Abstract
Bergeyella zoohelcum is an uncommon zoonotic pathogen typically associated with cat or dog bites. Previously, only five cases of B. zoohelcum infection have been reported. We report the isolation and characterization of a fastidious Bergeyella species from acute cellulitis in the upper extremity of a 60-year-old woman. The organism was too fastidious for identification and susceptibility testing with traditional culture methods. The isolate was characterized further by PCR amplification and sequencing of the 16S rRNA gene with broad-range eubacterial primers. Phylogenetic analysis of the 16S ribosomal DNA sequence indicated that this isolate was a member of the species B. zoohelcum (previously Weeksella zoohelcum), a gram-negative bacillus that is rarely associated with infections in humans. Despite sharing a close genetic relationship with other B. zoohelcum strains, this isolate was extremely fastidious in nature, raising the possibility that similar strains from cat or dog bite wound infections have been underreported.
Visits to the emergency room (ER) due to animal bites are not uncommon. In fact, 1% of all visits to the ER are related to bites by animals. Even though the majority of animal bites are due to dogs, cat bites represent 3 to 15% of all cases (24). The risk of an infection after an animal bite is always high, and for a cat bite, it is between 5 and 15% (26). Infections following animal bites tend to be polymicrobial in nature and include both aerobes and anaerobes. At least 30 different known infectious agents have been reported to be transmitted from a dog or a cat bite (4, 5). In a study conducted by Talan et al., the major groups of pathogens isolated from 57 cases of cat bites were species of Pasteurella, Streptococcus, Staphylococcus, Neisseria, Corynebacterium, and Moraxella. Anaerobes included Fusobacterium, Bacteroides, Prevotella, and Porphyromonas species (6, 23). Pasteurella multocida is present as normal flora in 50 to 70% of otherwise healthy cats, and not surprisingly 43% of cat bite wounds had this pathogen as well (3).
Bergeyella zoohelcum is an underreported zoonotic pathogen afflicting humans after an animal bite, even though this microorganism can be found in 38 to 90% of nasal and oral fluids and gingival scrapings of dogs (20). Historically, this species was referred to as a Centers for Disease Control and Prevention group IIj organism. However, in 1986 Holmes et al. proposed the name Weeksella zoohelcum for the group IIj bacteria. Weeksella virosa is the only other species in this genus (10). Finally, in 1994 Vandamme et al. proposed a new genus, Bergeyella, and renamed W. zoohelcum B. zoohelcum based on the genetic differences with W. virosa (25). B. zoohelcum is a nonfermentative, gram-negative, rod-shaped, non-spore-forming, nonmotile aerobic bacterium that typically grows well on blood agar but not on MacConkey's agar. Colonies are circular, shiny, and smooth with entire edges (11). B. zoohelcum is oxidase, catalase, and indole positive. At present, B. zoohelcum is the only representative of the genus Bergeyella. B. zoohelcum has been previously reported in a wound from a 35-year-old man following a bite from a Siberian tiger (12). It has also been linked to a leg abscess in a 10-year-old boy after a dog bite (20). Besides these reports, unidentified Weeksella species have been isolated from oral rinses of patients who were treated with radiation following nasopharyngeal carcinoma (15). A variety of food sources have been linked to Weeksella- and Bergeyella-like bacteria as well (1). A related pathogen, W. virosa, on the other hand, is a female urogenital pathogen and has been reported with an incidence rate of 2% in healthy women and up to 15% in women with a high rate of sexual activity (16). Here, we report a case of cellulitis due to zoonotic transmission of a fastidious B. zoohelcum in an elderly woman after a cat bite. The organism was compared with several other zoonotic B. zoohelcum strains, and their ecological and clinical significance is discussed.
Case history.
A 60-year-old previously healthy female employed as a veterinary technician presented to the ER after being bitten on the hand by a stray cat while administering medication. Physical examination revealed two puncture wounds on the dorsal and ventral aspect of the second metacarpal head. Samples were taken for culture, and the patient was given amoxicillin-clavulanic acid, 875 mg twice a day. The patient returned 24 h later with fever, severe left arm pain, and cellulitis and lymphangitis extending to the left axilla. The leukocyte count was 10,500/mm3 with 71% neutrophils, 22% lymphocytes, and 5% monocytes. After incision and drainage, the patient was begun on ampicillin-sulbactam, 3 g intravenously every 6 h. Her symptoms resolved completely within 1 week.
MATERIALS AND METHODS
Strains.
