Abstract
We describe here the characterization of five isolates of Mycobacterium simiae-like organisms representing a novel group based on whole-cell fatty acid analysis and genotypic evaluation. Two of the five isolates in this study, W55 and W58, were previously considered to belong to M. simiae serotype 2. Analysis of cellular fatty acids by gas-liquid chromatography indicated a close clustering of this group, which was well differentiated from the other M. simiae-like species. Molecular characterization was performed by nucleic acid sequencing of the small subunit rRNA gene and the gene encoding the 65-kDa heat shock protein and genomic DNA hybridization. Sequence analysis of the entire 16S rRNA gene showed a unique sequence most closely related to those of M. triplex and M. simiae. The hsp65 partial gene sequence was identical for the five isolates, with 97% identity to the M. simiae type strain. However, qualitative whole genomic DNA hybridization analysis confirmed that this group is genetically distinct from M. simiae and M. triplex. Antimicrobial susceptibilities for this group resemble those of M. simiae and M. lentiflavum. We conclude that this group represents a unique Mycobacterium species for which we propose the name Mycobacterium sherrisii sp. nov.
The genus Mycobacterium consists of a diverse group of acid-fast bacilli that exist in the environment and cause infections in humans and animals (22). Conventional methods for identification of these organisms are well established and inexpensive, but these methods are also time-consuming and labor-intensive and can provide inconsistent results, leading to species identifications based on a “best-fit” method. The introduction of several molecular tools and improved techniques in the last two decades has not only revolutionized the identification process by reducing the time to identification but also has provided clues to the presence of novel strains. Mycolate analysis has proven to be a powerful phenotypic method for determining the presence of new species based on novel patterns (9, 25). Fatty acid analysis techniques such as high-performance liquid chromatography (HPLC) and gas-liquid chromatography (GLC) were developed to analyze the mycolic acid and whole-cell fatty acid profiles, respectively, to identify characteristic patterns of established mycobacterial species (7, 10, 12, 30).
The advent of nucleotide probes designed to hybridize to specific rRNA gene sequences allowed rapid identification of the most common mycobacterial species (16). Lately, genotypic analyses aimed at identifying nucleotide sequences of several chromosomal genes in the Mycobacterium genus coding for 16S rRNA, internal transcribed spacer 1, and 65-kDa heat shock protein (hsp65) have proven to be useful in the molecular characterization of well-established species and in identifying new ones, including previously misidentified groups (8, 15, 17, 26). Analysis of the hypervariable region in the 16S rRNA gene sequence has a high species specificity and also has been useful to assign new species status (15). The hsp65 gene is useful for identifying the rapidly growing species and certain slow-growing nontuberculous mycobacteria (8, 24, 29). Proposal of a new species based on the above molecular testing usually included qualitative whole-cell DNA-DNA hybridization studies to provide evidence for the heterogeneity at the genomic level of the new isolates.
In the present study we characterized a novel group of five isolates which in HPLC analysis produced a trimodal mycolic acid profile (TMAP) closely resembling that of M. simiae (25). We undertook a polyphasic approach to differentiate these novel organisms from M. simiae and the other TMAP species Mycobacterium genavense, Mycobacterium lentiflavum, and Mycobacterium triplex. The study also included related species such as Mycobacterium interjectum, Mycobacterium intermedium, and Mycobacterium heidelbergense. The methods for further characterization of these novel isolates included the determination of growth patterns, conventional biochemical properties, whole-cell short-chain fatty acid profiles by GLC, and mycolic acid profiles by HPLC. DNA studies included sequencing of the 16S rRNA and hsp65 genes and qualitative whole-cell DNA-DNA hybridization analysis.
MATERIALS AND METHODS
Strains.
Clinical isolates were provided by the following investigators: E. C. Bottger (strain 4773), G. D. Cage (strain 283416), and N. G. Warren (strain Va2). A. Y. Tsang and P. J. Brennan provided strains W55 and W58. The type strain M. simiae (ATCC 25275T) was obtained from the American Type Culture Collection (ATCC; Manassas, Va.). The type strains provided by E. C. Bottger included M. lentiflavum ATCC 51985T, M. intermedium ATCC 51848T, M. interjectum ATCC 51457T, and M. heidelbergense ATCC 51253T. The M. triplex strain 4382 was also from E. C. Bottger. The M. genavense type strain ATCC 51234T originated as a clinical isolate in the Harborview Mycobacteriology Laboratory of the University of Washington. The investigation of biochemical and growth characteristics was carried out by conventional techniques (22). The biochemical tests included nitrate reduction, catalase at 68°C and semiquantitative catalase, 7-day pyrazinamidase, Tween 80 hydrolysis, 14-day aryl sulfatase, and urease tests (14). The growth study included analysis of the growth rate; pigmentation type; growth at 25, 30, 37, and 42°C; and colonial morphology.
Chromatographic analysis. (i) HPLC.
