Abstract
Fluorescent amplified fragment length polymorphism (FAFLP) analysis was applied to 276 Campylobacter jejuni strains and 87 Campylobacter coli strains isolated from humans, pigs, cattle, poultry, and retail meats to investigate whether certain FAFLP genotypes of C. jejuni and C. coli are associated with a particular host and to determine the degree of association between FAFLP-defined genotypes and heat-stable serotypes and/or phage types. Within C. coli, the poultry strains clustered separately from those of porcine origin. In contrast, no evidence of host specificity was detected among C. jejuni strains. While C. coli strains show host specificity by FAFLP genotyping, C. jejuni strains that are genotypically similar appear to colonize a range of hosts, rather than being host adapted. Some serotypes and/or phage types (C. jejuni serotype HS18, phage type PT6, and serophage type HS19/PT2 and C. coli HS66, PT2, and HS56/PT2) were the most homogeneous by FAFLP genotyping, while others were more heterogeneous (C. jejuni HS5 and PT39, and C. coli HS24 and PT44) and therefore poor indicators of genetic relatedness between strains. The lack of host specificity in C. jejuni suggests that tracing the source of infection during epidemiological investigations will continue to be difficult. The lack of congruence between some serotypes and/or phage types and FAFLP genotype underlines the need for phenotypic testing to be supplemented by genotyping. This study also demonstrates how, in general, FAFLP generates “anonymous” genetic markers for strain characterization and epidemiological investigation of Campylobacter in the food chain.
Campylobacter is the most common cause of bacterial gastrointestinal infection in both developed and developing countries (13). In recent years the number of laboratory-confirmed cases of Campylobacter in England and Wales has been high and has continued to increase, and the reason for this is not fully understood. In 2001, 56,420 laboratory-confirmed infections were reported (Health Protection Agency website, http://www.hpa.org.uk). Effective intervention strategies to limit the spread of campylobacters in the food chain are still lacking. Although the major route of infection in humans is thought to be consumption of contaminated poultry, campylobacters are also part of the gastrointestinal flora of many other animals, such as cows, pigs, sheep, and domestic pets (17). It is unclear whether certain strains of Campylobacter spp. are specific to particular hosts, and the relative contribution that each of these sources makes to human infection is not yet adequately determined. The ubiquitous nature of campylobacters in the environment and possibility of cross-contamination during food preparation make it difficult to trace the origin of Campylobacter infections.
A number of phenotypic methods have been used in epidemiological investigations of Campylobacter infection. Serotyping has been suggested as the practical solution for large-scale surveillance of Campylobacter (22) and is current practice for routine reference typing in England and Wales, along with phage typing (9). Serotyping detects soluble heat-stable (HS) antigens (23) or heat-labile antigens (15) exposed on the cell surface. Frost et al. (10) modified the Penner serotyping scheme by introducing absorbed antisera and using whole-cell agglutination to eliminate nonspecific agglutination reactions and reduce the number of cross-reactions. However, the majority of Campylobacter isolates belong to a limited number of HS serotypes (10, 26), so that phage typing is used to further discriminate between the predominant serotypes (9). However, the high number of untypeable strains and time-consuming and technically demanding protocols are limiting factors (29).
Molecular genotyping techniques may assist in epidemiological investigations as well as being useful in identifying strains or successful clones and their possible association to certain hosts. Comparison of pulsed-field gel electrophoresis, fla gene typing, ribotyping, and fluorescent amplified fragment length polymorphism analysis (FAFLP) for the genotyping of thermotolerant Campylobacter suggested that FAFLP is the most discriminatory of these methods (1, 14). A study comparing FAFLP with multilocus sequence typing (MLST) resulted in similar clustering of the strains in both; therefore, the two methods may be equally useful in disclosing genetic relationships (25). FAFLP has been suggested as the most promising method for global epidemiological studies due to its apparent insensitivity to the genetic instability of Campylobacter that complicates other typing methods (29). FAFLP samples the genome in an unweighted manner and has the potential to discover as yet unidentified genes. It has already proved to be a powerful tool for identifying molecular markers in the study of the epidemiology, pathogenicity, and genetic variation of various genera (8, 28).