B. zoohelcum BZ-1 was isolated from a 60-year-old woman with cellulitis after a cat bite wound. The strains G6669 (cellulitis) and H1890 and H1971D (wound) were isolated following dog bites. The strain G8498 was from an unspecified infection from an adult male. The last four strains mentioned were from the Centers for Disease Control and Prevention.
Microbiology (strain BZ-1).
Wound swab samples were used to inoculate chocolate, blood, and MacConkey agar media. The plate contents were incubated at 37°C for 48 h under aerobic and anaerobic conditions. The growth of this strain was too poor to be characterized by standard biochemical methods. Strains G6669, G8498, H1890, and H1971 were characterized by methods described earlier (27).
16S rDNA sequence analysis.
Molecular methods for species identification have been described previously (21, 22). Briefly, the isolates were characterized by amplifying, sequencing, and analyzing the 16S rRNA gene. The template DNA for 16S ribosomal DNA (rDNA) PCR was prepared by boiling a few isolated colonies in 200 μl of sterile water for 10 min. The sample was centrifuged at 15,300 × g for 2 min to pellet the cell debris. The supernatant was collected and used directly as the template for PCR amplification. The broad-range eubacterial primers, FD1 (5′ AGA GTT TGA TCC TGG CTC AG 3′) and RD1 (5′ AAG GAG GTG ATC CAG CC3′), were used to amplify an ∼1,500-bp product from the 16S rRNA gene. A 500-μl PCR mixture contained 5 μl of 1× PCR buffer; 1.5 mM MgCl2; 200 μM dATP, dCTP, dGTP, and dTTP; 2.5 U of Taq polymerase; 20 pmol each of FD1 and RD1 primers; and 3 μl of the template DNA. The single PCR product was column purified and was then sequenced directly by cycle sequencing with the Thermosequenase kit (Amersham Pharmacia Biotech, Piscataway, N.J.) and Cy5-labeled nested primers. Both sense and antisense strands were sequenced. The sequencing reaction was run overnight in the ALF Express DNA sequencer (Amersham Pharmacia Biotech). The DNA sequence was aligned with DNAsis (Hitachi Software Engineering Co., Ltd., San Bruno, Calif.) and was edited manually to remove ambiguities. The consensus sequence was compared to all bacterial sequences available from the GenBank database by using the Blast 2.0 program (National Center for Biotechnology Information, Bethesda, Md.).
The rDNA sequence of the various Bergeyella species and closely related genera was aligned with a database of archaeal, bacterial, and eucaryal SSU rRNA sequences (ca. 10,000 sequences in total) by using the ARB software package (http://www.mikro.biologie.tu-muenchen.de/). Both BLAST analysis and the parsimony insertion tool of ARB tentatively placed the sequence of Bergeyella sp. isolate BZ-1 within the group Flavobacteria and specifically associated it with B. zoohelcum. Consequently, a subset of the ARB alignment, including the Bergeyella sp. isolate BZ-1 sequence, related sequences (which represent the known breadth of the genus, as well as representatives of the Bacteroides, Cytophagales, and Prevotella and other members of Flexibacteriaceae), and the sequence of Escherichia coli (used as an outgroup), was selected for more detailed phylogenetic analysis (1,262 to 1,542 nucleotide positions were sampled). The presented dendrogram was constructed by evolutionary distance analysis (weighted least-squares mean with GTR correction) with the PAUP software package (PAUP* version 4.0b2). The robustness of this tree was assessed by bootstrap resampling (1,000 replicates with resampling of all positions) of evolutionary distance trees (PAUP* version 4.0b2) (weighted least-squares mean with GTR correction). Parsimony (1,000 replicates) and maximum-likelihood analyses (100 replicates) provided results that were substantially similar to those of the evolutionary distance algorithm. Reported percent sequence identities are uncorrected.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rRNA sequence of the B. zoohelcum strains are as follows: AY289204 (BZ-1), AY398695 (G6669), AY398697 (H1890), AY398698 (H1971D), and AY398696 (G8498).
RESULTS AND DISCUSSION
Light growth of BZ-1 strain was observed only on the chocolate agar plates after 48 h of incubation at 37°C. The organism did not grow on blood or MacConkey agar plates. The strain stained was gram negative and oxidase and indole positive. The organism was too fastidious for identification and susceptibility testing with traditional methods. The remaining four strains were positive for catalase, oxidase, urease, indole, H2S production, and gelatin hydrolysis and were negative for esculin hydrolysis, citrate, and nitrate reduction. A 1,415-nucleotide consensus sequence of the 16S rRNA gene of strain BZ-1 was submitted to GenBank to search for matching bacterial sequences by BLAST. The closest match was B. zoohelcum ATCC 43767, with only 96.6% sequence identity. We observed that the 16S rDNA sequence of the ATCC strain (GenBank accession no. M93153) had a high number of ambiguous bases. In order to further determine whether this pathogen was a novel species in the genus Bergeyella, we sequenced the 16S rRNA genes of four additional B. zoohelcum strains linked to cellulitis and dog bite wounds and performed a detailed phylogenetic analysis.
