Abstract
Determination of the hepatitis C virus (HCV) genotype for infected patients increasingly has become accepted as the standard of care. Genotype assignment helps in assessing disease prognosis and assists in establishing the appropriate duration of treatment. The great genetic diversity of HCV, with 11 major genotypes and >70 subtypes, contributes to the technical difficulty of genotype testing. While the “gold standard” for testing is nucleic acid sequencing, a variety of hybridization assays, including the line probe assay, have been developed to provide more rapid and accessible forms of testing. The aim of this study was to determine whether denaturing high-performance liquid chromatography (dHPLC) could be used as a clinical method for distinguishing HCV genotypes 1, 2, 3, and 4. A portion of the 5′ untranslated region of the HCV genome was amplified by heminested multiplex reverse transcription PCR. The two amplicons then were analyzed by dHPLC analysis and compared to the genotypes determined by sequence analysis. After 115 specimens were analyzed as standards, 200 masked specimens (specimens whose identity was not known before testing) were analyzed to determine the concordance of the assay. The assay had a concordance of 96% at the genotype level and a concordance of 87% at the subtype level. However, the dHPLC method was not as accurate as other reported methods of HCV genotyping. This is the first time that HCV genotyping has been performed by dHPLC.
Hepatitis C virus (HCV) is a single-stranded RNA virus of the family Flaviviridae that causes both acute and chronic hepatitis. Approximately 85% of acute cases progress to chronic infection. Patients with chronic infection are at substantial risk for the development of cirrhosis and eventually hepatocellular carcinoma. The Centers for Disease Control and Prevention estimates that in the United States alone, approximately 3.8 million people are chronically infected. Chronic HCV infection is the most common disease leading to liver transplant in the United States. HCV has a positive-sense genome of approximately 9.4 kb and which encodes a polyprotein of 300 amino acids (15, 20). Like the genomes of other single-stranded RNA viruses, the HCV genome is vulnerable to high rates of mutational change.
Eleven genotypes have been identified for HCV, with approximately 70 subtypes, based upon the sequence variability identified within its 5′ untranslated region (UTR) (11). The most common subtypes of HCV seen within the continental United States are 1a, 1b, 2a, 2b, and 3a. There does appear to be a correlation between the type of HCV and the efficacy of treatment with pegylated alpha interferon and ribavarin (16, 23). HCV genotypes 2 and 3 are more sensitive to treatment, making HCV genotyping important (1, 5, 8).
Denaturing high-performance liquid chromatography (dHPLC) is a method of separating nucleic acids based upon their sequence composition. It is primarily a tool to identify mutations or polymorphisms based upon separation of heteroduplexes from homoduplexes under partially denaturing conditions (21). For example, a common application is screening for heterozygosity in organisms that have pairs of chromosomes (6, 14, 17, 22). However, it is more difficult to identify or genotype microorganisms involved in infectious disease. The unpaired genome of microorganisms requires an exogenous template to form heteroduplexes. Exogenous templates have been used to discriminate between different bacterial species (7) and to genotype a meningococcus outbreak (19).
HCV genotyping usually is performed by a line probe assay or direct sequencing (3, 4, 12, 15). Additional methods include a microarray assay (24), modifying the HCV Amplicor monitor test for genotyping (13), heteroduplex analysis (20), and probe melting curve analysis (2, 18). Here we describe a novel assay that uses a heminested multiplex reverse transcription (RT)-PCR to distinguish HCV genotypes 1, 2, 3, and 4 by dHPLC analysis without heteroduplex formation.
MATERIALS AND METHODS
Human plasma samples.
The samples used in this study were submitted to ARUP Laboratories for HCV genotyping. Serum or EDTA-treated plasma was centrifuged, separated from cells, frozen at −20°C within 2 h of collection, and then shipped to ARUP Laboratories. A total of 315 samples were obtained, and any identifying material was removed according to Health Insurance Portability and Accountability Act regulations. Of those samples, 115 were used to determine parameters for genotyping masked samples (samples whose identity was not known before testing). The remaining 200 samples were masked so that the concordance of the assay could be determined.
HCV genotyping by PCR and sequencing.
