Abstract
Between 1992 and 2000, a total of 4,173 rotavirus-positive samples were collected from two areas of Hungary. Of these, 2,020 specimens (48.4%) were analyzed for G serotype, using monoclonal antibody-based immunoassay and reverse transcription-PCR. By the two methods, 1,789 samples were specified as G1 (62%), G2 (12.2%), G3 (1.4%), G4 (6.4%), G6 (1.0%), G9 (2.9%), or mixed infection (2.6%), and the remaining 231 (11.4%) could not be G typed. The linkage between G and P type, subgroup specificity, and RNA profile was investigated with a sample subset. Among these specimens, we identified both the four globally common strains (P[8],G1 subgroup II (sgII); P[4],G2 sgI; P[8],G3 sgII; and P[8],G4 sgII) and six uncommon strains (P[6],G4 sgII; P[9],G3 sgI; P[9],G6 sgI; P[14],G6 sgI; P[8],G9 sgII; and P[8],G9 sgI). All strains with P[8], P[6], P[9], and P[14] specificities had a long electropherotype, whereas most of those carrying a P[4] specificity were associated with a short electropherotype. Although once considered to be rare, P[9],G6 and P[8],G9 rotavirus strains represent potentially important new serotypes in Hungary.
Rotavirus, a member of the family Reoviridae, has an icosahedral, nonenveloped, triple-layered capsid and a genome of 11 double-stranded RNA segments. Rotaviruses are classified into seven serogroups, A to G. Group A rotaviruses, which are the single most important cause of severe dehydrating diarrhea in infants and young children, are responsible for 20 to 40% of diarrhea-associated hospitalizations worldwide and about 20% of diarrhea-associated deaths, mostly in the developing countries (12, 26). Development of an efficacious rotavirus vaccine is therefore a high priority for developing countries and would have important socioeconomic benefits for industrialized countries, as well (31, 44).
Although the mechanism of immunity to rotavirus is not completely understood, the knowledge that the surface antigens, VP7 and VP4, elicit neutralizing and protective antibodies, respectively (22, 33), had a fundamental impact on vaccine development (5, 6, 32). VP7 is a glycoprotein and determines G serotypes, while VP4 is a protease-sensitive protein and determines P serotypes. On the basis of sequence and antigenic differences, 15 G types and 21 P types have been described (23, 39). During the last two decades, 10 G types and 11 P types in at least three dozen different combinations have been recovered in association with human infections (15, 23, 34), but only a few of these have been demonstrated to be epidemiologically important. Worldwide, P[8],G1, P[4],G2, P[8],G3, P[8],G4, and more recently, P[8],G9 and P[6],G9 strains have been shown to be responsible for most rotavirus infections, although temporal and geographic variations can be significant (15). Thus, for example, the prevalence of P[8],G5 and P[6],G8 strains is high in Brazil and in some parts of Africa, respectively (2, 10, 30), and G9 strains are very common in India (38).
Because rotaviruses have a segmented genome, many surveillance studies have recently included subgrouping and RNA profile analysis in addition to G and P typing assays in order to identify novel strains that can arise from gene reassortment between different strains. Subgrouping is used to study antigenic features of the inner capsid protein VP6, encoded by gene segment 6, while RNA profile analysis (or electropherotyping) is suitable for examining the migration pattern differences of the rotaviral genome, particularly gene segment 11, which can lead to “short” and “long” patterns. This complex approach in epidemiologic surveys is important in order to identify and trace the geographic and temporal spread of novel rotavirus strains arising as human-animal and human-human reassortants (46).
In Hungary, epidemiologic data on the prevalence of G serotypes have been collected since the middle of the 1980s. A large survey demonstrated the predominance of serotypes G1 to G4 during an 8-year period from 1984 to 1991 (42), and we recently detected novel human G6 rotaviruses circulating in two regions (3, 4). To provide molecular epidemiologic data for a future program of vaccination against group A rotaviruses planned in Hungary, we performed G typing on samples collected between 1992 and 2000. In addition, to examine the relationship among antigenic and genomic features and to identify novel reassortant strains, subgrouping and P and G genotyping were done with a subset of samples selected on the basis of RNA profile differences.
