Abstract
Drug resistance in Mycobacterium tuberculosis complex strains is solely due to chromosomal mutations that could affect bacterial virulence. Molecular epidemiology studies have shown that resistant strains are less likely to be clustered than susceptible strains. However, a few multidrug-resistant (MDR) M. tuberculosis complex strains have been described as causing outbreaks, suggesting that they have restored virulence or increased transmission. One of the biggest MDR tuberculosis outbreaks documented to date was caused by the B strain of M. bovis. Restriction fragment length polymorphism fingerprinting revealed that the B strain contains two copies of IS6110. Here, we mapped and sequenced the regions flanking the two copies of IS6110 in the B strain. Ligation-mediated PCR showed that one of these IS6110 copies is located within the promoter region of phoP, a transcriptional regulator that is essential for M. tuberculosis virulence. We used PCR to screen 219 MDR M. tuberculosis complex strains (90.4% of all MDR isolates) isolated in Spain between 1998 and 2002 and found that the B strain was the only strain that contained a copy of IS6110 in the phoP promoter. To determine whether IS6110 affects phoP promoter activity in the B strain, we individually cloned the phoP gene and its promoter region (including IS6110 from the B strain and the equivalent region from M. tuberculosis without IS6110 as a control) into a mycobacterial replicative plasmid and transformed M. smegmatis with the resulting plasmid. Primer extension analysis showed that phoP transcription was strongly upregulated when the promoter region contained IS6110, as in the case of the B strain.
Tuberculosis (TB) is currently one of the leading causes of mortality throughout the world (8, 25, 27). The human immunodeficiency virus-AIDS pandemic, the deterioration of public health systems in developing countries, and the emergence of multidrug-resistant (MDR) Mycobacterium tuberculosis complex strains have further contributed to the spread of TB. Knowledge of the molecular mechanisms involved in the bacillus-host cell interaction is essential for developing adequate strategies for TB control. Recent advances in the genetic manipulation of mycobacteria (2, 29) combined with the publication of the complete M. tuberculosis genome sequence (6) have made it possible to study the contribution of individual genes to M. tuberculosis virulence (5, 7). However, little is known about the regulatory and expression mechanisms that determine the virulence of clinical isolates of M. tuberculosis.
Insertion sequence (IS) 6110 has been extensively used for molecular typing of M. tuberculosis strains. Restriction fragment length polymorphism (RFLP) analysis using IS6110 as a probe is currently the most common molecular method used to type M. tuberculosis complex strains. However, the physiological role and impact of specific IS6110 insertions on the biology of bacilli are not well known. IS6110 fingerprinting studies have demonstrated heterogeneity between virulence and transmissibility of different M. tuberculosis clinical isolates, as judged by rates of skin test conversion and onset of clinical disease among exposed individuals (34, 43). In some strains of M. tuberculosis, drug resistance is associated with low growth rates and reduced persistence in mice and guinea pigs (19). Although drug-resistant M. tuberculosis strains appear to be less transmissible than susceptible isolates (45), MDR strains of M. tuberculosis have caused several outbreaks (4, 38). For example, the widely distributed W strain, which belongs to the Beijing family of isolates (1, 4), and the MDR M. bovis B strain (35, 38, 39) have caused large outbreaks in the United States and Spain, respectively. The high transmission rate of some MDR strains suggested that their fitness and virulence are unaffected, making these strains especially adapted to their hosts. The study of the genetic mechanisms involved in the transmissibility of these epidemic MDR strains could provide valuable information about the genes implicated in M. tuberculosis virulence.
Many IS elements affect the expression of neighbor genes by generating new promoter sequences capable of driving their expression. The presence of IS6110 in M. tuberculosis clinical isolate 210, which belongs to the Beijing family, provides a promoter sequence that enhances gene transcription (3).
MDR TB strains isolated in Spain have been systematically typed since 1998. The typing results showed that most MDR M. tuberculosis complex strains are not further transmitted to other patients (reference 38 and our unpublished data). In contrast, the B strain was found to cause severe MDR TB outbreaks (35, 39). RFLP analysis showed that the B strain contains two copies of IS6110 (39).
An M. tuberculosis phoP mutant was recently constructed by gene disruption (30). This mutant exhibits impaired growth in vitro within mouse-grown bone marrow-derived macrophages, and it was also attenuated in vivo in a mouse model of infection. Thus, phoP may act as a virulence regulator in M. tuberculosis. The phoP locus from pathogenic bacterial species is a member of the PhoP/PhoQ two-component regulatory system (12, 42). This system is required in Salmonella for the expression of virulence factors in mice and for survival within macrophages and confers resistance to killing by certain defensins (13).