The aim of this study was to apply a standardized genome sequence-based FAFLP to a well-defined collection of Campylobacter jejuni and Campylobacter coli strains from animal hosts, retail meat, and human infection and to investigate the possibility that certain genotypes are associated with a particular host. FAFLP genotypes were also compared with HS serotypes and phage types to determine the degree of association between them.
MATERIALS AND METHODS
Bacterial strains, phenotypic typing, and culture conditions.
A recent and diverse collection, predominantly from the United Kingdom, comprised 276 strains of C. jejuni and 87 strains of C. coli. They were obtained from live poultry, retail chicken portions, live cattle, retail bovine liver, milk, live pigs, retail porcine liver, and humans (see Table 1). Strains were cultured microaerobically on blood agar plates for 48 h at 37°C and preserved at −80°C in Microbank cryovials (Pro-Lab Diagnostics, Cheshire, United Kingdom). All strains were HS serotyped using the direct agglutination method and phage typed according to the standard protocols employed by the Campylobacter Reference Unit (9, 10).
TABLE 1.
Strain collection: origin and source
| Origin (na) | Source | No. of C. jejuni strains | No. of C. coli strains |
|---|---|---|---|
| Poultry (132) | Live poultry | 67 | 1 |
| Chicken portionsb | 52 | 12 | |
| Cattle (85) | Live cattle | 41 | 0 |
| Bovine liverb | 30 | 1 | |
| Milk | 13 | 0 | |
| Pigs (83) | Live pigs | 0 | 33 |
| Porcine liverb | 10 | 40 | |
| Human (56) | Fecal samples | 56 | 0 |
| NCTCc (7) | 7 | 0 | |
| Total (363) | 276 | 87 |
n, total no. of strains.
Strains from a previous study (12).
NCTC, National Collection of Type Cultures, Health Protection Agency, London, United Kingdom.
FAFLP.
A standardized DNA extraction and genome sequence-based FAFLP was performed as described previously (2). Briefly, genomic DNA was digested sequentially with HindIII and HhaI endonucleases (New England BioLabs, Hertfordshire, United Kingdom), and HindIII-specific and HhaI-specific adapters were then ligated to the double-digested DNA. Touchdown PCR was performed using a HindIII adapter-specific primer labeled with the blue fluorescent dye 5-carboxyfluorescein and a HhaI adapter-specific primer, both primers having an extra selective base A at the 3′ end (Genosys Biotechnologies, Cambridge, United Kingdom) (6). Touchdown PCR cycling conditions were as described previously (3). FAFLP products were separated on a 377 automated DNA sequencer (Applied Biosystems [AB], Warrington, Cheshire, United Kingdom) as described previously (2).
Fragment analysis.
Fluorescent amplified fragments (AFs) were sized with GeneScan 3.1.0 software (AB). The data were transferred to GenoTyper 2.5 software (AB), and AFs were scored as present or absent in a binary matrix, using a tolerance level of ±0.5 bp. Binary data were exported for storage in Excel version 6.0 (Microsoft) before being imported into Bionumerics software version 3.0 (Applied Maths, Kortrijk, Belgium). Cluster analysis was performed and a dendrogram was constructed in Bionumerics using the Dice similarity coefficient, bootstrap analysis, and the unweighted pair group method with arithmetic averages. Group separation statistical analysis was performed to determine the internal stabilities (significance) of the defined groups (origin, HS serotype, phage type, and serophage type) with five or more strains. This was performed within Bionumerics using the Jacknife method with the maximum similarity setting and equal distribution over the groups when identical values were found for different groups.
RESULTS
FAFLP analysis: general features.
Among the 363 strains examined, 333 distinct FAFLP profiles were detected; 311 were unique to individual strains. The FAFLP profiles consisted of 45 to 122 AFs ranging in size from 60 to 600 bp (beyond this range the accuracy of AF sizing decreases (2). No single AF was common to all Campylobacter strains. Cluster analysis was performed, and a dendrogram was constructed (see Materials and Methods), relating C. jejuni and C. coli strains as defined by FAFLP. Two distinct clusters were generated with a linkage level between the two species of only 22% (data not shown). Thirteen AFs were specific to C. coli, and two AFs were C. jejuni specific. Since our FAFLP was genome sequence based (2), the last two highly conserved AFs could be mapped to a putative lipoprotein gene and to the 50S ribosomal protein L10 gene in the C. jejuni genome (21).