Mycolic acids were prepared as described previously (6, 32). They were then dissolved in 200 μl of chloroform containing 200 μg of 4 bromomethyl-6,7-dimethoxycoumarin and 200 μg of 18-crown-6 ether and transferred to a 2-ml autosampler vial into which 100 μl of a 2% potassium bicarbonate solution had been evaporated previously. The vial was heated at 60°C for 15 min, and the mixture was allowed to evaporate. The mycolic esters were dissolved in 500 μl of chloroform containing internal standards and then analyzed by fluorescence detection HPLC as described previously (6, 32).
(ii) GLC.
Whole-cell fatty acid analysis was performed by GLC by using profiles in the Sherlock Microbial Identification System (MIDI, Inc., Newark, Del.). Mycobacterial cells were grown and harvested according to the manufacturer's protocols. The extracted fatty acids were profiled by using a Hewlett-Packard 5890 series II gas chromatograph with electronic pulse control, a sample controller, and a Vectra computer (Hewlett-Packard, Palo Alto, Calif.) with MIDI's mycobacterial library (v3.8) and Library Generation Software (vl.06).
The data were analyzed with the Library Generation Software of MIDI, which is a program that provides a two-dimensional cluster plot based on cluster analysis. The two-dimensional plots are based on principle component analysis. Principle component 1 is the component responsible for the greatest degree of variability among the samples tested and is represented on the horizontal axis. Principle component 2 is responsible for the second greatest degree of variability and is displayed on the vertical axis. The scale for both axes is the Euclidian distance.
Antimicrobial susceptibility testing.
Susceptibility testing was performed by a previously described broth microdilution MIC testing method for slow-growing mycobacteria (33). The method and the MIC breakpoints (for ciprofloxacin, clarithromycin, ethambutol, isoniazid, rifampin, rifabutin, and sulfamethoxazole) for mycobacteria have been approved by the NCCLS in the M24-A monograph, which replaced the M24-T2 version (23). MIC breakpoints for the remaining drugs were chosen based on the knowledge of success of therapy of M. tuberculosis and/or achievable levels in serum. The drugs tested were isoniazid, ethambutol, rifampin, ciprofloxacin, clarithromycin, gatifloxacin, minocycline, moxifloxacin, rifabutin, and sulfamethoxazole.
Nucleotide sequencing and analysis:.
DNA for 16S rRNA and hsp65 gene sequencing were prepared after mechanical disruption of bacterial cells (15). Briefly, one loopful of bacteria was suspended in 300 μl of Tris-EDTA (10 mM-Tris, 1 mM EDTA [pH 7.4]), and 100 μl of acid-washed glass beads was added to the tubes, followed by heating at 94°C for 15 min and shaking in a mechanical disrupter. A 5-μl volume of the supernatant was used for PCR amplification.
The reaction solution was composed of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 200 μM (each) deoxynucleoside triphosphates, 1 μM (each) primer, and 2.5 U of Taq DNA polymerase (Perkin-Elmer Cetus). The complete 16S rRNA gene was amplified by using primer pair 8FPL and DG74 (corresponding to positions 8 to 27 and positions 1522 to 1540, respectively, according to Escherichia coli 16S rRNA gene numbering) as described previously (11). Several primer pairs designed to amplify internal regions of the 16S rRNA gene were used. Each thermal cycle consisted of a 10-min initial denaturation at 95°C, followed by 30 cycles of amplification (30 s at 95°C, 30 s at 68°C, and 45 s at 72°C) and a final 10-min extension at 72°C. The PCR products were column purified (Microcon 100; Amicon, Beverly, Mass.), and cycle sequencing was subsequently performed with the Big Dye terminator kit (Perkin-Elmer Applied Biosystems) as recommended by the manufacturer. Sequencing products were column purified (CentriSep, Adelphia, N.J.) and analyzed on an ABI Avant 3100 capillary electrophoresis unit (Applied Biosystems). Sequences assembled and edited with the Sequencher software version 3.1 (Gene Codes Corp., Ann Arbor, Mich.) were used to generate the complete16S rRNA gene sequence. The consensus sequences were searched against the GenBank database by using the BLAST tool (2). Sequence alignments in the region corresponding to gene positions 129 to 236 (hypervariable region A) of the E. coli 16S rRNA gene were used for identification and to determine the phylogenetic relatedness of the strains.
The Hsp65 protein gene was amplified by using primer TB11 (ACCAACGATGGTGTGTCCAT) and primer TB12 (CTTGTCGAACCGCATACCCT) as described by Telenti et al. (29). The PCR was 50 thermal cycles (1 min at 94°C, 1 min at 60°C, and 1 min at 72°C), followed by a final 10-min extension at 72°C. The PCR product was column purified and sequenced as described above. Sequences were assembled and edited with the Sequencher software. The sequences were aligned in MultAlin (http://prodes.toulouse.inra.fr/multalin/multalin.html) and manually trimmed in Microsoft Word. The lengths of the trimmed sequence for hsp65 and 16S rRNA gene sequences used for constructing the phylogenetic tree were 254 and 1,405 bp, respectively.