The survey was performed by screening rotavirus-positive stool samples collected from mostly hospitalized (97.6%) children with diarrhea who were ≤14 years of age (range, 0.1 to 14.7 years; mean, 2.3 years; with the preponderance of children [94.2%] being <5 years). All samples were obtained between July 1992 and June 2000 from two areas of Hungary: Budapest, including the metropolitan area (BP), and Baranya County (BaC), which is located on the Hungarian-Croatian border. Rotaviruses were identified by latex agglutination (Rotalex; Orion Diagnostica, Espoo, Finland) both in Budapest (Laboratory for Diagnostic Virology of “St. Laszlo” Central Hospital for Infectious Diseases, Budapest; BP samples) and in BaC (Regional Laboratory of Virology, Baranya County Institute of State Public Health Service, Pécs; BaC samples). Rotavirus-positive specimens were analyzed by polyacrylamide gel electrophoresis (PAGE) on 10% slab gels (acrylamide/bis-acrylamide, 30:0.8) and silver staining (13, 29, 40). If samples were available in sufficient volume, they were further characterized by VP7 (G) serotyping monoclonal antibody (MAb)-based enzyme immunosorbent assay (MAb-EIA) using antibodies and procedures described previously (7, 43). In general, MAbs specific to the G1, G2, G3, and G4 serotypes (19, 35, 43) were included in the first round, and serotype G6-, G9-, and, in some instances, serotype G5-, G8-, and G10-specific MAbs (28, 41, 48) were included for those specimens that were reactive with the type-common VP7-binding MAb but not reactive with G1- to G4-specific antibodies. A subset of samples that remained untypeable by this method was subjected to G genotyping, using G1- to G4-, G6-, and G9-specific typing primers (4, 11, 14) in reverse transcription (RT) followed by seminested multiplex PCR (11, 14, 17) or, alternatively, RT followed by single-round multiplex PCR (RT-PCR) (11). Distinct electropherotypes were subsequently correlated with particular G serotypes. Representatives of strains with distinct electropherotypes, particularly those that appeared to be unusual on the basis of their electropherotype and G type profile, were selected for subgrouping and VP4 (P) genotyping to investigate whether they might represent novel reassortants. VP6 (subgrouping) was assayed with MAb-EIA (18), but only occasionally, because of the limited amount of sg I and sg II antibodies. VP4 genotyping was carried out by multiplex RT-PCR using P[4]-, P[6]-, P[8]-, and P[9]-specific primers (14).
During the study period, 4,173 rotavirus-positive stool specimens were obtained, of which 2,020 samples (48.4%) were G serotyped or genotyped at annual rates that varied from 14.8 to 80.2% (BaC specimens) and from 29.9 to 60.7% (BP samples) due to differences in availability of sufficient amounts of sample for further testing (Table 1). Common serotypes (G1 to G4) were found in 87 and 81% of all cases in BaC and BP, respectively, consistent with previously described findings (23, 42). Serotype G1 rotaviruses were the most prevalent strains (average prevalence, 62%; range, 18.6 to 100%), followed by serotype G2 (in BaC, 14.1%; in BP, 11.8%) and serotype G4 (in BaC, 13.1%; in BP, 5.1%) rotaviruses. In certain years, serotypes G2 and G4, respectively, emerged to become predominant (G2 in Budapest in 1997-1998, 46.1%; G4 in Baranya County in 1999-2000, 42.7%), while serotype G3 rotaviruses circulated at low frequency throughout the study period (BaC, 1%; BP, 1.5%). The highest prevalence of G3 strains (5.8%) was observed in Budapest in 1995-1996. Serotype G6 rotaviruses were identified at an overall prevalence of 1% (BaC, 1.3%; BP, 1%). Of interest, G6 strains were the second- and third-most-frequent serotype in certain seasons (BP, 1998-1999; BaC, 1997-1998). Such prevalence of human G6 rotaviruses has been rarely recognized (27), although single, sporadic cases associated with serotype G6 infection were reported from Italy, Australia, and the United States (16, 21, 37). In contrast, the emergence of G9 rotaviruses has recently been demonstrated worldwide (1, 9, 20, 24, 36, 45). In Hungary, the first case associated with this serotype was identified in 1998, and in the 1999-2000 season G9 strains represented the second- and fourth-most-prevalent serotypes in the two areas (BP, 15%; BaC, 6.7%), respectively. The emergence of G9 rotaviruses in 1999-2000 raised questions on whether this new strain had the capability of evading the immunity of older children, as was reported recently by Cubitt et al. from the United Kingdom (8), or, as we found with G6 strains in Hungary (3), there were age differences among cases. Thus, we studied the age distribution of children infected with G9 strains, as well. However, we found that the ages of children infected with G9 strains was similar to the ages we determined for all rotavirus-infected children (serotype G9 cases [mean age, 2.2 years; range, 0.1 to 6.8 years] versus all rotavirus positive cases [mean age, 2.3 years; range, 0.1 to 14.7 years]).