HCV sequencing was performed at the Sequencing Laboratory at ARUP Laboratories by standard methods. HCV RNA was extracted by using an Amplicor HCV preparation kit, version 2.0 (Roche, Indianapolis, Ind.), and a 245-bp region of the 5′ UTR was amplified by using an Amplicor HCV amplification kit, version 2.0 (Roche). The amplified nucleic acid was sequenced bidirectionally by using dye terminator chemistry with an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, Calif.). Genotyping results were based on a comparison with a database derived from GenBank sequences, published information, and in-house sequencing (11).
RNA extraction.
HCV RNA was extracted from plasma samples by using a QIAamp viral RNA extraction kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. In brief, virions were lysed, and the RNA genomes were captured in the spin columns provided. The HCV RNA was washed and then eluted by incubation of the spin columns for 1 min in 60 μl of sterile molecular-grade water followed by 1 min of centrifugation at 6,000 × g and room temperature. The HCV RNA was collected in a clean 1.5-ml Eppendorf tube and stored at −20°C until used.
Heminested multiplex RT-PCR.
The extracted HCV RNA was amplified by a heminested multiplex RT-PCR approach with a Qiagen OneStep RT-PCR kit. A heminested approach was implemented because it was simple, and the use of two amplicons with different melting characteristics made it possible to differentiate genotypes that were not resolvable with either amplicon alone. The sequence of forward primer HCV5UTR01F was 5′-GTGAGTACACCGGAAT-3′, the sequence of reverse primer HCV5UTR02R was 5′-ATCCAAGAAAGGACCC-3′, and the sequence of reverse primer KY78 (10) was 5′-CTCGCAAGCACCCTATCAGGCAGT-3′ (Fig. 1). The heminested RT-PCR leads to both 45- and 153-bp amplicons. Reaction mixtures contained Qiagen OneStep RT-PCR buffer, 2 mM GeneAmp deoxynucleoside triphosphate blend (0.4 mM dATP, dCTP, and dGTP and 0.8 mM dUTP) (Applied Biosystems), 3.0 mM MgCl2, 1 μM HCV5UTR01F, 1 μM HCV5UTR02R, 0.25 μM KY78, 0.01 U of uracil DNA glycosylase (Roche)/μl, 0.3 U of RNase inhibitor (Roche)/μl, Qiagen OneStep RT-PCR enzyme mix, 2 μl of extracted HCV RNA, and molecular-grade water to 25 μl. Thermal cycling was performed with a 96-well GeneAmp PCR system 9700 (Applied Biosystems) as follows: an initial 50°C hold for 10 min; a 60°C hold for 30 min; a 95°C hold for 15 min; 40 cycles of 95°C for 30 s, 54°C for 30 s, and 72°C for 1 min; and a 72°C hold for 10 min.
FIG. 1.
Locations and sequences of primers used in the HCV heminested multiplex RT-PCR. (A) Sequence of HCV 5′ UTR (9) around amplicons of interest, showing the positions of primers HCV5UTR01F, HCV5UTR02R, and KY78. The primer locations are outlined in the boxes, and the numbers above the sequences are nucleotide positions with respect to the starting nucleotide of the translated region.
dHPLC.
dHPLC was performed by using a WAVE system (Transgenomic, Omaha, Nebr.) equipped with a DNASep high-throughput cartridge, an L-761 online degasser, a D-7000 interface module, an L-7100 pump, an L-7250 autosampler, a PCR7250 Peltier cooling rack, a WAVE accelerator, an L-7300+ column oven, and an L-7400 UV detector. All buffers were supplied by Transgenomic. Ten microliters of RT-PCR amplicon was loaded without treatment into the cartridge by an autosampler. The amplicon was eluted from the column with a linear acetonitrile gradient in 0.1 mM triethylammonium acetate buffer (TEAA; pH 7) (Transgenomic) at a constant flow rate of 0.9 ml/min. The acetonitrile gradient was formed by mixing 0.1 mM TEAA and 0.1 mM TEAA containing 25% (vol/vol) acetonitrile. The 45-bp amplicon was eluted from the column with a gradient of 7.5 to 10% (vol/vol) acetonitrile. The 153-bp amplicon was eluted from the column with a gradient of 11.5 to 13.75% (vol/vol) acetonitrile. Acetonitrile gradients lasted for 4.5 min at a flow rate of 0.9 ml/min. The column then was cleaned with 75% (vol/vol) acetonitrile for 0.5 min with the WAVE accelerator and equilibrated with loading conditions for 0.9 min before the next sample was loaded. Carryover contamination did not appear to be a problem with this cleaning procedure. The temperature of the oven was fixed at 55°C for analysis of the 45-bp amplicon and at 63.4°C for analysis of the 153-bp amplicon. These temperatures were determined by a combination of evaluation with melting simulation software (Wavemaker 4.1) and empirical experiments. Elution of the amplicon was detected by measuring the absorbance at 260 nm. HPLC System Manager software regulated every parameter of the WAVE system during analysis and stored the data.