A phylogenetic tree was constructed by using PHYLIP (version 3.573; J. Q. Felsenstein, Department of Genetics, University of Washington, Seattle [http://evolution.genetics.washington.edu/phylip.html]). The branching order of the neighbor-joining dendrograms was evaluated with 1,000 bootstrap analyses by using the SEQBOOT program in the PHYLIP software package. Nucleotide sequences from M. tuberculosis were used as an outgroup in the construction of the dendrograms for 16S rRNA and hsp65 gene sequences.
Qualitative whole-cell DNA-DNA hybridization experiment:.
Chromosomal DNA purification and the hybridization studies were done exactly as previously described (28), except that lysates were digested with proteinase K instead of pronase. As described earlier, the DNA-DNA hybridization reactions were carried out under conditions that permitted a 29 to 31% mismatch (28).
Nucleotide sequence accession numbers.
The DNA sequence for the M. sherrisii type strain 4773 has been deposited in the GenBank; the accession number for the 16S rRNA gene is AY353699, and the accession number for the hsp65 gene is AY365190.
RESULTS
Phenotypic analysis. (i) Growth and biochemical characteristics.
The novel group of isolates grew as white to buff colonies within 28 days at 37°C. Cultures regularly produced both raised and flat colony types. As seen in Table 1 two of the isolates failed to grow at 42°C, while the other three demonstrated minimal growth at this temperature. The strains were uniformly urease positive. Two strains were pyrazinamidase positive (strains 283416 and 4773). The Tween 80 hydrolysis, nitrate reductase, and 3-day arylsulfatase tests were negative for all of the isolates. The results of the 14-day arylsulfatase test varied from 1+ to 3+. Although the heat-stable catalase was positive for all of the isolates, variable results were observed for the semiquantitative catalase test. Our novel group can be differentiated from M. simiae-related organisms such as M. heidelbergense, M. intermedium, and M. interjectum by the inability to hydrolyze Tween 80. The following biochemical properties separate these novel isolates from the individual TMAP species. Unlike M. triplex, these strains lack nitrate reductase and are differentiated from M. lentiflavum by their production of urease and arylsulfatase. Their eugonic growth distinguishes them from M. genavense. Unlike M. simiae, only one of the five novel isolates, Va2, is photochromogenic.
TABLE 1.
Comparison of phenotypic characteristics of the novel isolates and related Mycobacterium species
| Species or isolate | Growth ata:
|
Test result
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25°C | 30°C | 37°C | 42°C | Pigmentationb | Niacin | Urease | Pyrazinamidase | Tween hydrolysis (10 day) | Nitrate reductase | Arylsulfatase (14 day)c | Catalase at 68°C | Semiquantitative catalase | |
| Novel group | |||||||||||||
| 4773 | ± | + | + | − | N | − | + | + | − | − | 3+ | + | − |
| 283416 | + | + | + | − | N | − | + | + | − | − | 2+ | + | − |
| W55 | + | + | + | ± | N | + | + | − | − | − | 1+ | + | + |
| W58 | + | + | + | ± | N | + | + | − | − | − | 1+ | + | + |
| Va2 | + | + | + | ± | P | + | + | − | − | − | 3+ | + | − |
| M. simiae | + | + | + | + | P | ± | + | + | − | − | + | + | + |
| M. genavensed | ± | + | + | + | N | − | + | + | − | − | − | + | + |
| M. lentiflavum | ± | + | + | − | S | − | − | ± | − | − | − | ± | ± |
| M. triplex | − | + | + | + | N | − | + | + | − | + | 4+ | + | − |
| M. heidelbergense | − | + | + | − | N | − | + | + | + | − | 1+ | + | − |
| M. intermediume | + | + | + | + | P | − | + | − | + | − | + | + | + |
| M. interjectum | − | + | + | − | N | − | + | + | + | − | − | + | + |
+, positive; −, negative; ±, variable.
P, photochromogenic; S, scotochromogenic; N, nonchromogenic.
The 3-day arylsulfatase test was negative for all species tested in our laboratory
Growth on solid media requires mycobactin J supplement and prolonged incubation for up to 60 days.
Data are from reference 20.
(ii) HPLC.
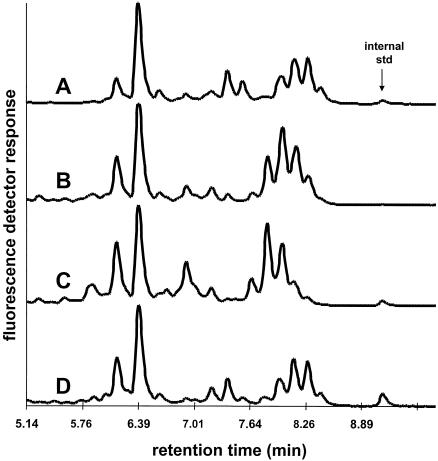
HPLC chromatograms from isolate 4773 of the novel group and from three of the four TMAP species are presented in Fig. 1. The chromatograms do not show the low-molecular-weight internal standard since it eluted at approximately 2.5 min. The study isolates did not produce any significant peaks between 2.5 and 6 min. The first cluster of peaks includes two major peaks, with the second peak relatively greater than the first. The second cluster of peaks is not prominent in M. simiae, M. triplex, M. lentiflavum, or in our novel group, and it elutes closer to the first in M. triplex. The third cluster shows variation within species and is not uniformly distinct. The novel group produced an HPLC mycolic acid pattern most closely resembling that of M. simiae. The presence of a prominent peak at approximately 8.29 min serves to differentiate our novel group and M. simiae from the other species studied by HPLC.