TABLE 1.
Distribution of rotavirus G types in two areas of Hungary
| Area, serotype, or category | No. (%) of strains positive
|
Total no. (%) positive | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1992-1993 | 1993-1994 | 1994-1995 | 1995-1996 | 1996-1997 | 1997-1998 | 1998-1999 | 1999-2000 | ||
| BaCa | |||||||||
| G1 | 6 (66.7) | 18 (100) | 4 (100) | 9 (100) | 52 (57.8) | 46 (66.7) | 32 (84.2) | 17 (22.7) | 184 (59.0) |
| G2 | 27 (30.0) | 3 (4.3) | 1 (2.6) | 13 (17.3) | 44 (14.1) | ||||
| G3 | 1 (1.1) | 1 (1.4) | 1 (1.3) | 3 (1.0) | |||||
| G4 | 3 (33.3) | 4 (4.4) | 2 (5.3) | 32 (42.7) | 41 (13.1) | ||||
| G6 | 3 (4.3) | 1 (1.3) | 4 (1.3) | ||||||
| G9 | 5 (6.7) | 5 (1.6) | |||||||
| Mixed | 9 (13.0) | 3 (4.0) | 12 (3.8) | ||||||
| Nontypeable | 6 (6.7) | 7 (10.1) | 3 (7.9) | 3 (4.0) | 19 (6.1) | ||||
| BPb | |||||||||
| G1 | 95 (67.4) | 114 (65.5) | 194 (85.9) | 119 (57.5) | 79 (28.7) | 130 (81.8) | 141 (78.8) | 197 (56.8) | 1069 (62.6) |
| G2 | 1 (0.7) | 12 (6.9) | 2 (0.9) | 38 (18.3) | 127 (46.2) | 2 (1.3) | 3 (1.7) | 17 (4.9) | 202 (11.8) |
| G3 | 4 (2.8) | 1 (0.4) | 12 (5.8) | 1 (0.4) | 1 (0.6) | 3 (1.7) | 4 (1.1) | 26 (1.5) | |
| G4 | 30 (21.3) | 38 (21.8) | 12 (5.3) | 1 (0.5) | 2 (0.7) | 2 (1.3) | 1 (0.6) | 2 (0.6) | 88 (5.1) |
| G6 | 5 (2.4) | 2 (0.7) | 1 (0.6) | 6 (3.3) | 3 (0.8) | 17 (1.0) | |||
| G9 | 1 (0.6) | 52 (15.0) | 53 (3.1) | ||||||
| Mixed | 4 (2.8) | 5 (2.9) | 1 (0.4) | 2 (1.0) | 6 (2.2) | 5 (3.1) | 7 (3.9) | 11 (3.2) | 41 (2.4) |
| Nontypeable | 7 (5.0) | 5 (2.9) | 16 (7.1) | 30 (14.5) | 58 (21.1) | 17 (10.7) | 18 (10.0) | 61 (17.6) | 212 (12.4) |
For BaC, the total numbers of rotavirus-positive stool samples that were G typed were as follows: for 1992-1993, 9 of 32 (28.1%); for 1993-1994, 18 of 37 (48.7%); for 1994-1995, 4 of 27 (14.8%); for 1995- 1996, 9 of 24 (37.5%); for 1996-1997, 90 of 141 (63.8%); for 1997-1998, 69 of 86 (80.2%); for 1998-1999, 38 of 109 (34.9%); for 1999-2000, 75 of 142 (52.8%); and for all years combined, 312 of 598 (52.2%).
For BP, the total numbers of rotavirus-positive stool samples that were G typed were as follows: for 1992-1993, 141 of 266 (53.0%); for 1993-1994, 174 of 582 (29.9%); for 1994-1995, 226 of 481 (47.0%); for 1995-1996, 207 of 341 (60.7%); for 1996-1997, 275 of 476 (57.8%); for 1997-1998, 159 of 474 (33.5%); for 1998-1999, 179 of 351 (50.9%); for 1999-2000, 347 of 604 (57.5%); and for all years combined, 1,708 of 3,575 (47.7%).