One specimen each of genotypes 1a, 1b, 2a, 2b, 3a, and 4 was included in every separate dHPLC run of masked samples to control for between-experiment variations. If the retention time of the standards was not within 0.15 min of the expected value, the results were not used for genotyping. In addition, these standards were used routinely to check for carryover contamination by examining the traces to determine whether any of the previously injected standard could be seen in the following trace.
Data analysis of masked samples.
The majority of samples were tested only once. However, if a genotype was ambiguous, then the sample was retested. If a sample had characteristics that did not fit the parameters for defining the HCV genotypes described in Table 1, it was deemed to be ambiguous. If the repeat data still were difficult to interpret, the assigned genotype was based upon the repeat run. The second run was used as the basis for the genotype assignment because it produced better-quality data.
TABLE 1.
Parameters used to genotype unknown HCV specimensa
| Retention time (min) for the following amplicon: | Genotype | |
|---|---|---|
| 153 bp | 45 bp | |
| <3.5b | <5.0 | 2b |
| 3.5-4.5 | >5.3 | 3a |
| 4.2-4.5c | <5.0 | 2a |
| 4.4-5 | 5.0-5.3 | 1a |
| >4.5 | 5.0-5.3 | 1b |
| >4.7 | >5.3 | 4 |
Parameters are described for each amplicon separately but must both be present in the same sample for the sample to be allocated into a specific genotype.
The trace usually has a high absorbance and a shoulder.
The trace sometimes has a sharp spike associated with the trailing edge of the peak.
RESULTS
Genotyping of HCV standards by dHPLC.
The RT-PCR approach used in this study resulted in two products being amplified from a single reaction. These two products, 45- and 153-bp amplicons, could be visualized by dHPLC under nondenaturing conditions at 50°C (Fig. 2). However, because the optimal elution characteristics for the amplicons were so different, the data for the two amplicons were acquired separately with different temperatures and different acetonitrile gradients.
FIG. 2.
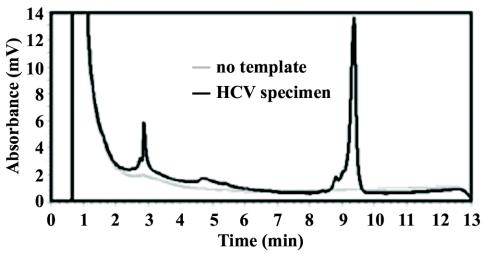
dHPLC trace of HCV heminested multiplex RT-PCR products under nondenaturing conditions at 50°C. The HCV specimen was type 1b. The elution of nucleic acid is based upon size in this analysis, so the first peak, at 2.9 min, is the 45-bp amplicon, and the peak at 9.3 min is the 153-bp amplicon.
The 115 specimens used as standards were assigned a genotype based on sequence analysis. There were 30 type 1a, 21 type 1b, 12 type 2a, 18 type 2b, 22 type 3a, and 12 type 4 HCV specimens. Genotyping appeared possible when both the retention time and the shape of the dHPLC trace were considered.
153-bp amplicon standards.
No data were obtained for 3 of 115 specimens because the specimens had a relatively low HCV RNA template concentration (≤3.9 log IU/ml). Mean retention times and standard deviations for the genotypes were as follows: 1a, 4.75 ± 0.23 min; 1b, 5.20 ± 0.22 min; 2a, 4.48 ± 0.17 min; 2b, 3.25 ± 0.17 min; 3a, 4.21 ± 0.24 min; and 4, 4.99 ± 0.27 min. The standard deviations of the retention times were <0.3 min and appeared to correlate with sequence variations seen among specimens of the same genotype. Based on the retention time results, the 153-bp amplicon could be used to discriminate type 2b from types 1a, 1b, 2a, 3a, and 4. Types 1a, 1b, 2a, 3a, and 4 could not be completely discriminated by retention time alone.