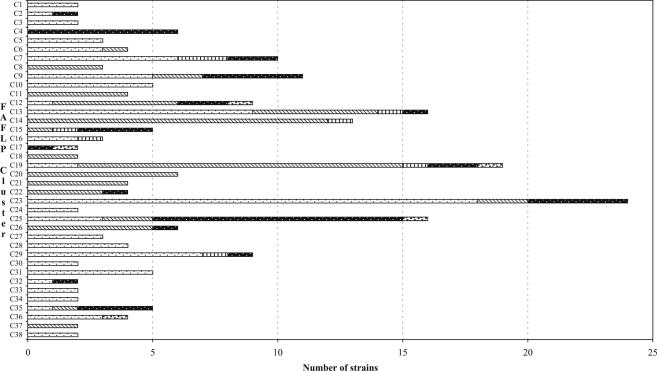
Previous studies have shown that FAFLP profiles sharing ≥90% similarity are genetically highly related and often represent epidemiologically related strains (6, 25). Within the C. coli strains, 16 clusters (two or more strains) were defined at ≥90% similarity, and 63.2% of the C. coli strains were assigned to these clusters (Fig. 1). Within the C. jejuni strains, 38 clusters (two or more strains) were defined at ≥90% similarity and 81.5% of C. jejuni strains fell into these clusters (data not shown). To simplify analysis, the number of strains of C. jejuni in each of the 38 FAFLP-defined clusters (C1 to C38), together with their source, was used to construct a frequency distribution graph (Fig. 2).
FIG. 1.
Dendrogram showing the relationship between FAFLP genotypes and host in C. coli. Cluster analysis was performed, and the dendrogram was constructed with Bionumerics version 3.0, using the unweighted pair group method with arithmetic averages and the Dice similarity coefficient. The horizontal scale indicates percent genetic similarity between strains, with 90% genetic similarity indicated by a dotted line. Symbols represent the source: ▪, live poultry; □, chicken portion; ▴, bovine liver; ○, live pig; •, porcine liver.
FIG. 2.
Frequency distribution of C. jejuni FAFLP-defined clusters (containing ≥2 strains defined at ≥90% similarity) with respect to source: [ ], live poultry or chicken portions; ▧, live cattle, milk or bovine liver; │, live pigs or porcine liver;
, human fecal samples; [
], live poultry or chicken portions; ▧, live cattle, milk or bovine liver; │, live pigs or porcine liver;
, human fecal samples; [ ], National Collection of Type Culture strains.
], National Collection of Type Culture strains.
FAFLP analysis of C. coli strains and relationship to host specificity, HS serotype, and phage type.
Cluster analysis of C. coli strains showed that with one exception, the C. coli pig strains clustered separately from the poultry C. coli strains (Fig. 1). Group separation analysis was applied to determine whether the C. coli FAFLP profiles could be separated into host-specific groups. This showed that both the pig strains and the poultry strains were most closely related to other strains from the same host group: 92% of poultry FAFLP profiles were most closely related to other poultry FAFLP profiles, and 99% of pig FAFLP profiles were most closely related to other pig FAFLP profiles. When the occurrence of every single AF in the poultry and pig C. coli profiles was compared, no host-specific AFs responsible for this delineation were found.
Group separation analysis was also applied to determine whether C. coli FAFLP profiles could be separated into groups based on HS serotype, on phage type, and on serophage type (Table 2). Serotypes HS56 and 66 were the most statistically significant groups, related to >60% of other profiles with the same HS serotype (Table 2). None of the HS24 profiles were most closely related to other HS24 profiles. The phage type 2 (PT2) group was the most significant: 75% of FAFLP profiles from PT2 strains were most closely related to other profiles of PT2 strains (Table 2), while the PT44 group had the least internal stability: only 16.7% of profiles were most closely related to profiles of other PT44 strains. As expected, a serophage type comprising an HS serotype and a phage type that were highly significant when analyzed separately gave a high internal stability in combined form (Table 2). One hundred percent of HS56/PT2 profiles were most closely related to other HS56/PT2 profiles.
TABLE 2.