FIG. 1.
Comparison of representative HPLC phenotypes of M. simiae (A), M. lentiflavum (B), M. triplex (C), and isolate 4773 (D).
(iii) GLC.
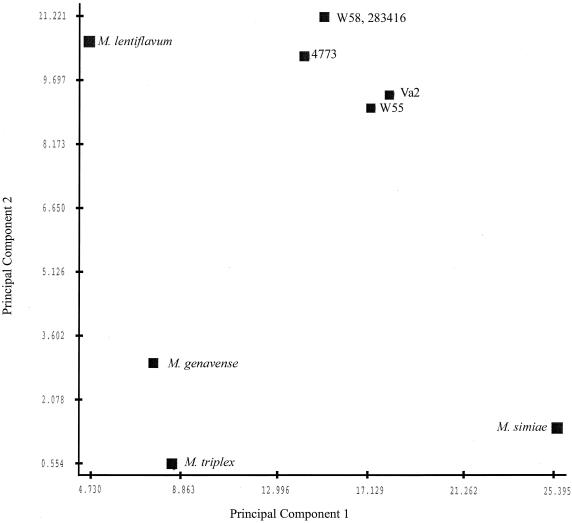
The fatty acid patterns obtained by GLC analysis of the novel isolates appeared indistinguishable and similar to that of the TMAP species. However, our isolates could be distinguished from these closely related species by a few qualitative and quantitative differences (Table 2). Qualitative differences from M. genavense, M. triplex, and M. lentiflavum included the presence of cis-11-hexadecenoic acid (16:1ω5c) and absence of cis-10-hexadecenoic acid (16:1ω6c). Quantitatively, our group differed from M. simiae by containing less of 16:0 and more of tuberculosteric acid (10Me-18:0). Interestingly, although fatty acid methyl ester (FAME) analysis was able to satisfactorily differentiate the isolates from all strains except M. simiae, the cluster analysis method closely grouped all five strains and distinguished them from the other taxa (Fig. 2).
TABLE 2.
Microbial identification system whole-cell fatty acid analysis of the novel group of isolates and related species
| Species (no. of isolates) | Fatty acid content (%)a
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12:0 | 14:0 | 2-Me-14:0 | 15:0 | 16:1ω9c | 16:1ω7c | 16:1ω6c | 16:1ω5c | 16:0 | 17:1ω8c | 17:0 | 18:1ω9c | 18:0 | 10Me-18:0 (TBSA) | 20:0 | |
| Novel isolates (5) | 7.4 | 0.7 | 2.5 | 1.5 | 2.9 | 37.7 | 0.5 | 0.8 | 30.7 | 3.2 | 10.8 | 1.0 | |||
| M. simiae (17) | 8.2 | 0.7 | 2.1 | 1.3 | 2.8 | 41.4 | 0.9 | 27.7 | 5.4 | 7.2 | 1.0 | ||||
| M. genavense (25) | 8.6 | 0.8 | 0.7 | 10.2 | 35.9 | 0.6 | 31.1 | 2.3 | 8.6 | ||||||
| M. lentiflavum (7) | 6.9 | 0.7 | 7 | 3.8 | 31.8 | 0.9 | 34.5 | 4.3 | 8.4 | 1.1 | |||||
| M. triplex (4) | 8.1 | 0.6 | 2.1 | 2.3 | 10.5 | 32.2 | 0.7 | 28 | 3.1 | 10.8 | 0.7 | ||||
| M. heidelbergense (1) | 1.0 | 5.5 | 1.3 | 0.6 | 1.7 | 3.4 | 4.6 | 0.5 | 43.0 | 0.6 | 22.1 | 5.1 | 9.1 | 0.7 | |
| M. intermedium (3) | 1.2 | 6.6 | 1.5 | 0.4 | 0.8 | 5.4 | 4.4 | 0.6 | 44.5 | 0.5 | 21.6 | 4.1 | 7.1 | 0.5 | |
| M. interjectum (1) | 5.6 | 1.2 | 4.8 | 4.0 | 23.7 | 1.2 | 23.5 | 7.9 | 15.6 | 0.4 | |||||
Values are average peak areas represented in percentages. For each fatty acid designation, the number to the left of the colon is the number of carbon atoms and the number to the right of the colon is the number of double bonds. TBSA, tuberculosteric acid (10-methyloctadecanoic acid).
FIG. 2.
Microbial identification system whole-cell fatty acid cluster analysis of the novel isolates and related species. Prinicipal-component plots based on fatty acid profiles of the individual mycobacterial strains. The axes are in Euclidian distance.
(iv) Antimicrobial susceptibility.