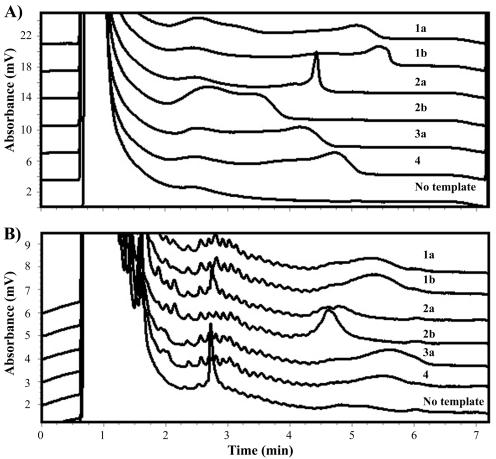
The dHPLC traces often had more than one peak (Fig. 3A). The last peak was used to determine the retention time. The traces usually had broad peaks due to partial denaturation and melting of the amplicon at the temperature chosen for analysis. At nondenaturing temperatures or more denaturing temperatures, the amplicons appeared as single well-defined peaks but were not adequate for genotyping because of a loss of resolution between different HCV types.
FIG. 3.
Representative dHPLC traces for each genotype and a no-template control. (A) Analysis of the 153-bp amplicon carried out at 63.4°C with a 11.5 to 13.75% (vol/vol) acetonitrile gradient. (B) Analysis of the 45-bp amplicon carried out at 55°C with a 7.5 to 10%(vol/vol) acetonitrile gradient.
The shape of the dHPLC traces was used to aid in discrimination of the genotypes. For example, some specimens of type 2a had a sharp spike at the trailing edge of the peak, compared to other genotypes, which all had round broad peaks (Fig. 3A). This sharp spike was not observed in the other genotypes tested. In addition, specimens of type 2b usually had a much higher absorbance and a shorter retention time than all of the other genotypes. Type 3a specimens were generally the next genotype to elute. However, some type 3a and 1a specimens had overlapping retention times and therefore were not distinguishable by the shape of the dHPLC trace. Furthermore, some type 1a, 1b, and 4 specimens were difficult to discriminate with the 153-bp amplicon. Usually, the retention times of the type 1b and 4 specimens were shorter than those of the type 1a specimens (Fig. 3A).
45-bp amplicon standards.
No data were obtained for 3 of 115 specimens tested. As for the 153-bp amplicon, this result was due to a relatively low HCV RNA template concentration (<3.9 log IU/ml). Mean retention times and standard deviations for the genotypes were as follows: 1a, 5.35 ± 0.11 min; 1b, 5.36 ± 0.15 min; 2a, 4.85 ± 0.11 min; 2b, 4.58 ± 0.14 min; 3a, 5.7 ± 0.05 min; and 4, 5.62 ± 0.11 min. The retention times measured for this amplicon, in comparison to those obtained for the 153-bp amplicon, had smaller standard deviations. This finding correlates with the smaller size of the amplicon and less sequence variation found within a genotype. From the retention times, types 2a and 2b could not be distinguished from each other but could be distinguished from types 1a, 1b, 3a, and 4. Type 3a could be distinguished from types 1a and 1b but not from type 4. In contrast to the results obtained with the 153-bp amplicon, types 1a and 1b were indistinguishable.
The shapes of the traces obtained with the 45-bp amplicon were more uniform among all of the genotypes and were not as informative as the shapes obtained with the 153-bp amplicon (Fig. 3B). More information was obtained from the actual retention times. Even though the retention times for types 1a and 1b were indistinguishable, it was possible to distinguish these types from type 3a. However, types 3a and 4 were not distinguishable with the 45-bp amplicon.
Combined results for genotyping.
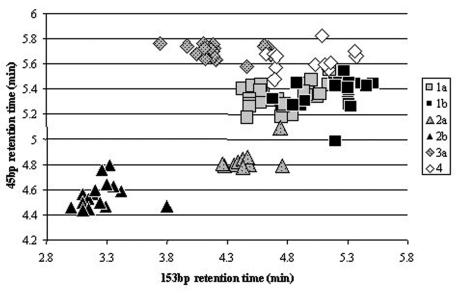
Each of the amplicons by itself could discriminate some of the HCV types, but combining the results improved the discrimination considerably (Fig. 4). Type 2a and 2b specimens were well discriminated from the other HCV types, except for one type 2a specimen that was very close to a type 1a specimen. Type 3a specimens were relatively well clustered away from the other HCV types, apart from a few specimens that overlapped with some type 4 specimens. Type 4 specimens were not easy to identify, with some being very close to types 1a and 1b. Type 1a and 1b specimens clustered together as a large group, with significant overlap.