In this work, we found that one copy of IS6110 is located within the phoP promoter (6) in the MDR epidemic B strain. We used PCR to screen more than 200 MDR M. tuberculosis complex clinical isolates and showed that the B strain is the only strain to contain IS6110 in its phoP promoter region. Primer extension (PE) analysis showed that IS6110 increased the expression of the phoP virulence gene in the B strain causing MDR tuberculosis outbreaks.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
All MDR M. tuberculosis complex clinical isolates were sent to our laboratory as part of the M. tuberculosis genotyping network of the University of Zaragoza (38). The M. bovis B strain was selected for this study (39). M. tuberculosis Mt14323, M. bovis BCG Pasteur, and M. smegmatis mc2155 (41) were also used. Mycobacterial strains were routinely grown in Middlebrook 7H10 Bacto agar supplemented with oleic acid-albumin-dextrose-catalase or in Middlebrook 7H9 broth supplemented with albumin-dextrose-catalase (Difco Laboratories, Detroit, Mich.) and 0.05% Tween 80 (17). M. smegmatis mc2155 was grown in Dubos broth containing 0.1% Tween 80 and albumin (Difco Laboratories) for RNA extraction.
Escherichia coli XL1-blue (37) was grown on solid medium or in liquid Luria-Bertani medium for cloning and plasmid propagation.
All the strains were grown at 37°C. When required, kanamycin (20 μg/ml) or hygromycin (50 μg/ml for M. smegmatis and 200 μl/ml for E. coli) was added to the culture medium.
IS6110 fingerprinting.
MDR M. tuberculosis strains isolated in Spain since 1998 have been systematically typed by standard RFLP and spoligotyping in our laboratory (38, 44, 46, 47). For RFLP, cetyltrimethylammonium bromide-NaCl and chloroform-isoamyl alcohol are used as described previously to extract chromosomal DNA from the isolates (46). Extracted DNA is digested with PvuII (Boehringer-Mannheim), separated in 0.8% agarose gels in Tris-borate-EDTA buffer, and vacuum blotted onto nylon membranes. Blotted DNA was hybridized with a 0.8-kb fragment corresponding to the PCR-amplified fragment between the PvuII restriction site and the 3′ end of IS6110. The probe was labeled and RFLP patterns were visualized by use of an ECL direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England). Autoradiograms of IS6110 fingerprints were scanned, analyzed, and compared by use of Bionumerics software (version 3.0; Applied Maths, Kortrijk, Belgium).
Mapping of the regions flanking IS6110 in the B strain.
Ligation-mediated PCR (LM-PCR) was used as described by Prod'hom et al. (32) to determine the location of the two copies of IS6110 in the M. bovis B strain. Briefly, genomic DNA was digested with SalI (Boehringer-Mannheim) and ligated to a linker containing a SalI restriction site at its 3′ end and the Salgd primer sequence (5′-tagcttattcctcaaggcacgagc-3′). The resulting template was then digested with SalI. PCR was performed using ISA1 (5′-cctgacatgaccccatcctttcc-3′) and ISA3 (5′-gaggctgcctactacgtcaacg-3′), specific primers for IS6110 directed outwards from this element (22), and the linker primer Salgd (1 μM). The template was initially denatured by incubation at 95°C for 9 min and amplified by 35 cycles of PCR (95°C for 30 s, 70°C for 30 s, and 72°C for 90 s) followed by a final extension at 72°C for 10 min in a Gene Amp PCR System 9700 thermocycler (PE Applied Biosystems, Foster City, Calif.). Amplified products were separated by standard horizontal gel electrophoresis in a 1.5% agarose gel in Tris-borate-EDTA buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA) and stained with ethidium bromide. PCR products were purified using GFX PCR DNA and a gel band purification kit (Amersham Pharmacia Biotech). The amplified products were sequenced on an ALF DNA sequencer using a Thermo Sequenase fluorescent labeled primer cycle sequencing kit (Amersham Pharmacia Biotech).
Detection of IS6110 upstream from phoP in M. tuberculosis complex isolates.
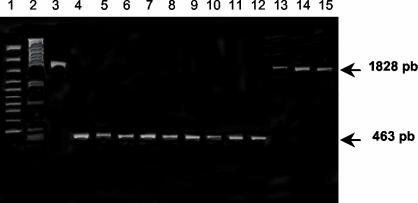
We used PCR (10) to seek IS6110 in the phoP promoter region of 219 of the 241 MDR M. tuberculosis complex strains previously genotyped by RFLP or spoligotyping. We used the BCG2B (5′-ccatgttcaaaccggtgtc-3′) and BCG2A (5′-gccgtccatcccgggcatc-3′) primers, which annealed 136 bp upstream and 293 bp downstream from IS6110 in the B strain, respectively. The PCR conditions were as follows: 35 cycles of 95°C for 60 s, 62°C for 60 s, and 72°C for 60 s followed by a final extension at 72°C for 10 min. When IS6110 was present upstream of phoP, an 1,828-bp amplification product was obtained. However, when the insertion element was absent, a 463-bp PCR product was obtained.
Construction of pSO5 and pSO7.