Among the 10 antimicrobial agents tested, only clarithromycin, rifabutin, and sulfamethoxazole appeared to have activity at clinically achievable levels against all five isolates (Table 3). The antimicrobial susceptibilities for this novel group resemble those of M. simiae and M. lentiflavum (R. J. Wallace, Jr., unpublished data).
TABLE 3.
Antimicrobial susceptibility pattern of novel group of isolates
| Antimicrobial agent | MIC (μg/ml)a for strain:
|
||||
|---|---|---|---|---|---|
| 4773 | W55 | W58 | 283416 | Va2 | |
| Ciprofloxacin | >16 (r) | 4 (r) | >16 (r) | 8 (r) | 16 (r) |
| Clarithromycin | 0.25 (s) | 0.25 (s) | 4 (s) | 2 (s) | 4 (s) |
| Ethambutol | 8 (r) | 4 (s) | 16 (r) | >16 (r) | >16 (r) |
| Gatifloxacin | 8 (r) | 4 (i) | 8 (r) | 1 (s) | 4 (i) |
| Isoniazid | 1 (s) | 2 (s) | 4 (s) | >16 (r) | 8 (r) |
| Minocycline | >16 (r) | 16 (r) | >16 (r) | >16 (r) | >16 (r) |
| Moxifloxacin | 1 (s) | 2 (i) | 2 (i) | 1 (s) | 1 (s) |
| Rifabutin | ≤0.12 (s) | ≤0.12 (s) | 0.5 (s) | 0.25 (s) | 0.25 (s) |
| Rifampin | 4 (r) | 4 (r) | 32 (r) | 8 (r) | 32 (r) |
| Sulfamethoxazole | 32 (s) | 16 (s) | 16 (s) | 32 (s) | 32 (s) |
r, resistant; s, susceptible, i, intermediate.
Genotypic analysis.
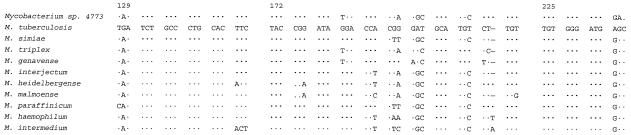
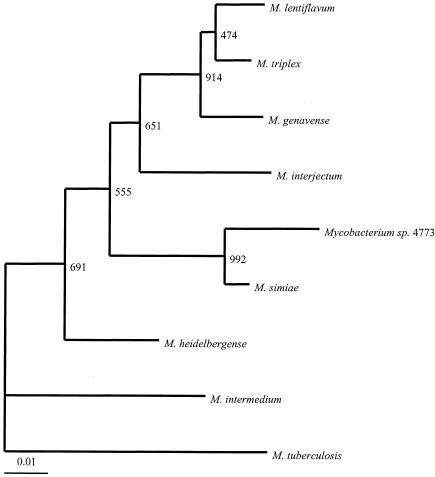
Nucleotide sequencing of the entire16S rRNA gene (1,510 bp) of the proposed type strain 4773 was performed, and the sequence was compared to other published mycobacterial sequences in the GenBank database and interpreted by using the BlastN algorithm. The closest matches were obtained with M. simiae (99%) and M. triplex (99%). Since hypervariable region A has been demonstrated to be a useful locus for identifying new mycobacterial species, this region from our isolate was compared to that of the other mycobacterial species (Fig. 3). There were a total of eight nucleotide differences in the hypervariable region A of isolate 4773 when it was compared to sequences of M. simiae and related species. Hence, the hypervariable region in our isolate was unique and allowed differentiation from other species (15).
FIG. 3.
Sequence alignment of the hypervariable region A within the 16S rRNA gene of the novel isolates and related species. M. tuberculosis was used as the reference sequence. Nucleotides different from those of M. tuberculosis are indicated; dashes indicate deletions, and dots indicate identity. The first nucleotide corresponds to E. coli 16S rRNA gene position 129.
Sequence diversity in the hsp65 gene has been demonstrated to allow identification to the species level of slow-growing and fast-growing mycobacteria (8, 29). Sequences of the hsp65 gene (441 bp) from all five isolates were identical and unique compared to other published sequences. Homology with the sequence from the M. simiae type strain was 97%. The closest match was 99% homology with the hsp65 gene sequence recently described from a novel M. simiae allelic variant (18).