Internal stability (significance) of FAFLP-defined C. coli strain clusters in association with HS serotype, phage type, and serophage type
| HS serotype and/or phage type | % closest relative by FAFLPa |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HS serotypeb |
Phage typeb |
Serophage typeb |
||||||||||||
| HS24 | HS48 | HS56 | HS66 | UT | PT1 | PT2 | PT44 | UT | HS56/PT2 | HS66/PT2 | UT/PT1 | UT/PT2 | UT/PT44 | |
| HS24 | 0 | 0 | 12.5 | 7.1 | 5.4 | |||||||||
| HS48 | 20 | 16.7 | 0 | 0 | 10.8 | |||||||||
| HS56 | 20 | 0 | 62.5 | 7.1 | 8.1 | |||||||||
| HS66 | 20 | 0 | 12.5 | 64.3 | 16.2 | |||||||||
| UT | 40 | 83.3 | 12.5 | 21.4 | 59.5 | |||||||||
| PT1 | 48.5 | 16.7 | 41.7 | 30.8 | ||||||||||
| PT2 | 21.2 | 75 | 33.3 | 7.7 | ||||||||||
| PT44 | 9.1 | 4.2 | 16.7 | 7.7 | ||||||||||
| UT | 21.2 | 4.2 | 8.3 | 53.9 | ||||||||||
| HS56/PT2 | 100 | 0 | 6.3 | 0 | 0 | |||||||||
| HS66/PT2 | 0 | 66.7 | 25 | 22.2 | 20 | |||||||||
| UT/PT1 | 0 | 33.3 | 43.8 | 11.1 | 60 | |||||||||
| UT/PT2 | 0 | 0 | 18.8 | 55.6 | 20 | |||||||||
| UT/PT44 | 0 | 0 | 6.3 | 11.1 | 0 | |||||||||
The percentages of correct identifications for members of a group are given in the respective rows in bold text.
Included in the analyses were the following: only serotypes represented by five or more strains (equivalent to 80.5% of C. coli strains), only phage types represented by five or more strains (equivalent to 94.2% of C. coli strains) and only serophage types represented by five or more strains (equivalent to 50.6% of C. coli strains).
FAFLP analysis of C. jejuni strains and relationship to host specificity, HS serotype, and phage type.
Overall, FAFLP-derived clustering did not indicate host specificity in C. jejuni (Fig. 2), where all isolates from the same host would be expected to group together. However, certain FAFLP genotypic clusters showed evidence of host association, where a cluster was restricted to one host: 12 contained only poultry strains, six were cattle-only clusters, and one cluster had only human strains (Fig. 2).
Group separation analysis revealed that the poultry, cattle, and human FAFLP profiles were most closely related to other profiles of the same host group (Table 3). Pig FAFLP profiles appeared to be less host specific and were more closely related to cattle and poultry profiles than to other pig profiles (Table 3). In addition, less than 3% of human, poultry, or cattle profiles were closely related to pig profiles. The occurrence of every single AF was compared in each host to determine whether host specificity could be attributed to a single AF or subset of AFs or if there were any AFs that were predominantly present in a particular host and not in others. However, none of the C. jejuni-derived AFs was specific for a particular host, i.e., they also occurred at least occasionally in strains from other hosts.
TABLE 3.
Significance of FAFLP-defined C. jejuni strain clusters in association with host specificity
| Host | % closest relative by FAFLPa |
|||
|---|---|---|---|---|
| Pig | Cattle | Poultry | Human | |
| Pig | 10 | 0 | 0.8 | 2.7 |
| Cattle | 40 | 78.6 | 5 | 19.1 |
| Poultry | 40 | 6 | 83.6 | 15.5 |
| Human | 10 | 15.5 | 10.5 | 62.7 |
The percentages of correct identifications for members of a group are given in the respective columns in bold text.
Most HS serotypes were related to 50 to 75% of other FAFLP profiles with the same HS serotype by group separation analysis (Table 4). HS18, HS19, and HS50 were the most discrete groups and were related to more than 80% of other members of the same group. However, HS5 was not related to any other HS5 profiles and was more closely related to HS6 and UT profiles. Most phage type groups were also related to 50 to 75% of other FAFLP profiles with the same phage type (Table 5). PT6 and PT33 were the most significant groups, related to 87.5 and 85.5% of other group members, respectively, while the phage type PT34, PT39, and UT groups were the least significant (Table 5). In general, those serophage types comprising a serotype and a phage type that were highly significant when analyzed separately gave a high internal stability in combined form for C. jejuni, for example, the HS18/PT2, HS19/PT2, and HS50/PT6 groups (Table 6). However, only 6.7% of HS50/PT33 profiles were related to other members of that group, and instead they were more closely related to UT/PT33 profiles (Table 6), even though the HS50 group and PT33 group were highly significant when analyzed separately (Tables 4 and 5).