FIG. 4.
dHPLC retention times for the 153- and 45-bp amplicons of the 5′ UTR of HCV. The data are from the specimens used as standards.
In order to standardize the assignment of HCV genotypes, the following method was used (Table 1). To make a genotype designation, parameters from both amplicons were used. First, the unknown traces were compared to the standards used in the same experimental run, to examine them for overlap and shape. If a genotype could not be assigned from trace comparisons, then the retention time cutoffs were used to assign a genotype.
Masked specimens.
Out of the 200 masked specimens, 3 could not be genotyped because of a low HCV load. For specimens that were amplified successfully, the results for 190 of 197 specimens (96%) were concordant with the sequence analysis at the major genotype level. Seventeen percent (34 of 200) of specimens had to be retested. For about half of these (15 of 34), the first and second runs did not agree, and the second run was used. The remaining specimens were confirmed by the subsequent run. The discrepant results are described in Table 2.
TABLE 2.
Masked specimens that were incorrectly identified at the major genotype level
| Specimen | Genotype determined by:
|
Source of discrepancya | |
|---|---|---|---|
| Sequencing | dHPLC | ||
| 1 | 4 | 3a | First repeat, type 4; second repeat, 3a; used second repeat data |
| 2 | 4 | 3a | Sample in type 3a- type 4 gray area; second repeat, 3a; used second repeat data |
| 3 | 3a | 1a | First repeat, second type 1a; not subsequently repeated |
| 4 | 3a | 1a | Sequence variant; G159A |
| 5 | 1b | 2a | Sequence variant; C158T |
| 6 | 1b | 2a | Sequence variant; T143G |
| 7 | 1b | 2a | Sequence variant; C162T |
Sequence variants are numbered starting from the first nucleotide in the translated region and then counted negatively.
The sources of error fell into two categories. Some specimens were difficult to assign, and although they were correctly identified on the first run, they were misidentified on the second run (specimens 1 and 2). However, one specimen that appeared to be unambiguous was misidentified and was not retested (specimen 3). The second category of error resulted from unexpected sequence variants not observed in the training set (specimens 4 to 7).
At the subtype level, the number of specimens for which results were concordant dropped to 171 of 197 (87%). The majority of errors were made between types 1a and 1b. One type 2a specimen was incorrectly identified by dHPLC as type 2b. Analysis of the sequence data for this specimen identified a single base change (C117T) in the 153-bp amplicon. This was another sequence variant not identified in the population of standards used.
DISCUSSION
One advantage of HCV genotyping by dHPLC is that no external probes or dyes are required. The method is simple, does not require heteroduplex formation, and is partially automated, reducing hands-on time and cost. However, several disadvantages and limitations were found in this study. Every dHPLC run must include a sample of each genotype as a reference. There is a contamination risk because tubes must be opened in order to inject the amplicon into the dHPLC cartridge. Perhaps most importantly, the current protocol is only 96% specific at the major genotype level and 87% specific at the subtype level. Consistent with what another group of investigators found when comparing the line probe assay to sequencing (3), types 1a and 1b are difficult to distinguish. It is also difficult to distinguish correctly types 1 and 3 from type 4. This study also did not include any examples of type 5 or 6, so it is unknown whether these particular genotypes could be distinguished from the other genotypes. In addition, as in other assays that are not based on sequencing, polymorphisms or mutations that have not been observed previously may contribute to incorrect genotype assignment.