Phylogenetic analysis.
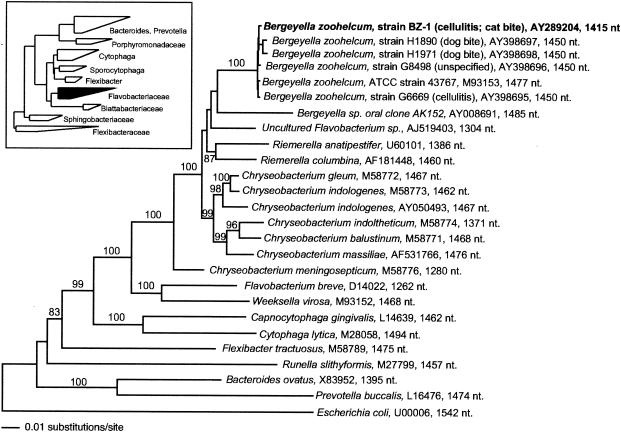
A representative phylogenetic tree, inferred by evolutionary distance methods (see Materials and Methods for details) is shown as Fig. 1. The robustness of this tree was assessed by bootstrap resampling with evolutionary distance (1,000 replicates), maximum parsimony (1,000 replicates), and maximum likelihood (100 replicates) analyses. The phylogenetic grouping of all B. zoohelcum strains reported here demonstrated 100% bootstrap values for all three types of phylogenetic reconstruction. The 16S rRNA gene of the Bergeyella isolate BZ-1 is, on average, 99.7% identical (range, 99.6 to 99.8%) to the five other B. zoohelcum rDNA sequences included in this analysis. Although no clear criteria exist to establish species identity on the basis of rDNA sequence similarity, a value of 99.7% is well within the range of intraspecific percent sequence identities that are observed for the 16S rRNA genes of other microbial species (e.g., E. coli). Moreover, the average pairwise percent sequence identity for the five previously characterized B. zoohelcum isolates is 99.5% (range, 99.4 to 99.9%). We therefore conclude that the isolate BZ-1 is probably a subspecies of B. zoohelcum.
FIG. 1.
Evolutionary distance phylogram of selected flavobacterial 16S rRNA sequences, including that of the newly isolated Bergeyella sp. isolate BZ-1. rRNA sequences of more distant relatives, including representatives of the genera Bacteroides, Cytophagales, Flexibacteriaceae, and Prevotella, are also included for comparison. The 16S rRNA sequence of E. coli (U00006) was chosen as an outgroup for this phylogenetic analysis. Sequences are identified by species name and length of sequence analyzed. Branch points supported by bootstrap values that are ≥90% (1000 bootstrap replicates with resampling) and their respective values are mentioned. Unlabeled branches have <70% bootstrap values. The inset depicts the larger-scale phylogenetic relationships among the major clades of the Bacteroides-Cytophagales-Flexibacterium domain of bacteria. The heights of the triangles in this figure represent the relative numbers of taxa in the 16S rRNA sequence database. nt, nucleotides.
Clinical relevance.
B. zoohelcum is a zoonotic pathogen indigenous to dogs and cats. That pathogen is reported to be present in 38 to 90% of the gingival scrapings and nasal and oral fluids of dogs (3, 20). It is transmitted to humans after a bite from a dog or a cat or from close contact with cats (13). Whether this bacterium is pathogenic to the hosts itself is unknown (19). Only one published report linked this pathogen to respiratory disease in a cat, since it was isolated from necrotic lung tissue of the cat (3). B. zoohelcum has been associated with several clinical syndromes in humans subsequent to dog or cat bites or repeated association or contact with them. This is not surprising because this pathogen has been found in up to 90% of the oral and nasal fluids and gingival scrapings of dogs (20). It caused pneumonia in a patient who had exposure to a dog suspected to be a carrier of this pathogen and caused meningitis after a dog bite (2, 9). It has also caused cases of septicemia in (i) an 80-year-old diabetic woman, (ii) in a 77-year-old woman suffering from cat associated-severe skin infections, and (iii) in a 33-year-old man after a dog bite (13, 17, 18). Other cases included a leg abscess in a 10-year-old after a dog bite and tenosynovitis in a 35-year-old healthy man after a Siberian tiger bite (12, 20). The tenosynovitis case was polymicrobial in nature and also had a Comamonas species, P. multocida, and other gram-negative bacteria (12). These clinical cases suggest that B. zoohelcum is an opportunistic pathogen. Even though this bacterium is considered to be of low virulence (13), infections due to this bacterium should be treated promptly to avoid complications such as endocarditis.