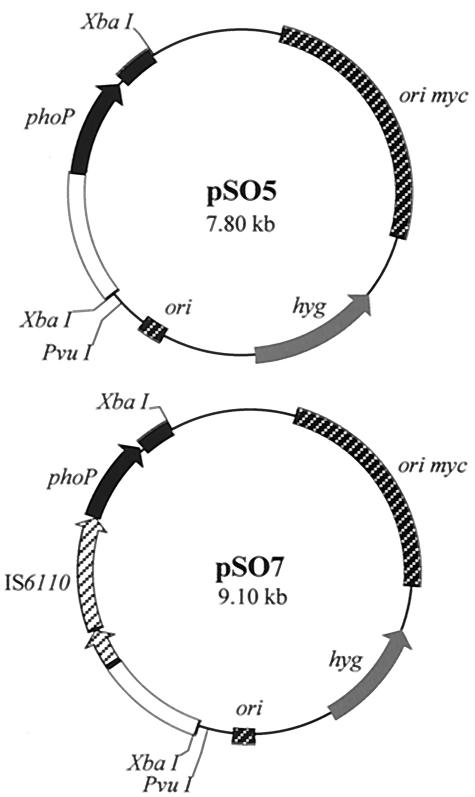
pSO5 has been described previously (30). This recombinant plasmid contains a 2-kb genomic fragment containing both the promoter region and the entire phoP coding region of M. tuberculosis MT103. The phoP nucleotide sequence was cloned into pNBV1 (kindly provided by W. R. Bishai) (16). pNBV1 is a shuttle plasmid that contains the origin of replication from E. coli and the origin of replication from M. fortuitum pAL5000 (33). pSO7 contains the phoP promoter region from the B strain, including a complete copy of IS6110. To construct pSO7, primers phoPF (5′-aatctagatcaagcatcagccgaggtac-3′) and phoPR (5′-aatctagacccgaacgtagaacc) were used to amplify a 3,411-bp fragment from the M. bovis B strain. PhoPF annealed 1,000 and 929 bp upstream from the phoP start codon and the IS6110 insertion site, respectively. PhoPR annealed 305 and 1,117 bp downstream from the phoP termination codon and IS6110, respectively. As PhoPF and PhoPR contained an XbaI restriction site, the PCR product was ligated to pNBV1 that had been cut with XbaI. The purified plasmids (pSO5 and pSO7) were then electroporated into M. smegmatis mc2155 (17). The pSO5 and pSO7 plasmids were sequenced using an ABI 3700 DNA sequencer with a BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems). Primers SePho1 (5′-gagagcacccgctcgataag-3′), BCG2B (5′-ccatgttcaaaccggtgtc-3′), PhoExp (5′-cccatatgcggaaaggggttgatctct-3′), T3, and T7 were used for sequencing of pSO5. Primers BCG2A, ISA3 (see Fig. 2), PhoRF (5′-aatctagagggcaagggcaacaaggaac-3′), PhoPR (5′-aactagagatcacccgaacgtagaagg-3′), SePho8 (5′-gttccttgttgcccttgccc-3′), T3, and T7 were used for sequencing of pSO7. DNA sequences were used to search the TubercuList database.
FIG. 2.
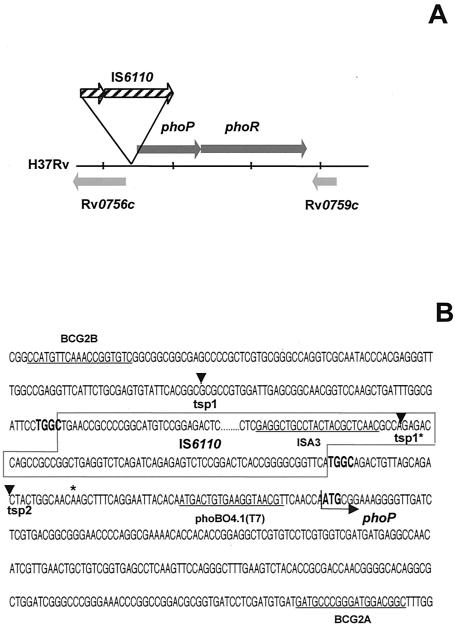
Location of IS6110 upstream from phoP in the M. bovis B strain. (A) Schematic representation showing the position of IS6110 in the phoP promoter region. (B) Nucleotide sequence surrounding the second copy of IS6110. The IS6110 nucleotide sequence is boxed. The TGGC DRs flanking the IS6110 insertion site are shown in boldface characters. The ATG start codon of phoP is indicated by an arrow. The nucleotide sequences of primers BCG2A, BCG2B, phoBO4.1(T7), and ISA3 are underlined. ▾, transcription starting points tsp2 and tsp1 from pSO5 and pSO7 (tsp1*). tsp1*, tsp1 position in pSO5 wild-type M. tuberculosis. The nucleotide change (A for G) 46 bp upstream from the phoP ATG start codon of M. bovis with respect to M. tuberculosis H37Rv is indicated by an asterisk.
Isolation of total RNA.
RNA was isolated from the M. smegmatis mc2155 wild-type strain, and transformants were grown to middle exponential phase (optical density at 600 nm, 0.4 to 0.6) as described previously (11). Further DNase treatment was performed for the RNase protection assays.
Identification of transcription starting site.
PE and RNase protection (RPA) experiments were carried out as described previously (24). Briefly, PE studies was carried out using primer phoBO4.1 (5′-aacgttaccttcacagtcat-3′), which anneals immediately upstream from the phoP ATG start codon (see Fig. 2). PhoBO4.1 was end labeled with [γ-32P]ATP by a T4 polynucleotide kinase reaction. A total of 100 fmol of labeled primer was annealed to 30 μg of total RNA and extended by using avian myeloblastosis virus reverse transcriptase (11, 24). The polymerization products were ethanol precipitated and resuspended in an appropriate volume.