TABLE 4.
Internal stability (significance) of FAFLP-defined C. jejuni strain clusters in association with HS serotype
| HS serotypea | % closest relative by FAFLPb |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HS2 | HS5 | HS6 | HS11 | HS12 | HS13 | HS16 | HS18 | HS19 | HS31 | HS37 | HS44 | HS50 | UT | |
| HS2 | 55.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.3 | 0 | 5.4 |
| HS5 | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.7 | 2.7 |
| HS6 | 0 | 28.6 | 75 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.8 | 1.8 |
| HS11 | 0 | 0 | 0 | 75 | 0 | 0 | 0 | 0 | 0 | 8.3 | 0 | 0 | 0 | 1.8 |
| HS12 | 0 | 0 | 0 | 0 | 71.4 | 0 | 0 | 0 | 0 | 0 | 8.3 | 0 | 0 | 1.8 |
| HS13 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 16.7 | 25 | 0 | 1.8 | 3.6 |
| HS16 | 0 | 14.3 | 0 | 0 | 0 | 15.6 | 54.6 | 0 | 0 | 0 | 0 | 5.3 | 1.8 | 1.8 |
| HS18 | 0 | 14.3 | 0 | 0 | 0 | 0 | 0 | 84.6 | 0 | 0 | 8.3 | 0 | 0 | 0 |
| HS19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 81.3 | 0 | 0 | 0 | 1.8 | 8.9 |
| HS31 | 0 | 0 | 0 | 5.6 | 0 | 6.3 | 0 | 0 | 0 | 66.7 | 0 | 0 | 0 | 1.8 |
| HS37 | 0 | 0 | 0 | 0 | 14.3 | 6.3 | 0 | 7.7 | 0 | 0 | 41.7 | 0 | 3.6 | 5.4 |
| HS44 | 11.1 | 0 | 0 | 0 | 0 | 6.3 | 9.1 | 0 | 0 | 0 | 0 | 54.4 | 1.5 | 8.9 |
| HS50 | 22.2 | 14.3 | 0 | 5.6 | 14.3 | 9.4 | 27.3 | 0 | 0 | 0 | 8.3 | 9.7 | 80.1 | 5.4 |
| UT | 11.1 | 28.6 | 0 | 13.9 | 0 | 6.3 | 9.1 | 7.7 | 18.8 | 8.3 | 8.3 | 25.4 | 5.1 | 50.9 |
Included in the analyses were only serotypes represented by five or more strains (equivalent to 92% of C. jejuni strains).
The percentages of correct identifications for members of a group are given in the respective rows in bold text.
TABLE 5.
Internal stability (significance) of FAFLP-defined C. jejuni strain clusters in association with phage type
| Phage typea | % closest relative by FAFLPb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PT1 | PT2 | PT5 | PT6 | PT8 | PT33 | PT34 | PT35 | PT39 | PT44 | UT | |
| PT1 | 57.3 | 17.3 | 12.1 | 3.1 | 50 | 3.2 | 0 | 0 | 6.7 | 0 | 56.9 |
| PT2 | 14.7 | 69.1 | 9.1 | 0 | 0 | 0 | 14.3 | 0 | 80 | 25 | 20.8 |
| PT5 | 4.5 | 0 | 54.6 | 0 | 0 | 3.2 | 28.6 | 20 | 6.7 | 0 | 2.8 |
| PT6 | 0 | 0 | 0 | 87.5 | 0 | 6.5 | 0 | 0 | 0 | 0 | 0 |
| PT8 | 3.5 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 0 | 0 | 0 |
| PT33 | 0 | 0 | 0 | 6.3 | 0 | 85.5 | 14.3 | 20 | 0 | 0 | 0 |
| PT34 | 0 | 4.6 | 18.2 | 0 | 0 | 0 | 42.9 | 0 | 0 | 12.5 | 0 |
| PT35 | 1.7 | 0 | 0 | 0 | 0 | 1.6 | 0 | 60 | 0 | 0 | 8.3 |
| PT39 | 5.3 | 4.6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.8 |
| PT44 | 1.7 | 5.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 8.3 |
| UT | 11.4 | 1.8 | 3 | 3.1 | 0 | 0 | 0 | 0 | 6.7 | 12.5 | 0 |
Included in the analyses were only phage types represented by five or more strains (equivalent to 85.1% of C. jejuni strains).