A limited number of studies have been done to determine the antimicrobial sensitivities of Bergeyella species. The isolate from a bacteremia case in a 33-year-old was susceptible to β-lactams, macrolides, and tetracyclines (17). Goldstein et al. have extensively studied the susceptibility of aerobic and anaerobic pathogens isolated from soft tissue bite infections (7, 8). Specifically, they reported that W. zoohelcum (Bergeyella) isolates are sensitive to gatifloxacin at ≤0.016 μg/ml and to the linezolid at that drug's MIC (≤2 μg/ml), besides other standard antibiotics (8). However, one should know that there are no accepted NCCLS standards for antibiotic susceptibility testing or breakpoints for B. zoohelcum.
Although B. zoohelcum is well recognized as a zoonotic pathogen, the detection of this bacterium in human subgingival crevice by 16S rDNA sequence analysis (14) raises important questions regarding its host diversity, ecological significance, and pathogenesis. First, is this zoonotic pathogen also a commensal of human oral flora as described by Kroes et al. (14)? Second, it is not known if the unidentified Weeksella species from oral rinses of patients with nasopharyngeal carcinoma (15) were related to W. virosa-like organisms or were related to B. zoohelcum. Interestingly, Woo et al. reported that B. zoohelcum could be misidentified as Riemerella anatipestifer if the MicroSec 500 16S rDNA-based identification system, which uses the first 527 bases of the 16S rRNA gene, is used (28). Third, the ecological habitat of B. zoohelcum seems to be related to the oral environment, regardless of the host. These questions can be answered only when more clinical cases involving this pathogen are thoroughly characterized by biochemical and molecular means and are reported. In conclusion, it is possible that fastidious strains of B. zoohelcum from animal bite wounds could have been underreported.
Acknowledgments
We thank Marshfield Clinic Research Foundation for its support through the assistance of Alice Stargardt in the preparation of the manuscript.
REFERENCES
- 1.Botha, W. C., P. J. Jooste, and C. J. Hugo. 1998. The incidence of Weeksella- and Bergeyella-like bacteria in the food environment. J. Appl. Microbiol. 84:349-356. [DOI] [PubMed] [Google Scholar]
- 2.Bracis, R., K. Seibers, and R. M. Julien. 1979. Meningitis caused by group II J. following a dog bite. West. J. Med. 131:438-440. [PMC free article] [PubMed] [Google Scholar]
- 3.Decostere, A., L. A. Devriese, R. Ducatelle, and F. Haesebrouck. 2002. Bergeyella (Weeksella) zoohelcum associated with respiratory diseases in a cat. Vet. Rec. 151:392. [DOI] [PubMed] [Google Scholar]
- 4.Elliot, D. L., S. W. Tolle, L. Goldberg, and J. B. Miller. 1985. Pet-associated illness. N. Engl. J. Med. 313:985-995. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein, E. J. 1991. Household pets and human infections. Infect. Dis. Clin. N. Am. 5:117-130. [PubMed] [Google Scholar]
- 6.Goldstein, E. J. 1995. Bites, p. 2068-2070. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.). Principles and practice of infectious diseases. Churchill Livingstone, Inc., New York, N.Y.