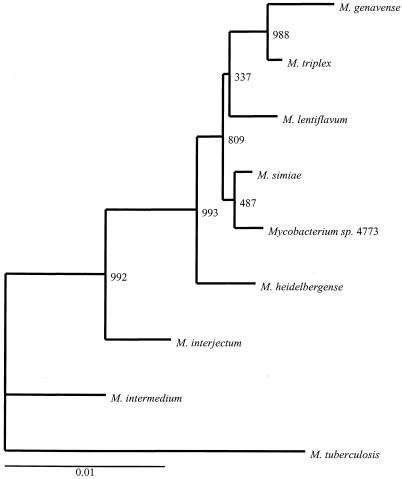
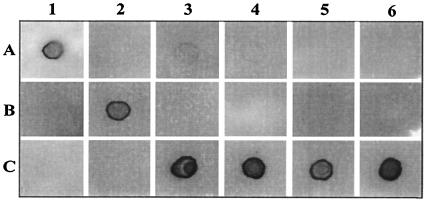
A phylogenetic tree based on the 16S rRNA and hsp65 gene sequences placed these isolates in a distinct clade closely related to M. simiae (Fig. 4 and 5). Isolates were further analyzed by qualitative DNA-DNA hybridization studies with M. simiae and M. triplex. Whole chromosomal DNA from strain 4773 was used as a probe to hybridize with DNA from M. simiae, from M. triplex, and from the four available isolates of the novel group. The probe hybridized with DNA from the four isolates in the new group and failed to hybridize to DNA from M. simiae or M. triplex. In addition, probes prepared from M. simiae and M. triplex hybridized with their autologous DNAs but exhibited little to no hybridization with DNA from isolates of the novel group (Fig. 6). Thus, the result from the solid-phase hybridization procedure provided supportive evidence that the new group was distinct from both M. simiae and M. triplex.
FIG. 4.
16S rRNA gene sequence-based phylogenetic tree. A consensus neighbor-joining dendrogram with 1,000 bootstrap replicates was based on alignment of the 1,405 aligned bases of the 16S rRNA gene. The numbers at the nodes indicate bootstrap values. The lower bar indicates the relative genetic distance.
FIG. 5.
hsp65 sequence-based phylogenetic tree. A consensus neighbor-joining dendrogram with 1,000 bootstrap replicates was based on alignment of the 254-bp sequence of the hsp65 gene. The numbers at the nodes indicate bootstrap values. The lower bar indicates the relative genetic distance.
FIG. 6.
DNA-DNA homology study in a solid-phase hybridization analysis. Whole-cell genomic DNA was used to prepare the probe and target DNA. Probes: A, M. simiae; B, M. triplex; C, isolate 4773. Target DNA: 1, M. simiae; 2, M. triplex; 3, isolate 283416; 4, isolate 4773; 5, isolate W55; 6, isolate W58.
DISCUSSION
We describe here a novel group of isolates that by HPLC analysis resembled M. simiae and M. lentiflavum but not the other members of the TMAP group, including M. triplex, M. genavense, or closely related organisms such as M. intermedium, M. interjectum, and M. heidelbergense. Analysis by GLC identified few qualitative differences between our isolates and M. simiae or M. triplex. Interestingly, the cluster analysis by GLC tightly grouped the five isolates and differentiated them from all other TMAP species. Biochemical and growth studies did not provide any unique property useful in differentiation of this group from M. simiae. Genetic evidence by sequencing of the 16S rRNA and the hsp65 gene identified these isolates as a novel group. Phylogenetic analysis based on 16S rRNA and hsp65 gene sequences placed these novel isolates in a close cluster well differentiated from other species. DNA-DNA hybridization studies provided further evidence for genetic diversity between this group and the other closely related species. Subcultures of the flat and raised colonial variants of the novel isolates revealed no differences in biochemical reactions, chromatographic profiles, or DNA sequences.
Interestingly, the sequence of the hypervariable region A from our type strain was identical to the “genotype 3” profile of a recently published study on a few mycobacterial isolates phylogenetically related to M. simiae (31). The authors of that study concluded that the genotype 3 isolates (n = 2) had an HPLC phenotype similar to that of M. lentiflavum, but conventional tests identified them as M. simiae.
M. simiae was originally isolated from rhesus monkeys (34). It is an opportunistic mycobacterial species occasionally associated with lung infections and also implicated in disseminated infections in patients with AIDS (1, 19, 27). HPLC analysis of this species produces characteristic triple clusters of peaks. Molecular characterization by genetic and fatty acid analysis in the last decade has led to descriptions of several new slow-growing mycobacterial species similar to M. simiae. M. lentiflavum was described as a new species that is similar in biochemical properties to M. avium but closely resembles M. simiae (99.7% identity) by 16S rRNA gene analysis (28). Although mycolate patterns for M. heidelbergense and M. simiae were distinct as determined by HPLC, the 16S rRNA gene analysis showed a 99.6% homology with M. simiae (13). Based on its 16S rRNA gene sequences, M. intermedium was similar to M. simiae and phylogenetically placed between fast- and slow-growing mycobacteria (20). Although the HPLC mycolic acid pattern of M. triplex closely resembles that of M. simiae, it was classified as a new species based on a distinctive 16S rRNA gene hypervariable region (9). The sequence of the gene for 16S rRNA showed that M. genavense also is closely related to M. simiae (5).
When Meissner and Schröder did seroagglutination studies of the newly described species M. simiae, these researchers found two serotypes and assigned isolates W55 and W58 to serotype 2 (21). A positive niacin reaction is rare outside of the species M. tuberculosis and M. simiae, which probably explains why strain W55 and W58 originally were identified as M. simiae. Before the advent of molecular methods, slow-growing mycobacteria that were photochromogenic and niacin positive were considered members of the species M. simiae (34). However, a laboratory that routinely uses HPLC and sequencing for identification of mycobacteria found many isolates of M. simiae that were neither photochromogenic nor niacin positive (27). Conversely, molecular and chromatographic analyses in the present study revealed that two strains, W55 and W58, with biochemical reactions similar to those of M. simiae are in fact members of a novel species.