Mixed infections were found in 2.6% of all specimens. A total of 378 samples (18.7%) were nontypeable by MAb-EIA. Because it was not feasible to further characterize all these untypeable strains, 155 specimens were selected for VP7 genotyping. Each specimen shared RNA profile identity with those samples for which a G serotype could previously be assigned. Of these 155 specimens, 147 (94.8%) could be serotyped as G1 (32.2%), G2 (12.9%), G3 (0.6%), G4 (10.3%), G6 (7.1%), G9 (24.5%), and mixed infections (7.1%), reducing the overall rate of G-nontypeable samples to 11.4%. Since the remaining 223 untyped strains had ratios of the individual electropherotypes similar to those of the 147 genotyped strains and to those of the 1,642 MAb-EIA serotyped strains (Fig. 1), these results strongly suggest that their G type prevalence profile would be similar. Thus, had we been able to genotype these remaining untypeable strains by RT-PCR, it is likely that the final percentage of untyped strains would have been as low as 1% based on extrapolation from the percentage of the 155 strains that could be genotyped by RT-PCR (94.8%). The lack of type-specific reactivity with the MAb panel used in this study may be due to antigenic drift of the common VP7 serotypes (25, 45) or, in a few cases, to the circulation of novel G type specificities. Both possibilities would need to be confirmed by molecular analysis. Of the 11 mixed infections we identified by RT-PCR among the MAb-EIA-nontypeable samples, only 1 specimen had extra double-stranded RNA bands identifiable by PAGE. The inability to identify multiple infections by PAGE for the remaining 10 samples is consistent with the high sensitivity of RT-PCR compared to the sensitivity of PAGE.
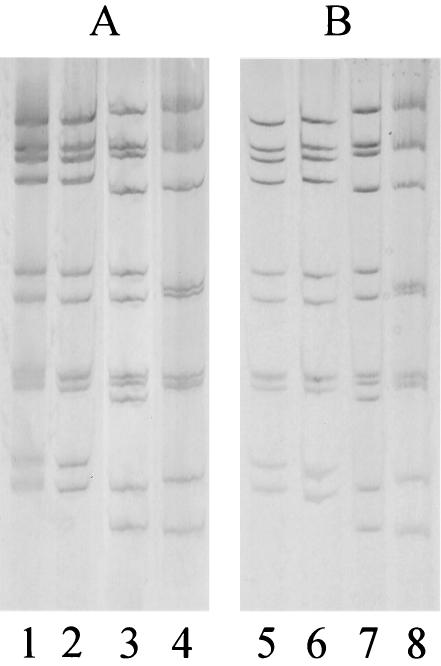
FIG. 1.
A set of RNA profiles of the 147 MAb-EIA-untypeable strains for which VP7 genotyping was successfully done (A) in comparison with RNA profiles of some of those remaining 223 MAb-EIA-untypeable strains for which we did not determine the G type specificity (B). Identities can be seen among strains in lanes 1, 2, 5, and 6 (short profile), in lanes 3 and 7, and in lanes 4 and 8 (long profile). The VP7 genotype profiles of strains in panel A were G2 (lanes 1 and 2), G1 (lane 3), and G9 (lane 4).
We then examined the results of genotyping, subgrouping, and electropherotyping together (Table 2). From each season, 30 to 44 samples, (284 total samples, 39 from BaC and 245 from BP) were subjected to both G and P genotyping, with the sample selection based in part on our intention of characterizing samples representative of each different RNA pattern to gain insight into strain diversity. This approach helped us identify unusual strains, although with this selection bias, it was not suitable for describing the true prevalence of the VP4 genotype. Common genotypes included P[8],G1, P[8],G3, P[8],G4, P[8],G9, and P[4],G2 combinations, of which P[8] strains were usually associated with sg II specificity and the long (L) electropherotype, whereas P[4] strains were usually linked to sg I specificity and the short (S) electropherotype. A variety of strains with unusual linkages were identified, such as P[6],G4 sg II L, P[8],G9 sg I L, P[9],G3 sg I L, P[4],G2 sg I L, P[9],G6 sg I L, and P[14],G6 sg I L. Genotype P [14] was determined by nucleic acid sequencing (4). In this list of unusual genotype combinations, a subgroup was assigned only for those specimens for which there was clear reactivity with one subgroup-specific MAb and low reactivity with the other subgroup-specific MAb (i.e., samples that met the definition of a subgroup-specific reaction). Thus, samples that reacted with both sg I and sg II MAbs or with neither sg I nor sg II MAbs were specified as subgroup nontypeable until further characterization. With the exception of the single P[14],G6 strain and eight P[8],G3 strains detected in the BP area, each surface antigen combination was identified in both regions.