The dHPLC method reported here is not as specific as other techniques for HCV genotyping. Zhao et al. (24) reported a 93.3% concordance rate at the subtype level when comparing microarray genotyping with sequencing of the 5′ UTR for a population of 60 samples of types 1b, 2a, 2b, 3a, and 3b. Schröter et al. (18) reported a 100% concordance rate when amplifying the 5′ UTR and genotyping with three pairs of hybridization probes by using the LightCycler compared to sequencing of the 5′ UTR for a population of 190 samples of types 1, 2, 3, and 4. Bullock et al. (2) reported that 110 of 111 samples (99%) were concordant at the genotype level for types 1, 2, 3, and 4 and that 108 of 110 samples (98%) were concordant at the subtype level. Their study compared a LightCycler assay amplifying the 5′ UTR with a single set of fluorescence energy transfer probes in the line probe assay. The lowest viral titer that could be successfully genotyped in this study was approximately 4 log IU/ml. This titer is comparable to that in other assays because one study reported detection to 3 log IU/ml (18) and another study reported detection to 4 log IU/ml with their assay and the line probe assay (2).
Despite its current limitations, HCV genotyping by dHPLC is a good screening tool at the major genotype level. For example, types 2 and 3 can be distinguished from type 1 with a false-positive rate of 2.5% and a false-negative rate of 1%. Furthermore, the current assay could be improved by including other regions of the HCV genome that discriminate genotypes and subtypes better. Ideally, the assay would be refined to a single amplicon that would require analysis at a single temperature. These parameters present a challenge for all HCV genotyping assays, as the HCV genome is so variable.
The results reported in this study provide a novel approach to the genotyping of HCV and potentially other viruses with single-stranded genomes. As a screening tool, dHPLC can be used to determine the major genotype but not the viral subtype. This approach does not require external heteroduplex formation, reducing the chance of contamination and eliminating the effort of mixing genotypes either before or after amplification. However, compared to other reported methods of HCV genotyping, the dHPLC method reported here is not as specific. dHPLC genotyping without heteroduplex formation is most applicable to infectious agents that do not have much genetic variation.
Acknowledgments
We thank Misi Robinson (Transgenomic) and Rebecca Margraf for valuable input and Melissa Seipp, Nora Arias, and Jason Metz for technical assistance.
REFERENCES
- 1.Akuta, N., F. Suzuki, A. Tsubota, Y. Suzuki, T. Hosaka, T. Someya, M. Kobayashi, S. Saitoh, Y. Arase, K. Ikeda, and H. Kumada. 2003. Association of amino acid substitution pattern in nonstructural protein 5A of hepatitis C virus genotype2a low viral load and response to interferon monotherapy. J. Med. Virol. 69:376-383. [DOI] [PubMed] [Google Scholar]
- 2.Bullock, G. C., D. E. Bruns, and D. M. Haverstick. 2002. Hepatitis C genotype determination by melting curve analysis with a single set of fluorescence resonance energy transfer probes. Clin. Chem. 48:2147-2154. [PubMed] [Google Scholar]
- 3.Chen, Z., and K. E. Weck. 2002. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 40:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbet, S., J. Bukh, A. Heinsen, and A. Fomsgaard. 2003. Hepatitis C virus subtyping by a core envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J. Clin. Microbiol. 41:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121-S127. [DOI] [PubMed] [Google Scholar]
- 6.Gross, E., K. Seck, S. Neubauer, J. Mayr, H. Hellebrand, A. Ratanaphan, V. Lutz, H. Stockinger, and M. Kiechle. 2003. High-throughput genotyping by DHPLC of the dihydropyrimidine dehydrogenase gene implicated in (fluoro)pyrimidine catabolism. Int. J. Oncol. 22:325-332. [PubMed] [Google Scholar]
- 7.Hurtle, W., D. Shoemaker, E. Henchal, and D. Norwood. 2002. Denaturing HPLC for identifying bacteria. BioTechniques 33:386-388, 390-391. [DOI] [PubMed]
- 8.Jessner, W., R. Stauber, F. Hackl, C. Datz, T. Watkins-Riedel, H. Hofer, A. Gangl, H. Kessler, and P. Ferenci. 2003. Early viral kinetics on treatment with pegylated interferon-alpha-2a in chronic hepatitis C virus genotype 1 infection. J. Viral Hepatol. 10:37-42. [DOI] [PubMed] [Google Scholar]
- 9.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 10.Liou, T. C., T. T. Chang, K. C. Young, X. Z. Lin, C. Y. Lin, and H. L. Wu. 1992. Detection of HCV RNA in saliva, urine, seminal fluid, and ascites. J. Med. Virol. 37:197-202. [DOI] [PubMed] [Google Scholar]
- 11.Maertens, G., and L. Stuyver. 1997. Genotypes and genetic variation of hepatitis C virus, p. 183-234. In T. G. Harrison and A. J. Zuckerman (ed.), The molecular medicine of viral hepatitis. John Wiley & Sons Ltd., Chichester, England.