A number of publications have noted heterogeneity within M. simiae. Wolinsky commented that it was not clear whether serotype 2 belonged in the same species as serotype 1 (35). He further noted that serotype 2 is a heterogeneous group. Boisvert and Truffot found diversity in phenotypic characteristics of serotype 2, including the production of pigment and PZA (4). In an elegant study Baess and Magnusson analyzed strains of M. simiae serotypes 1 and 2 by seroagglutination and quantitative DNA-DNA hybridization (3). Both methods showed that serotype 1 strains were closely related to each other and only moderately related to strains of serotype 2. Interestingly, these researchers found that the two tested serotype 2 strains were not closely related to each other. Neither strain W55 nor W58 was included in the study.
Since the niacin-positive test was a major reason that isolates W55, W58, and Va2 were included in our laboratory's large study of M. simiae-like organisms, it is difficult to predict its true incidence in this novel taxon. In contrast, isolates 4773 and 283416 were analyzed simply because initial studies in their original laboratories indicated that they were novel, which suggests that they represent a more common biotype of this taxon.
By using a polyphasic approach, we provide evidence that our group of isolates, including the W55 and W58 strains previously considered to be M. simiae serotype 2, are phylogenetically related but distinct from M. simiae. Hence, we propose the name M. sherrisii sp. nov.
Description of Mycobacterium sherrisii sp. nov.
M. sherrisii (named in honor of John C. Sherris for many significant contributions to the field of clinical microbiology) is an acid-fast organism. Visible growth of a dilute inoculum requires >7 days, and the colonies on 7H10 media appear smooth to dry and 1 to 2 mm in diameter. Colonies are white to buff and produce both raised and flat colony types. Photochromogenic strains can occur. Best growth occurs at temperatures between 30 and 37°C. The organism is resistant to most of the tested antibiotics except clarithromycin, rifabutin, moxifloxacin, and sulfamethoxazole. Biochemical properties uniformly positive for this group are urease, catalase at 68°C, and 14-day arylsulfatase. Some isolates of this group are positive for pyrazinamidase and semiquantitative catalase. All are negative for Tween hydrolysis nitrate reduction and 3-day arylsulfatase. Phylogenetic analyses indicate that these isolates are closely related to M. simiae and M. triplex. The type strain of M. sherrisii is strain 4773. Cultures of this strain and strains Va2 and W55 have been deposited in the American Type Culture Collection as strains BAA-832, BAA-833, and BAA-834, respectively. The DNA sequence information from the type strain has been deposited in GenBank, and the accession number for the 16S rRNA gene is AY353699 and hsp65 gene is AY365190.
Acknowledgments
We thank Uyen Bui for computer assistance in building the 16S phylogenetic tree from the 16S sequences and Grace Koon for computer assistance with Fig. 6. We also thank Eric C. Bottger for providing the strains and for support over the years in our research on mycobacterial taxonomy.
REFERENCES
- 1.Al Abdely, H. M., S. G. Revankar, and J. R. Graybill. 2000. Disseminated Mycobacterium simiae infection in patients with AIDS. J. Infect. 41:143-147. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baess, I., and M. Magnusson. 1982. Classification of Mycobacterium simiae by means of comparative reciprocal intradermal sensitin testing on guinea-pigs and deoxyribonucleic acid hybridization. Acta Pathol. Microbiol. Immunol. Scand.[B] 90:101-107. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, H., and C. Truffot. 1979. [Relationships between “Mycobacterium simiae” and the “M. avium-intracellulare-scrofulaceum” complex (author's transl.)]. Ann. Microbiol. 130B:457-466. (In French.) [PubMed] [Google Scholar]
- 5.Bottger, E. C., B. Hirschel, and M. B. Coyle. 1993. Mycobacterium genavense sp. nov. Int. J. Syst. Bacteriol. 43:841-843. [DOI] [PubMed] [Google Scholar]
- 6.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, Jr., S. H. Chiu, G. O. Onyi, E. C. Bottger, and R. J. Wallace, Jr. 1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49(Pt. 4):1493-1511. [DOI] [PubMed] [Google Scholar]
- 7.Butler, W. R., K. C. Jost, Jr., and J. O. Kilburn. 1991. Identification of mycobacteria by high-performance liquid chromatography. J. Clin. Microbiol. 29:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devallois, A., K. S. Goh, and N. Rastogi. 1997. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. Clin. Microbiol. 35:2969-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floyd, M. M., L. S. Guthertz, V. A. Silcox, P. S. Duffey, Y. Jang, E. P. Desmond, J. T. Crawford, and W. R. Butler. 1996. Characterization of an SAV organism and proposal of Mycobacterium triplex sp. nov. J. Clin. Microbiol. 34:2963-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glickman, S. E., J. O. Kilburn, W. R. Butler, and L. S. Ramos. 1994. Rapid identification of mycolic acid patterns of mycobacteria by high-performance liquid chromatography using pattern recognition software and a Mycobacterium library. J. Clin. Microbiol. 32:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrant, G. O., M. A. Lambert, and C. W. Moss. 1981. Gas-chromatographic analysis of mycolic acid cleavage products in mycobacteria. J. Clin. Microbiol. 13:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas, W. H., W. R. Butler, P. Kirschner, B. B. Plikaytis, M. B. Coyle, B. Amthor, A. G. Steigerwalt, D. J. Brenner, M. Salfinger, J. T. Crawford, E. C. Bottger, and H. J. Bremer. 1997. A new agent of mycobacterial lymphadenitis in children: Mycobacterium heidelbergense sp. nov. J. Clin. Microbiol. 35:3203-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, Atlanta, Ga.