TABLE 2.
Linkage among P and G genotype, electropherotype, and subgroup specificities of 284 rotavirus strains circulated between 1992-1993 and 1999-2000 in Hungarya
| Strain type | RNA profile | No. of samples in category or with subgroup specificityb
|
Season(s) of detectionc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | Non- groupable | Not done | 1992-1993 | 1993-1994 | 1994-1995 | 1995-1996 | 1996-1997 | 1997-1998 | 1998-1999 | 1999-2000 | ||
| Usual strains | |||||||||||||
| P[8],G1 | Long | 100 | 6 | 27 | + | + | + | + | + | + | + | + | |
| P[4],G2 | Short | 25 | 17 | 1 | + | + | + | + | + | + | + | ||
| P[8],G3 | Long | 6 | 2 | + | + | + | + | ||||||
| P[8],G4 | Long | 21 | 3 | 8 | + | + | + | + | + | ||||
| Unusual strains | |||||||||||||
| P[4],G2 | Long | 1 | 1 | + | + | ||||||||
| P[6],G4d | Long | 5 | 1 | 3 | + | + | + | + | |||||
| P[9],G3 | Long | 2 | 3 | + | + | + | + | ||||||
| P[9],G6 | Long | 11 | 4 | + | + | + | |||||||
| P[14],G6 | Long | 1 | + | ||||||||||
| P[8],G9 | Long | 3 | 6 | 3 | 7 | + | + | ||||||
| Mixed infections (n = 17)e | NAf | NA | NA | NA | NA | + | + | + | + | + | + | ||
Please note that due to our sample selection, data in this table do not reflect a true P type prevalence; also, there is a strong bias toward the prevalence of less frequent G types, such as G3, G6, and G9.
Subgroup was assigned only for those specimens where there was clear reactivity with one sg MAb and low reactivity with the other sg MAb. Those specimens that reacted with both sg I and sg II MAbs or with neither sg I nor sg II MAb were specified as nongroupable.
Seasons when strains with the indicated P-G combinations circulated are marked with a plus sign.
All patients infected with P[6],G4 strains developed symptoms and were of an age (mean age, 2.0 years; range, 0.3 to 5.8 years) similar to that described for all rotavirus infected patients in this study (mean age, 2.3 years; range, 0.1 to 14.7 years; see the text). These data contrast with early reports, in which P[6],G4 strains were isolated from neonates without symptoms of gastroenteritis.
P[8]+P[4],G1; P[8]+P[4],G2; P[8]+P[4],G4+G2; P[8],G1+G3; P[8],G1+G4; P[8],G1+G9; P[8],G2+G4; P[9],G2+G6; P[4],G1+G2; P[4],G2+G3; P[4],G2+G4; all combinations were represented by one or two specimens.
NA, not applicable.
Eleven different mixed infections were identified: P[8]+P[4],G1; P[8]+P[4],G2; P[8]+P[4],G4+G2; P[8],G1+G3; P[8],G1+G4; P[8],G1+G9; P[8],G2+G4; P[9],G2+G6; P[4],G1+G2; P[4],G2+G3; and P[4],G2+G4. Mixed infections are significant because they may contribute to the generation of unusual antigen combinations by means of gene reassortment in vivo. Such novel surface antigen combinations may allow rotaviruses to escape preexisting immunity and thus cause infections in immunologically naive populations. It has been postulated that the high rates of mixed infections found in most developing countries may be responsible for the increased strain diversity compared with that in developed countries, where, in general, a lower rate of mixed infections and less strain diversity can be detected (15). Exceptions to this observation have been recently reported; for example, 15 and 12 different P-G combinations were recovered from Great Britain and the United States, respectively, despite the fact that the relative frequency of mixed infections was virtually as low as that observed in the present study (20, 21, 24). This variability was due primarily to the identification of unusual combinations of common VP7 and VP4 specificities rather than to the extraordinary diversity of serotypes.