The RPA method required the construction of a minigene corresponding to the region under analysis. This minigene was used as a template for the synthesis of a radiolabeled antisense RNA probe complementary to the RNA transcript. Minigenes were synthesized by using PCR to amplify the region upstream from the phoP gene of the M. bovis B strain and of M. tuberculosis. The phoBO4.1-T7 (phoBO4.1 plus T7 promoter sequence) and ISA3 primers were used for M. bovis, and the phoBO4.1-T7 and BCG2B primers were used for M. tuberculosis (see Fig. 2). Minigenes of 245 and 181 bp were obtained, respectively. Transcription products from minigenes were radiolabeled by using a Riboprobe kit (Promega, Madison, Wis.) according to the manufacturer's recommendations. The products were purified on a 6% (wt/vol) polyacrylamide gel; appropriate bands of the expected sizes were excised and eluted as described previously (24). Hybridization was carried out with 20 μg of DNA-free RNA and 2.5 × 105 cpm of radiolabeled samples and digested with an RNase cocktail containing both RNase T1 (20 U/μl) and RNase A (1 U/μl) (Ambion). The optimum RNase concentration for RPA was established.
Products obtained in both PE and RPA were separated on 8% (wt/vol) polyacrylamide-urea sequencing gels. Radioactive products were located by autoradiography using an intensifying screen. Radioactivity corresponding to the several transcriptional products detected by PE was measured by using an Instant Imager system (Packard-Izasa, Barcelona, Spain).
RESULTS
IS6110 genotyping of the MDR M. bovis B strain.
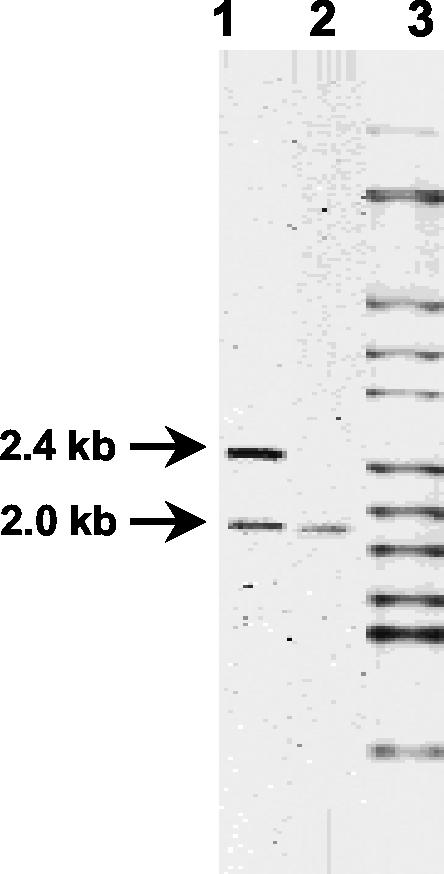
A total of 241 M. tuberculosis complex strains isolated in Spain between 1998 and 2002 from patients with MDR TB were analyzed by RFLP and spoligotyping. The IS6110 fingerprints are included in the Spanish MDR TB database (University of Zaragoza). A total of 26 (10.8%) of these isolates displayed a characteristic RFLP pattern, with two IS6110 bands of 2.4 and 2.0 kb (39) (Fig. 1, lane 1). These isolates belong to the previously described M. bovis B strain (39). The B strain was responsible for a severe nosocomial outbreak of MDR TB in Spain (35). The rate of reinfection with the B strain is high among human immunodeficiency virus-coinfected patients (35). The 2.4-kb IS6110 band was found only in B strains (whereas the 2.0-kb band was also found in M. bovis isolates [such as M. bovis BCG isolates] containing a single copy of IS6110) (Fig. 1, lane 2).
FIG. 1.
RFLP analysis showing the IS6110 fingerprint of the B strain. Fingerprints of strains B (lane 1), M. bovis BCG Pasteur (lane 2), and M. tuberculosis Mt14323 (lane 3) are shown. The 2.0- and 2.4-kb bands represent copies of IS6110 in DRs and upstream from phoP, respectively.
Localization of the two copies of IS6110 in the B strain.
LM-PCR was used to localize the two copies of IS6110 in the B strain genome. The regions flanking IS6110 were amplified by LM-PCR and sequenced.
The IS6110 corresponding to the 2.0-kb band of the B strain (Fig. 1) was inserted in a genomic region characterized by the presence of multiple direct repeats (DRs) (located between positions 3119182 and 3122372) in the M. tuberculosis H37Rv genome. (This region is composed of 36-bp DRs separated by 36- to 41-bp nonrepetitive segments. DRs are hotspots for IS6110 transposition in most M. tuberculosis and M. bovis strains.) In M. bovis BCG, which contains a single copy of IS6110 (Fig. 1), the insertion sequence is located in an identical position (3, 14).
