LL. Org Lett. 2002;4:2293–2296. doi: 10.1021/ol0259239. [DOI] [PubMed] [Google Scholar]
- 11.Love JA, Morgan JP, Trnka TM, Grubbs RH. Angew Chem Int Ed. 2002;41:4035–4037. doi: 10.1002/1521-3773(20021104)41:21<4035::AID-ANIE4035>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 12.Pontrello JK, Allen MJ, Underbakke ES, Kiessling LL. J Am Chem Soc. 2005;127:14536–14537. doi: 10.1021/ja053931p. [DOI] [PubMed] [Google Scholar]
- 13.Yang ZQ, Puffer EB, Pontrello JK, Kiessling LL. Carbohydr Res. 2002;337:1605–1613. doi: 10.1016/s0008-6215(02)00270-7. [DOI] [PubMed] [Google Scholar]
- 14.(a) Yang J, Chen HT, Vlahov IR, Cheng JX, Low PS. J Pharmacol Exp Ther. 2007;321:462–468. doi: 10.1124/jpet.106.117648. [DOI] [PubMed] [Google Scholar]; (b) Kaneko T, Willner D, Monkovic I, Knipe JO, Braslawsky GR, Greenfield RS, Vyas DM. Bioconjugate Chem. 1991;2:133–141. doi: 10.1021/bc00009a001. [DOI] [PubMed] [Google Scholar]
- 15.Kalia J, Raines RT. Intrinsic stability of common linkages used for bioconjugation. 234th ACS National Meeting, Division of Biological Chemistry #53; Boston, MA. August 2007. [Google Scholar]
- 16.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