Before the 1990s, MAb-EIA was the most important tool for collecting data on the distribution of rotavirus G serotypes. Studies from that era demonstrated that four serotypes (G1 to G4) were responsible for most rotavirus-associated hospitalizations worldwide (15, 47), and other serotypes were rarely detected, often in association with animal strains discussed in numerous studies. In this period, data on P type distribution was incomplete due to the lack of reliable type-specific MAbs to P serotypes suitable for MAb-EIA assays. With the development and use of nucleic acid-based genotyping methods (e.g., RT-PCR genotyping and probe hybridization), however, our knowledge about the molecular epidemiology of rotaviruses has been considerably enhanced. G and P types considered previously to be unusual were found to be present and even predominant in several countries. Most important in our study is the fact that we identified six G types and five P types in nine individual combinations. In addition to the globally important strains, P[8],G1, P[4],G2, P[8],G3, and P[8],G4 that were also shown to be common in Hungary, we found P[9],G6 strains to be endemic and P[8],G9 rotaviruses to be emerging in the recent years, suggesting that these strains may have the potential to become epidemiologically important in our country. Despite limitations of the current study, such as the inclusion of only two areas, the high percentage of untyped specimens, and the varying annual rate of specimens that were G typed, we believe that these findings contribute to our understanding of rotavirus epidemiology in Hungary and have implications for a future vaccination program. The first licensed human rotavirus vaccine (RotaShield rhesus rotavirus tetravalent vaccine) and the bovine reassortant vaccines currently in clinical trials were developed to protect against the four globally distributed G type specificities (G1 to G4) and one or more P type specificities (5, 6, 32). It remains unclear whether these vaccines would prevent disease caused by serotypes other than those included in the vaccine strains. Thus, after introduction of these vaccines, continuous strain surveillance will be required to monitor changes in serotype prevalence and to detect the emergence of novel rotavirus strains that could cause nationwide epidemics. If this happened, it would suggest that the vaccine strains did not evoke sufficient heterotypic protection against novel strains not present in the vaccine and would indicate the need to incorporate these specificities in additional reassortants to increase vaccine efficacy.
Acknowledgments
We thank Magdolna Preisz for her assistance in PAGE and John O'Connor for his valuable editorial work. We are very grateful to Dennis Lang and David Matson for providing serotype-specific MAbs.
This study was funded in part by the Hungarian Research Fund (OTKA T032933). The training visit of K.B. to the World Health Organization/Pan-American Health Organization Collaborating Center for Rotavirus and Other Agents of Viral Gastroenteritis at the Centers for Disease Control and Prevention (CDC) was supported by a grant from the CDC's Emerging Infectious Programs to the Collaborating Center.
REFERENCES
- 1.Araújo, I. T., M. S. R. Ferreira, A. M. Fialho, R. M. Assis, C. M. Cruz, M. Rocha, and J. P. G. Leite. 2001. Rotavirus genotypes P[4]G9, P[6]G9, and P[8]G9 in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 39:1999-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armah, G. E., C. T. Pager, R. H. Asmah, F. R. Anto, A. R. Oduro, F. Binka, and D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 3.Bányai, K., J. R. Gentsch, R. I. Glass, and G. Szücs. 2003. Detection of human rotavirus serotype G6 in Hungary. Epidemiol. Infect. 130:107-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bányai, K., J. R. Gentsch, D. D. Griffin, J. L. Holmes, R. I. Glass, and G. Szücs. 2003. Genetic variability among serotype G6 human rotaviruses: identification of a novel lineage isolated in Hungary. J. Med. Virol. 71:124-134. [DOI] [PubMed] [Google Scholar]
- 5.Clark, H. F., P. A. Offit, R. W. Ellis, J. J. Eiden, D. Krah, A. R. Shaw, M. Pichichero, J. J. Treanor, F. E. Borian, L. M. Bell, and S. A. Plotkin. 1996. The development of multivalent bovine rotavirus (strain WC3) reassortant vaccine for infants. J. Infect. Dis. 174(Suppl. 1):S73-S80. [DOI] [PubMed] [Google Scholar]