The nucleotide sequence of the genomic region flanking the second IS6110 copy in the B strain, corresponding to the 2.4-kb band (Fig. 1), showed that IS6110 was inserted at nucleotide 851531 upstream from open reading frame Rv0757 (annotated as phoP in the H37Rv genome) (6). This insertion site was located 75 bp upstream from the ATG start codon of phoP in the B strain (Fig. 2). IS6110 was flanked by a 4-bp DR (TGGC) on both sides (Fig. 2).
The copy of IS6110 located upstream from the phoP gene is specific to the M. bovis B strain from the MDR M. tuberculosis complex strains studied.
To determine whether the presence of the second copy of IS6110 in the promoter region of phoP was specific to the B strain, we used PCR to screen DNAs from different MDR M. tuberculosis complex strains.
We screened 218 of the 241 (90.4%) MDR TB strains isolated in Spain since 1998 for the presence of the IS6110 upstream from phoP. The specific primers BCG2A and BCG2B (10) were used to amplify the phoP promoter region from genomic DNA (Fig. 2B). A total of 21 (9.6%) of the MDR M. tuberculosis complex strains tested generated a 1,828-bp PCR product (Fig. 3, lanes 13 to 15), indicating that they contained IS6110 upstream from phoP as in the case of the B strain (Fig. 1 and 2A). According to our fingerprint databank, all of these strains were B strains.
FIG. 3.
The copy of IS6110 in the phoP promoter region is specific to B strains. Genomic DNA was PCR amplified with primers BCG2A and BCG2B (Fig. 2B). Lanes 3 and 13 to 15, B strains; lane 4, M. tuberculosis H37Rv; lanes 5 to 12, other MDR M. tuberculosis complex strains; lane 1, Gene Ruler 100-bp DNA Ladder Plus (MBI Fermentas, Vilnius, Lithuania); lane 2, λ/PstI. The presence of a 1,828-bp PCR product means that IS6110 was present in the promoter region of phoP in the analyzed strain.
A 463-bp amplification product (signifying that IS6110 was not present upstream from phoP) was obtained for 191 (87.6%) clinical isolates (Fig. 3, lanes 4 to 12). No amplification product was obtained with DNAs from five strains (1.8%), three of which were shown by RFLP to be B strains. We believe that the DNA preparations were contaminated in the two other cases (0.9%). All these results strongly suggest that only the outbreak-causing B strains contain a copy of IS6110 in the promoter region of phoP in the MDR TB strains present in our database.
Transcriptional level of phoP.
To study the effect of IS6110 on the transcription of phoP, we constructed the recombinant plasmids pSO5 and pSO7 (Fig. 4) in a pNBV1 background (16). We used M. smegmatis mc2155 as a surrogate host to study the role of IS6110 in phoP transcription, because it is difficult to manipulate the B strain due to its resistance to most antituberculosis drugs. The differences between the M. smegmatis and M. tuberculosis complex phoP gene sequences allowed the design of specific primers for PE.
FIG. 4.
pSO5 and pSO7 mycobacterial plasmids. The phoP genes (including the promoter region) of M. tuberculosis and M. bovis B were amplified by PCR and ligated to pNBV1 that had been cut with XbaI to obtain pSO5 and pSO7, respectively. pSO5 contained a 2,046-bp M. tuberculosis fragment containing a 1-kb region upstream from the ATG start codon plus the entire nucleotide sequence of phoP (30). pSO7 contained the equivalent region from the B strain (see text for more details). hyg, hygromycin-resistant gene; ori myc, mycobacterial origin of replication; ori, E. coli origin of replication.
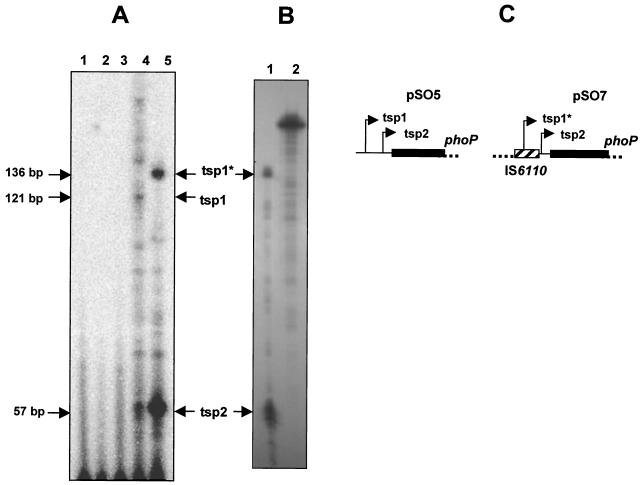
We carried out PE analysis with RNA isolated from M. smegmatis mc2155 harboring pSO5 and pSO7, which contain the phoP promoter regions of M. tuberculosis wild-type and B strains, respectively. We detected two putative transcription start points (tsp1 and tsp2) in both cases (Fig. 5 and 2B). In M. smegmatis harboring the wild-type promoter (pSO5), tsp1 and tsp2 were located 121 and 57 bp upstream from the ATG start codon of phoP, respectively; the position of tsp1 was confirmed by RPA (data not shown). In M. smegmatis harboring the B strain promoter (pSO7), tsp1 and tsp2 were located 136 and 57 bp upstream from the ATG starting codon of phoP, respectively (Fig. 5A and 2B). Thus, tsp1 from the B strain (tsp1*) was located inside IS6110 (Fig. 5A). The locations of tsp1* and tsp2 in pSO7 were confirmed by RPA (Fig. 5B).