The percentages of correct identifications for members of a group are given in the respective rows in bold text.
TABLE 6.
Internal stability (significance) of defined C. jejuni strain clusters in association with serophage type
| Serophage typea | % closest relative by FAFLPb |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HS11/PT1 | HS12/PT2 | HS13/PT1 | HS18/PT2 | HS19/PT2 | HS37/PT1 | HS37/PT2 | HS44/PT33 | HS50/PT5 | HS50/PT6 | HS50/PT33 | HS50/PT34 | UT/PT1 | UT/PT2 | UT/PT33 | UT/UT | |
| HS11/PT1 | 50 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14.3 | 0 | 20 |
| HS12/PT2 | 0 | 60 | 0 | 0 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 14.3 | 0 | 0 |
| HS13/PT1 | 0 | 0 | 0 | 0 | 0 | 40 | 20 | 0 | 0 | 0 | 0 | 0 | 30 | 0 | 0 | 0 |
| HS18/PT2 | 12.5 | 0 | 0 | 75 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HS19/PT2 | 0 | 0 | 0 | 0 | 90 | 0 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 14.3 | 0 | 0 |
| HS37/PT1 | 12.5 | 0 | 20 | 0 | 0 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HS37/PT2 | 0 | 20 | 0 | 12.5 | 0 | 0 | 60 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HS44/PT33 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 54.2 | 0 | 0 | 16.7 | 0 | 0 | 0 | 21.7 | 0 |
| HS50/PT5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 4.2 | 0 | 20 | 0 | 0 | 0 | 0 |
| HS50/PT6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 87.5 | 20 | 0 | 10 | 0 | 0 | 0 |
| HS50/PT33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22.9 | 0 | 4.2 | 6.7 | 20 | 0 | 0 | 31.7 | 20 |
| HS50/PT34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 4.2 | 0 | 60 | 0 | 0 | 0 | 0 |
| UT/PT1 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 14.3 | 0 | 40 |
| UT/PT2 | 12.5 | 20 | 10 | 12.5 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 42.9 | 0 | 0 |
| UT/PT33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22.9 | 0 | 0 | 56.7 | 0 | 0 | 0 | 46.7 | 20 |
| UT/UT | 12.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 |
Only serophage types represented by five or more strains (equivalent to 44.9% of C. jejuni strains) were included in the analysis.
The percentages of correct identifications for members of a group are given in the respective rows in bold text.
DISCUSSION
Though the incidence of Campylobacter infections continues to increase in England and Wales, microbiological evidence of sources of infection based on comparing isolates from patients and suspected sources is difficult to obtain. In addition, coinfection with more than one strain (12, 24, 25) and lack of widespread application of subtyping of Campylobacter species means that the source of a very large proportion of infections remains undetermined. In this study, a standardized genome sequence-based FAFLP was applied to C. jejuni and C. coli strains from humans, food animal hosts, and foods of animal origin to investigate the possibility that host-specific genotypes or anonymous genetic markers of host association could be detected and characterized. FAFLP was also compared with the HS serotype and phage type to determine the degree of congruence between these geno- and phenotypic methods.
In this study, two C. jejuni and 13 C. coli species-specific marker AFs were found, which may correlate with virulence or other strain characteristics. The two C. jejuni marker AFs, a putative lipoprotein gene and the 50S ribosomal protein L10 gene, may represent potential new species-specific markers, and further investigation of these genes is required. It has been shown in other studies that FAFLP analysis permits the identification of all Campylobacter spp., and therefore FAFLP has the advantage of concurrent species identification and genotyping (7, 18).