- 6.Clements-Mann, M. L., R. Dudas, Y. Hoshino, P. Nehring, E. Sperber, M. Wagner, I. Stephens, R. Karron, A. Deforest, and A. Z. Kapikian. 2001. Safety and immunogenicity of live attenuated quadrivalent human-bovine (UK) reassortant rotavirus vaccine administered with childhood vaccines to infants. Vaccine 19:4676-4684. [DOI] [PubMed] [Google Scholar]
- 7.Coulson, B., L. E. Unicomb, G. A. Pitson, and R. F. Bishop. 1987. Simple and specific enzyme immunoassay using monoclonal antibodies for serotyping human rotaviruses. J. Clin. Microbiol. 25:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cubitt, W. D., A. D. Steele, and M. Iturriza. 2000. Characterization of rotaviruses from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6]. J. Med. Virol. 61:150-154. [DOI] [PubMed] [Google Scholar]
- 9.Cunliffe, N. A., D. Winifred, J. E. G. Bunn, M. B. Ramadam, J. W. O. Nyangao, R. L. Riveron, L. E. Cuevas, and C. A. Hart. 2001. Expanding global distribution of rotavirus serotype G9: detection in Lybia, Kenya, and Cuba. Emerg. Infect. Dis. 7:890-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. M. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Zoysa, I., and R. G. Feachem. 1985. Interventions for the control of diarrhoeal diseases among young children: rotavirus and cholera immunization. Bull. W. H. O. 63:569-583. [PMC free article] [PubMed] [Google Scholar]
- 13.Dolan, K. T., E. M. Twist, P. Horton-Slight, C. Forrer, L. M. Bell, S. A. Plotkin, and H. F. Clark. 1985. Epidemiology of rotavirus electropherotypes determined by a simplified diagnostic technique with RNA analysis. J. Clin. Microbiol. 21:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 16.Gerna, G., A. Sarasini, M. Parea, S. Arista, P. Miranda, H. Brüssow, Y. Hoshino, and J. Flores. 1992. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J. Clin. Microbiol. 30:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z.-Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg, H., V. McAuliffe, J. Valdesuso, R. Wyatt, J. Flores, A. Kalica, Y. Hoshino, and N. Singh. 1983. Serological analysis of the subgroup protein of rotavirus using monoclonal antibodies. Infect. Immun. 39:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg, H. B., J. Valdesuso, K. van Wyke, K. Midthun, M. Walsh, V. McAuliffe, R. G. Wyatt, A. R. Kalica, J. Flores, and Y. Hosino. 1983. Production and preliminary characterization of monoclonal antibodies directed at two surface proteins of rhesus rotavirus. J. Virol. 47:267-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin, D. D., C. D. Kirkwood, U. D. Parashar, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and the National Rotavirus Strain Surveillance System Collaborating Laboratories. 2000. Surveillance of rotavirus strains in the United States: identification of unusual strains. J. Clin. Microbiol. 38:2784-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin, D. D., T. Nakagomi, Y. Hoshino, O. Nakagomi, C. D. Kirkwood, U. D. Parashar, R. I. Glass, J. R. Gentsch and the National Rotavirus Strain Surveillance System. 2002. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6], G12) and rare P3[9] strains related to bovine rotaviruses. Virology 294:256-269. [DOI] [PubMed] [Google Scholar]
- 22.Hoshino, Y., M. M. Sereno, K. Midthun, J. Flores, A. Z. Kapikian, and R. M. Chanock. 1985. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc. Natl. Acad. Sci. USA 82:8701-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshino, Y., and A. Z. Kapikian. 2000. Rotavirus serotypes: classification and importance in epidemiology, immunity, and vaccine development. J. Health Popul. Nutr. 18:5-14. [PubMed] [Google Scholar]
- 24.Iturriza-Gómara, M., J. Green, D. W. G. Brown, M. Ramsay, U. Desselberger, and J. J. Gray. 2000. Molecular epidemiology of human group A rotavirus infections in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 38:4394-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iturriza-Gómara, M., D. Cubitt, U. Desselberger, and J. Gray. 2001. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J. Clin. Microbiol. 39:3796-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. N. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Virology, vol. 2. Raven Press, New York, N.Y.