FIG. 5.
Mapping the phoP transcription starting points. The PE and RPA products were subjected to electrophoresis in 6% (wt/vol) polyacrylamide-8 M urea gels and then to autoradiography. (A) Products of PE analysis using total RNA from M. smegmatis mc2155 (lane 2), M. smegmatis mc2155:pNBV1 (lane 3), M. smegmatis mc2155:pSO5 (lane 4), M. smegmatis mc2155:pSO7 (lane 5) and using no RNA (lane 1). tsp1, tsp1*, and tsp2 are the putative transcription start points of potential phoP promoters for M. tuberculosis (pSO5) and the B strain (pSO7). The tsp1 for the B strain (tsp1*) lies within the IS6110 nucleotide sequence. No phoP transcripts were detected with the control strains (M. smegmatis mc2155 and the same strain transformed with pNBV1). (B) Products of RPA of M. smegmatis mc2155:pSO7 (lane 1) and a probe (lane 2). The locations of the transcription starting points (tsp1* and tsp2) are indicated. (C) Schematic figure showing the locations of tsp1, tsp1*, and tsp2 in the promoter regions of M. tuberculosis (pSO5) and the M. bovis B strain (pSO7).
The PE and RPA results showed that the B strain (pSO7) synthesized more phoP transcripts than M. tuberculosis (pSO5). Due to the use of end-labeled oligonucleotide in the PE experiments (see Materials and Methods), the relative radioactivity of the products reflects their relative abundance irrespective of their lengths. Thus, based on the amount of radioactivity incorporated into the cDNA, the B strain (pSO7) contained approximately 10-fold more phoP transcripts than the wild type (pSO5) (Fig. 5), indicating that differences in phoP expression occur at the transcription level. These results strongly suggest that phoP is overexpressed due to the presence of IS6110 in the gene promoter region.
DISCUSSION
A number of molecular epidemiology studies have shown that resistant and MDR M. tuberculosis strains are less transmissible than wild-type susceptible strains (references 38 and 45 and our unpublished data). The mutations responsible for drug resistance in bacteria are frequently associated with adaptive changes correlated with decreased growth rates in comparison with wild-type strains (40). Nevertheless, particular MDR TB strains cause outbreaks. Over the last 6 years, the M. bovis B strain was the strain most often isolated that caused MDR TB in Spain (35, 38, 39). Even though the sequences of M. bovis strains are very similar to those of M. tuberculosis, M. bovis is rarely transmitted between humans. No MDR M. bovis strains other than the B strain were isolated during this period. We focused on the B strain with the aim of elucidating the molecular mechanism of M. tuberculosis complex virulence.
RFLP fingerprinting showed that two copies of IS6110 are present in the B strain. Analysis of the nucleotide sequence of the genomic regions flanking the IS6110 in this strain revealed that an IS6110, corresponding to a 2.0-kb RFLP band, was inserted in a homologous region of DR, as is the case for most M. bovis isolates (including BCG isolates) (3). DRs are hotspots for IS6110 insertion that have been detected in 94% of M. tuberculosis clinical isolates (14). Common insertion of IS6110 in the DR region should represent an ancient IS6110 transposition event. The second copy of IS6110, corresponding to the 2.4-kb RFLP band in the B strain, was inserted 75 nucleotides upstream from the ATG start codon of a DR annotated as being phoP in M. tuberculosis H37Rv (6). The phoP/phoR locus of M. tuberculosis is a two-component system that is similar to the phoP/phoQ locus, a global virulence regulator with pleiotropic effects on gene expression in intracellular pathogens such as Salmonella spp. (15, 23). In the B strain, IS6110 was flanked on both sides by a 4-bp DR (TGGC). The generation of 3- to 4-bp DRs after a transposition is one of the characteristics of members of the IS3 family, particularly IS6110 in M. tuberculosis (22). Thus, the presence of 4-bp DRs flanking IS6110 strongly suggests that IS6110 was inserted upstream from phoP as a consequence of a transposition event in the B strain.
IS6110 transposes randomly within the M. tuberculosis genome, although it has a preference for particular sites such as DRs or the ipl locus (9, 14). We used PCR to search for IS6110 upstream from phoP in 221 MDR M. tuberculosis complex clinical isolates. IS6110 was found at this position in 21 strains, all B strains, indicating that this event is exclusive to this particular MDR isolate. Thus, the unusually high transmission rate of the B strain in comparison to those of most MDR M. tuberculosis complex clinical isolates suggests that the insertion of IS6110 in the promoter region of phoP confers some advantage allowing transmission.