- 12.Mitchell, P. S., L. M. Sloan, D. W. Majewski, P. N. Rys, P. J. Heimgartner, J. E. Rosenblatt, F. R. Cockerill III, T. F. Smith, and R. Patel. 2002. Comparison of line probe assay and DNA sequencing of 5′ untranslated region for genotyping hepatitis C virus: description of novel line probe patterns. Diagn. Microbiol. Infect. Dis. 42:175-179. [DOI] [PubMed] [Google Scholar]
- 13.Otagiri, H., Y. Fukuda, I. Nakano, Y. Katano, H. Toyoda, S. Yokozaki, K. Hayashi, T. Hayakawa, M. Kinoshita, and J. Takamatsu. 2002. Evaluation of a new assay for hepatitis C virus genotyping and viral load determination in patients with chronic hepatitis C. J. Virol. Methods 103:137-143. [DOI] [PubMed] [Google Scholar]
- 14.Pallares-Ruiz, N., P. Blanchet, M. Mondain, S. Low-Hong, J. Demaille, M. Claustres, and A. F. Roux. 2001. Evaluation of dHPLC for CX26 mutation screening in patients from southern France with sensorineural deafness. Genet. Test. 5:339-343. [DOI] [PubMed] [Google Scholar]
- 15.Roque-Afonso, A. M., M. P. Ferey, J. D. Poveda, E. Marchadier, and E. Dussaix. 2002. Performance of TRUGENE hepatitis C virus 5′ noncoding genotyping kit, a new CLIP sequencing-based assay for hepatitis C virus genotype determination. J. Viral Hepatol. 9:385-389. [DOI] [PubMed] [Google Scholar]
- 16.Saracco, G., A. Olivero, A. Ciancio, S. Carenzi, and M. Rizzetto. 2003. Therapy of chronic hepatitis C: a critical review. Curr. Drug Targets Infect. Disord. 3:25-32. [DOI] [PubMed] [Google Scholar]
- 17.Schaeffeler, E., T. Lang, U. M. Zanger, M. Eichelbaum, and M. Schwab. 2001. High-throughput genotyping of thiopurine S-methyltransferase by denaturing HPLC. Clin. Chem. 47:548-555. [PubMed] [Google Scholar]
- 18.Schroter, M., B. Zollner, P. Schafer, O. Landt, U. Lass, R. Laufs, and H. H. Feucht. 2002. Genotyping of hepatitis C virus types 1, 2, 3, and 4 by a one-step LightCycler method using three different pairs of hybridization probes. J. Clin. Microbiol. 40:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlush, L. I., D. M. Behar, A. Zelazny, N. Keller, J. R. Lupski, A. L. Beaudet, and D. Bercovich. 2002. Molecular epidemiological analysis of the changing nature of a meningococcal outbreak following a vaccination campaign. J. Clin. Microbiol. 40:3565-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White, P. A., X. Zhai, I. Carter, Y. Zhao, and W. D. Rawlinson. 2000. Simplified hepatitis C virus genotyping by heteroduplex mobility analysis. J. Clin. Microbiol. 38:477-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao, W., and P. J. Oefner. 2001. Denaturing high-performance liquid chromatography: a review. Hum. Mutat. 17:439-474. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida, N., Y. Nishimaki, M. Sugiyama, T. Abe, T. Tatsumi, A. Tanoue, A. Hirasawa, and G. Tsujimoto. 2002. SNP genotyping in the beta(2)-adrenergic receptor by electronic microchip assay, DHPLC, and direct sequencing. J. Hum. Genet. 47:500-503. [DOI] [PubMed] [Google Scholar]
- 23.Zein, C. O., and N. N. Zein. 2002. Advances in therapy for hepatitis C infection. Microbes Infect. 4:1237-1246. [DOI] [PubMed] [Google Scholar]
- 24.Zhao, W., W. Liu, Q. Liu, L. Zhang, Z. Zhou, X. Liu, and H. Zhang. 2002. Genotyping of hepatitis C virus by hepatitis gene diagnosis microarray. Zhonghua Yi Xue Za Zhi 82:1249-1253. [PubMed] [Google Scholar]