- 27.Kelkar, S. D., and V. L. Ayachit. 2000. Circulation of group A rotavirus subgroups and serotypes in Pune, India, 1990-1997. J. Health Popul. Nutr. 18:163-170. [PubMed] [Google Scholar]
- 28.Kirkwood, C., P. J. Masendycz, and B. S. Coulson. 1993. Characteristics and location of cross-reactive and serotype-specific neutralization sites on VP7 of human G type 9 rotaviruses. Virology 196:79-88. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Leite, J. P. G., A. A. Alfieri, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch. Virol. 141:2365-2374. [DOI] [PubMed] [Google Scholar]
- 31.Liddle, J. L. M., M. A. Burgess, G. L. Gilbert, R. M. Hanson, P. B. McIntyre, R. F. Bishop, and M. J. Ferson. 1997. Rotavirus gastroenteritis: impact on young children, their families and the health care system. Med. J. Aust. 167:304-307. [DOI] [PubMed] [Google Scholar]
- 32.Midthun, K., H. B. Greenberg, Y. Hoshino, A. Z. Kapikian, R. G. Wyatt, and R. M. Chanock. 1985. Reassortant rotaviruses as potential live rotavirus vaccine candidates. J. Virol. 53:949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Offit, P. A., and G. Blavat. 1986. Identification of the two rotavirus genes determining neutralization specificities. J. Virol. 57:376-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okada, J., T. Urasawa, N. Kobayashi, K. Taniguchi, A. Hasegawa, M. Keiji, and S. Urasawa. 2000. New P serotype of group A human rotavirus closely related to that of porcine rotavirus. J. Med. Virol. 60:63-69. [PubMed] [Google Scholar]
- 35.Padilla-Noriega, L., C. Arias, S. Lopez, F. Puerto, G. R. Snodgrass, K. Taniguchi, and H. B. Greenberg. 1990. Diversity of rotavirus serotypes in Mexican infants with gastroenteritis. J. Clin. Microbiol. 28:1114-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palombo, E. A., P. J. Masendycz, H. C. Bugg, N. Bogdanovic-Sakran, G. L. Barnes, and R. F. Bishop. 2000. Emergence of serotype G9 human rotaviruses in Australia. J. Clin. Microbiol. 38:1305-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palombo, E. A., and R. F. Bishop. 1995. Genetic and antigenic characterization of a serotype G6 human rotavirus isolated in Melbourne, Australia. J. Med. Virol. 47:348-354. [DOI] [PubMed] [Google Scholar]
- 38.Ramachandran, M., B. K. Das, A. Vij, R. Kumar, S. S. Bhambal, N. Kesari, H. Rawat, L. Bahl, S. Thakur, P. A. Woods, R. I. Glass, M. K. Bhan, and J. R. Gentsch. 1996. Unusual diversity of human rotavirus G and P genotypes in India. J. Clin. Microbiol. 34:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao, C. D., K. Gowda, and B. S. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276:104-113. [DOI] [PubMed] [Google Scholar]
- 40.Santos, N., and V. Gouvea. 1994. Improved method for purification of viral RNA from fecal specimens for rotavirus detection. J. Virol. Methods 46:11-21. [DOI] [PubMed] [Google Scholar]
- 41.Snodgrass, D. R., T. Fitzgerald, I. Campbell, F. M. M. Scott, G. F. Browning, D. L. Miller, A. J. Herring, and H. B. Greenberg. 1990. Rotavirus serotypes 6 and 10 predominate in cattle. J. Clin. Microbiol. 28:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szücs, G., D. O. Matson, M. Új, E. Kukán, I. Mihály, Z. Jelenik, and M. K. Estes. 1995. Group A rotavirus G type prevalence in two regions of Hungary. Arch. Virol. 140:1693-1703. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi, K., T. Urasawa, Y. Morita, H. B. Greenberg, and S. Urasawa. 1987. Direct serotyping of human rotavirus in stools using serotype 1-, 2-, 3-, and 4-specific monoclonal antibodies to VP7. J. Infect. Dis. 155:1159-1166. [DOI] [PubMed] [Google Scholar]
- 44.Tucker, A. W., A. C. Haddix, J. S. Bresee, R. C. Holman, U. D. Parashar, and R. I. Glass. 1998. Cost-effectiveness analysis of a rotavirus immunization program for the United States. JAMA 279:1371-1376. [DOI] [PubMed] [Google Scholar]
- 45.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. G. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type 9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, M., T. Nakagomi, Y. Koshimura, and O. Nakagomi. 2001. Direct evidence for genome segment reassortment between concurrently-circulating human rotavirus strains. Arch. Virol. 146:557-570. [DOI] [PubMed] [Google Scholar]
- 47.Woods, P. A., J. Gentsch, V. Gouvea, L. Mata, A. Simhon, M. Santosham, Z.-S. Bai, S. Urasawa, and R. I. Glass. 1992. Distribution of serotypes of human rotavirus in different populations. J. Clin. Microbiol. 30:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng, S., G. H. Woode, D. R. Melendy, and R. F. Ramig. 1989. Comparative studies of the antigenic polypeptide species VP4, VP6 and VP7 of three strains of bovine rotavirus. J. Clin. Microbiol. 27:1939-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]