Transposable elements frequently alter bacterial gene expression, either by inactivating the gene or by acting as a portable promoter for gene activation mediated by transcriptional regulation. This phenomenon can affect the regulation of the surrounding genes (20). For example, the insertion of IS1186 upstream from the cfiA gene induces resistance to carapenin in Bacteroides fragilis clinical isolates (31). The transcription of cfiA is driven from a promoter sequence located at the 3′ end of IS1186. Moreover, high- and low-level trimethoprim resistance in Staphylococcus aureus is mediated by genomic deletions adjacent to a copy of IS257 inserted upstream from the dihydrofolate reductase gene, dfrA. In this case, the insertion sequence encodes a −35 sequence that is necessary for full promoter activity (18). IS6110 can promote gene expression in M. tuberculosis 210 (3). This strain belongs to the Beijing family and has an increased capacity for transmission. According to reverse transcriptase PCR experiments, the insertion of IS6110 within ctpD generates a promoter sequence that upregulates the transcription of the gene (3).
The insertion of IS6110 upstream from phoP in the B strain may affect gene expression given that phoP is an important transcriptional regulator in intracellular pathogens such as Salmonella spp. and Yersinia spp. (13, 26, 28). The B strain grows slowly (mean growth time in Lowenstein medium, 57 days) and displays peculiar colony morphology (smaller colonies than other M. bovis strains) (35). The insertion of IS6110 at this particular locus could be responsible for these changes. As a consequence, the variation in the levels of phoP expression might affect the genes regulated by PhoP, which has pleiotropic effects in bacteria (13, 30). M. tuberculosis phoP mutants are a dramatically different shape and size compared to their wild-type parent when grown in liquid or on solid medium, which is consistent with PhoP being involved in global regulatory circuits in mycobacteria (reference 30 and our unpublished data).
In Corynebacterium diphtheriae (which is phylogenetically related to M. tuberculosis) iron depletion results in the derepression of virulence genes such as the diphtherial toxin (tox) gene mediated by DtxR (diphtheria toxin regulator). The corynebacterial DtxR has an homologue in M. tuberculosis, IdeR (iron-dependent regulator) (36). In six genes containing potential IdeR-binding sites, iron-box consensus sequences have been found. One of theses potential IdeR-binding sites is present immediately upstream from the ATG start codon of phoP in M. tuberculosis (21).
The molecular mechanisms possibly involved in the regulation of phoP transcription mediated by IS6110 in mycobacteria are unknown. The nucleotide sequences of the phoP promoter region in M. bovis and M. tuberculosis are well conserved, differing by just one nucleotide (Fig. 2B). The insertion of IS6110 into the phoP promoter region did not produce nucleotide sequence changes or deletions in the genomic region between the IS6110 insertion site and the phoP ATG start codon in the B strain. PE experiments detected two putative transcription start points (tsp1 and tsp2) for phoP in the B and M. tuberculosis strains. A new tsp1 promoter region (tsp1*) was detected in the B strain within the IS6110 nucleotide sequence. In addition, the amount of transcripts detected from tsp2 was much higher in the B strain than in the wild-type region, which does not contain IS6110. The increase in phoP transcription rates in M. bovis B is clearly a consequence of the IS6110 insertion. This suggests that the insertion of IS6110 upstream from tsp2 altered its promoter sequence, thus generating a new strong promoter region and increasing the phoP transcription rate from tsp2.
RFLP analysis using IS6110 as a probe is a powerful tool for differentiating M. tuberculosis strains, making it extremely useful for molecular studies of tuberculosis. Here, we show that IS6110 upregulates the transcription of a gene implicated in M. tuberculosis virulence in a particular clinical isolate that causes MDR TB outbreaks. Another example of an increase in gene transcription mediated by IS6110 has been shown for a Beijing strain (3). Further studies are necessary to determine how IS6110 mediates M. tuberculosis clinical-isolate physiological changes that can alter the expression of genes determining the infection or transmission rates of certain M. tuberculosis isolates. In addition to the most common use of IS6110 as an RFLP tool, results presented here show one example of the role of IS6110 in increasing expression of virulence genes in a clinical isolate. We suggest a new view of IS6110, according to which it plays a dynamic role in changing the expression of a gene or a network of genes that orchestrate transmissibility in conjunction with epidemiological, environmental, and host factors.
Acknowledgments
We thank Carmen Lafoz for technical assistance. We thank reviewers for comments that helped to clarify and focus the manuscript.
This work was supported by the Spanish Ministerio de Ciencia y Tecnologia (BIO2002-04133) and European Economic Community INCO (grant ICA4-CT2002 -10063). Both laboratories are members of the Red Latinoamericana y del Caribe de tuberculosis (RELACTB) supported by European Union INCO CA4-CT2001-10087.