- 15.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebrun, L., F. Espinasse, J. D. Poveda, and V. Vincent-Levy-Frebault. 1992. Evaluation of nonradioactive DNA probes for identification of mycobacteria. J. Clin. Microbiol. 30:2476-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leclerc, M. C., N. Haddad, R. Moreau, and M. F. Thorel. 2000. Molecular characterization of environmental Mycobacterium strains by PCR-restriction fragment length polymorphism of hsp65 and by sequencing of hsp65, and of 16S and ITS1 rDNA. Res. Microbiol. 151:629-638. [DOI] [PubMed] [Google Scholar]
- 18.Legrand, E., K. S. Goh, C. Sola, and N. Rastogi. 2000. Description of a novel Mycobacterium simiae allelic variant isolated from Caribbean AIDS patients by PCR-restriction enzyme analysis and sequencing of hsp65 gene. Mol. Cell. Probes 14:355-363. [DOI] [PubMed] [Google Scholar]
- 19.Matthews, J. H., and N. G. Warren. 1983. Mycobacterium simiae. Am. Rev. Respir. Dis. 127:788-789. [DOI] [PubMed] [Google Scholar]
- 20.Meier, A., P. Kirschner, K. H. Schroder, J. Wolters, R. M. Kroppenstedt, and E. C. Bottger. 1993. Mycobacterium intermedium sp. nov. Int. J. Syst. Bacteriol. 43:204-209. [DOI] [PubMed] [Google Scholar]
- 21.Meissner, G., and K. H. Schroder. 1975. Relationship between Mycobacterium simiae and Mycobacterium habana. Am. Rev. Respir. Dis. 111:196-200. [DOI] [PubMed] [Google Scholar]
- 22.Metchcock, B. G., F. S. Nolte, and R. J. Wallace III. 1999. Mycobacterium, p. 399-437. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 23.NCCLS. 2003. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, Pa. [PubMed]
- 24.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritter, D., L. D. Carlson, B. K. Logan, L. S. Ramos, J. O. Kilburn, and M. B. Coyle. 1996. Differentiation of Mycobacterium genavense and Mycobacterium simiae by automated mycolic acid analysis with high-performance liquid chromatography. J. Clin. Microbiol. 34:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rynkiewicz, D. L., G. D. Cage, W. R. Butler, and N. M. Ampel. 1998. Clinical and microbiological assessment of Mycobacterium simiae isolates from a single laboratory in southern Arizona. Clin. Infect. Dis. 26:625-630. [DOI] [PubMed] [Google Scholar]
- 28.Springer, B., W. K. Wu, T. Bodmer, G. Haase, G. E. Pfyffer, R. M. Kroppenstedt, K. H. Schroder, S. Emler, J. O. Kilburn, P. Kirschner, A. Telenti, M. B. Coyle, and E. C. Bottger. 1996. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J. Clin. Microbiol. 34:1100-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thibert, L., and S. Lapierre. 1993. Routine application of high-performance liquid chromatography for identification of mycobacteria. J. Clin. Microbiol. 31:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tortoli, E., C. Piersimoni, P. Kirschner, A. Bartoloni, C. Burrini, C. Lacchini, A. Mantella, G. Muzzi, C. P. Tosi, V. Penati, C. Scarparo, M. T. Simonetti, and E. C. Bottger. 1997. Characterization of mycobacterial isolates phylogenetically related to, but different from Mycobacterium simiae. J. Clin. Microbiol. 35:697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace, R. J., Jr., B. A. Brown-Elliott, L. Hall, G. Roberts, R. W. Wilson, L. B. Mann, C. J. Crist, S. H. Chiu, R. Dunlap, M. J. Garcia, J. T. Bagwell, and K. C. Jost, Jr. 2002. Clinical and laboratory features of Mycobacterium mageritense. J. Clin. Microbiol. 40:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace, R. J., Jr., D. R. Nash, L. C. Steele, and V. Steingrube. 1986. Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J. Clin. Microbiol. 24:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiszfeiler, J. G., V. Karasseva, and E. Karczag. 1981. Mycobacterium simiae and related mycobacteria. Rev. Infect. Dis. 3:1040-1045. [DOI] [PubMed] [Google Scholar]
- 35.Wolinsky, E. 1979. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 119:107-159. [DOI] [PubMed] [Google Scholar]