- 7.Goldstein, E. J. C., D. M. Citron, S. Hunt Gerardo, M. Hudspeth, and C. V. Merriam. 1998. Activities of HMR 3004 (RU 64004) and HMR 3647 (RU 66647) compared to those of erythromycin, azithromycin, clarithromycin, roxithromycin, and eight other antimicrobial agents against unusual aerobic and anaerobic human and animal bite pathogens isolated from skin and soft tissue infections in humans. Antimicrob. Agents Chemother. 42:1127-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein, E. J. C., D. M. Citron, and C. V. Merriam. 1999. Linezolid activity compared to those of selected macrolides and other agents against aerobic and anaerobic pathogens isolated from soft tissue bite infections in humans. Antimicrob. Agents Chemother. 43:1469-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimault, E., J. C. Glerant, P. Aubry, G. Laurans, J. P. Poinsot, and V. Jounieaux. 1996. Uncommon site of Bergeyella zoohelcum. Apropos of a case. Rev. Pneumol. Clin. 52:387-389. (In French.) [PubMed] [Google Scholar]
- 10.Holmes, B., A. G. Steigerwalt, R. E. Weaver, and D. J. Brenner. 1986. Weeksella zoohelcum sp. nov. (formerly group IIj) from human clinical specimens. Syst. Appl. Microbiol. 8:191-196. [Google Scholar]
- 11.Holt, J. G., N. R. Krieg, P. H. Sneath, J. T. Staley, and S. T. Williams. 1994. Weeksella, p. 99. In J. G. Holt, N. R. Krieg, P. H. Sneath, J. T. Staley, and S. T. Williams (ed.), Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
- 12.Isotalo, P. A., D. Edgar, and B. Toye. 2000. Polymicrobial tenosynovitis with Pasteurella multocida and other gram-negative bacilli after a Siberian tiger bite. J. Clin. Pathol. 53:871-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kivinen, P. K., M. R. Lahtinen, E. Ruotsalainen, I. T. Harvima, and M. L. Katila. 2003. Bergeyella zoohelcum septicemia of a patient suffering from severe skin infection. Acta Derm. Venereol. 83:74-75. [DOI] [PubMed] [Google Scholar]
- 14.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung, W. K., L. J. Jin, W. C. Yam, and L. P. Samaranayake. 2001. Oral colonization of aerobic and facultatively anaerobic gram-negative rods and cocci in irradiated, dentate, xerostomic individuals. Oral Microbiol. Immunol. 16:1-9. [DOI] [PubMed] [Google Scholar]
- 16.Mardy, C., and B. Holmes. 1988. Incidence of vaginal Weeksella virosa (formerly group IIf). J. Clin. Pathol. 41:211-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montejo, M., K. Aguirrebengoa, J. Ugalde, L. Lopez, J. A. Saez Nieto, and J. L. Hernandez. 2001. Bergeyella zoohelcum bacteremia after a dog bite. Clin. Infect. Dis. 33:1608-1609. [DOI] [PubMed] [Google Scholar]
- 18.Noell, F., M. F. Gorce, C. Garde, and C. Bizet. 1989. Isolation of Weeksella zoohelcum in septicaemia. Lancet ii:332. [DOI] [PubMed] [Google Scholar]
- 19.Quinn, P. J., M. E. Carter, B. Markey, and G. R. Carter. 1999. Glucose non-fermenting Gram-negative bacteria, p. 287-292. In P. J. Quinn, M. E. Carter, B. Markey, and G. R. Carter (ed.), Clinical veterinary microbiology, 3rd ed. Mosby, London, United Kingdom.
- 20.Reina, J., and N. Borrell. 1992. Leg abscess caused by Weeksella zoohelcum following a dog bite. Clin. Infect. Dis. 14:1162-1163. [DOI] [PubMed] [Google Scholar]
- 21.Shukla, S. K., and K. D. Reed. 2000. Desulfovibrio desulfuricans bacteremia in a dog. J. Clin. Microbiol. 38:1701-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla, S. K., K. A. Bernard, M. Harney, D. N. Frank, and K. D. Reed. 2003. Corynebacterium nigricans sp. nov.: proposed name for a black pigmented Corynebacterium species recovered from the human female urogenital tract. J. Clin. Microbiol. 41:4353-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talan, D. A., D. M. Citron, F. M. Abrahamian, G. J. Moran, E. J. Goldstein, et al. 1999. Bacteriologic analysis of infected dog and cat bites. N. Engl. J. Med. 340:85-92. [DOI] [PubMed] [Google Scholar]
- 24.Tan, J. S. 1997. Human zoonotic infections transmitted by dogs and cats. Arch. Intern. Med. 157:1933-1943. [PubMed] [Google Scholar]
- 25.Vandamme, P., J.-F. Bernardet, P. Segers, K. Kersters, and B. Holmes. 1994. New perspectives in the classification of the flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int. J. Syst. Bacteriol. 44:827-831. [Google Scholar]
- 26.Weber, D. J., and A. R. Hansen. 1991. Infections resulting from animal bites. Infect. Dis. Clin. N. Am. 5:663-680. [PubMed] [Google Scholar]
- 27.Weyant, R. S., C. W. Moss, R. E. Weaver, D. G. Hollis, J. G. Jordan, E. C. Cook, and M. I. Daneshvar. 1995. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria, 2nd ed. Williams & Wilkins, Baltimore, Md.
- 28.Woo, P. C. Y., K. H. L. Ng, S. K. P. Lau, K.-T. Yip, A. M. Y. Fung, K.-W. Leung, D. M. W. Tam, T.-L. Que, and K.-Y. Yuen. 2003. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J. Clin. Microbiol. 41:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]