REFERENCES
- 1.Anh, D. D., M. W. Borgdorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 5.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 8.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W. H. O. Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 9.Fang, Z., and K. J. Forbes. 1997. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J. Clin. Microbiol. 35:479-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fomukong, N. G., T. H. Tang, S. al-Maamary, W. A. Ibrahim, S. Ramayah, M. Yates, Z. F. Zainuddin, and J. W. Dale. 1994. Insertion sequence typing of Mycobacterium tuberculosis: characterization of a widespread subtype with a single copy of IS6110. Tuber. Lung Dis. 75:435-440. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-y-Merchand, J. A., M. J. Colston, and R. A. Cox. 1996. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology 142:667-674. [DOI] [PubMed] [Google Scholar]
- 12.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groisman, E. A., and F. Heffron. 1995. Regulation of Salmonella virulence by two-component regulatory systems, p. 319-332. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 14.Hermans, P. W., D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohmann, E. L., C. A. Oletta, and S. I. Miller. 1996. Evaluation of a phoP/phoQ-deleted, aroA-deleted live oral Salmonella typhi vaccine strain in human volunteers. Vaccine 14:19-24. [DOI] [PubMed] [Google Scholar]
- 16.Howard, N. S., J. E. Gomez, C. Ko, and W. R. Bishai. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181-182. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 18.Leelaporn, A., N. Firth, M. E. Byrne, E. Roper, and R. A. Skurray. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob. Agents Chemother. 38:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Z., C. Kelley, F. Collins, D. Rouse, and S. Morris. 1998. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. J Infect. Dis. 177:1030-1035. [DOI] [PubMed] [Google Scholar]
- 20.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manabe, Y. C., B. J. Saviola, L. Sun, J. R. Murphy, and W. R. Bishai. 1999. Attenuation of virulence in Mycobacterium tuberculosis expressing a constitutively active iron repressor. Proc. Natl. Acad. Sci. USA 96:12844-12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendiola, M. V., C. Martin, I. Otal, and B. Gicquel. 1992. Analysis of the regions responsible for IS6110 RFLP in a single Mycobacterium tuberculosis strain. Res. Microbiol. 143:767-772. [DOI] [PubMed] [Google Scholar]
- 23.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP/phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Movahedzadeh, F., J. A. Gonzalez-y-Merchand, and R. A. Cox. 2001. Transcription start-site mapping, p. 105-124. In T. Parish and N. G. Stoker (ed.), Mycobacterium tuberculosis protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 25.Murray, C. J., and A. D. Lopez. 1997. Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet 349:1269-1276. [DOI] [PubMed] [Google Scholar]
- 26.Oyston, P. C., N. Dorrell, K. Williams, S. R. Li, M. Green, R. W. Titball, and B. W. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pablos-Mendez, A., M. C. Raviglione, A. Laszlo, N. Binkin, H. L. Rieder, F. Bustreo, D. L. Cohn, C. S. Lambregts-van Weezenbeek, S. J. Kim, P. Chaulet, P. Nunn, and World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. 1998. Global surveillance for antituberculosis-drug resistance, 1994-1997. N. Engl. J. Med. 338:1641-1649. [DOI] [PubMed] [Google Scholar]
- 28.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169-181. [DOI] [PubMed] [Google Scholar]
- 29.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez, E., S. Samper, Y. Bordas, C. Guilhot, B. Gicquel, and C. Martin. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41:179-187. [DOI] [PubMed] [Google Scholar]
- 31.Podglajen, I., J. Breuil, and E. Collatz. 1994. Insertion of a novel DNA sequence, 1S1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol. Microbiol. 12:105-114. [DOI] [PubMed] [Google Scholar]
- 32.Prod'hom, G., B. Lagier, V. Pelicic, A. J. Hance, B. Gicquel, and C. Guilhot. 1998. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol. Lett. 158:75-81. [DOI] [PubMed] [Google Scholar]
- 33.Ranes, M. G., J. Rauzier, M. Lagranderie, M. Gheorghiu, and B. Gicquel. 1990. Functional analysis of pAL5000, a plasmid from Mycobacterium fortuitum: construction of a “mini” mycobacterium-Escherichia coli shuttle vector. J. Bacteriol. 172:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee, J. T., A. S. Piatek, P. M. Small, L. M. Harris, S. V. Chaparro, F. R. Kramer, and D. Alland. 1999. Molecular epidemiologic evaluation of transmissibility and virulence of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:1764-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivero, A., M. Marquez, J. Santos, A. Pinedo, M. A. Sanchez, A. Esteve, S. Samper, and C. Martin. 2001. High rate of tuberculosis reinfection during a nosocomial outbreak of multidrug-resistant tuberculosis caused by Mycobacterium bovis strain B. Clin. Infect. Dis. 32:159-161. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Samper, S., M. J. Iglesias, and O. Tello. 2000. The Spanish multidrug resistant tuberculosis network. Eurosurveillance 5:43-45. [DOI] [PubMed] [Google Scholar]
- 39.Samper, S., C. Martin, A. Pinedo, A. Rivero, J. Blazquez, F. Baquero, D. van Soolingen, and J. van Embden. 1997. Transmission between HIV-infected patients of multidrug-resistant tuberculosis caused by Mycobacterium bovis. AIDS 11:1237-1242. [DOI] [PubMed] [Google Scholar]
- 40.Sander, P., and E. C. Bottger. 1999. Mycobacteria: genetics of resistance and implications for treatment. Chemotherapy 45:95-108. [DOI] [PubMed] [Google Scholar]
- 41.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 42.Soncini, F. C., E. G. Vescovi, and E. A. Groisman. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valway, S. E., M. P. Sanchez, T. F. Shinnick, I. Orme, T. Agerton, D. Hoy, J. S. Jones, H. Westmoreland, and I. M. Onorato. 1998. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 338:633-639. [DOI] [PubMed] [Google Scholar]
- 44.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 46.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 47.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]