A variety of typing methods have been used in previous studies to investigate possible host specificity in C. jejuni. MLST studies have suggested differences in the frequency distribution by source of strains belonging to particular clonal complexes; certain complexes were overrepresented by strains from a particular source (4, 5, 25). Pulsed-field gel electrophoresis using three different restriction enzymes also identified clones found in cattle, poultry, and humans (19). However, restriction fragment length polymorphism analysis of the flaA gene using EcoRI and PstI showed that most flaA types were associated with several hosts (20).
Identification of markers of host specificity would enable human Campylobacter isolates to be screened to identify the most common sources of infection, and this information could then be used to introduce targeted intervention strategies. This study reports the first investigation of host specificity to be applied to both C. jejuni and C. coli strains. FAFLP cluster analysis and group separation suggested that C. coli poultry and pig strains are genetically distinct, and although the strain collection included relatively small numbers of poultry-derived strains, this may be evidence of host specificity in C. coli. Campylobacter coli accounts for only about 7 to 10% of human campylobacter infections (11, 12) and is therefore less of a disease burden than C. jejuni. However, the overall number of C. coli infections each year makes a significant contribution to the burden of foodborne disease in England and Wales (27). Nevertheless, data presented in this study may have significant implications for the epidemiology and control of C. coli. Genotypic differences identified by FAFLP between strains from different hosts could indicate a decreased likelihood of successful transmission between hosts and perhaps lead to a genetically based explanation for this.
FAFLP analysis failed to show evidence of a link between genotype and host specificity in C. jejuni strains, which is consistent with results of previous studies (6, 8, 25). However, group separation analysis of the FAFLP data, as presented here, can be used to study the relationships between the genotypes recovered from different hosts. In our study of the C. jejuni FAFLP data, cattle appeared to be as important a source of human infection as poultry. Only 15.5% of human strains were closely related to poultry strains, and only 10.5% of poultry strains were most closely related to human strains, compared to 19.1% of human strains that were closely related to cattle strains and 15.5% of cattle strains that were closely related to human strains. Although consumption of contaminated poultry products is believed to be a major route of infection, only a subset of poultry strains may be capable of infecting humans, and therefore, other sources of infection, such as cattle, may be equally important.
There were some contrasts between our results and those of a recent study by Schouls et al. (25). They noted an unexpected association between cattle and human strains. More than 50% of cattle FAFLP (and MLST) types had their closest relatives in human strains, and only 29% of cattle strains were most closely related to other cattle strains. We found, however, that 78.6% of cattle strains were most closely related to other cattle strains and only 15.5% were more closely related to human strains. This difference may be due to the relatively small number of cattle strains (31 strains) examined in the study by Schouls et al. (25) in comparison to the number analyzed in our study (85 strains), or it may be a feature of the particular strains in each study. The majority of cattle strains studied by Schouls et al. (25) were isolated in The Netherlands, and a possible explanation may be that cattle and humans were colonized from a common source that is absent in the United Kingdom, from where the majority of our cattle strains were obtained, or the environmental sources of C. jejuni in cattle may differ in the two countries.
Our earlier comparison between FAFLP genotype and serotype showed that some serotypes were genetically homogeneous and found in discrete clusters, while other serotypes were more genetically heterogeneous and exhibited a lack of congruence with the FAFLP genotype (2). In the present study, analysis of more strains and application of the group statistical method allowed a fuller comparison of FAFLP and serotyping data for C. jejuni and C. coli strains. Strains of C. coli serotypes HS66 and HS56 and C. jejuni HS18 and HS19 strains were genetically the most homogeneous, whereas C. coli HS24 and C. jejuni HS5 serotypes were genetically the most heterogeneous. Serotypes HS13, HS16, and HS50 form part of the HS4 complex and can cross-react during serotyping (10, 16). This finding is supported to an extent by group separation of the HS13 and HS16 FAFLP genotypes. Investigation of MLST type and serotype has found in some cases that membership in a lineage or sequence type correlated with possession of a particular Penner HS serotype, while members of other sequence type complexes were highly diverse for serotype (4, 5). In general, however, serotype was a poor indicator of a clonal complex. Campylobacter is capable of colonizing multiple hosts and environments; if the cell surface antigens responsible for determining serotype are also important in colonization of the host or adaptation to the environment, then the discrepancies between genotype and serotype may be the result of exposure to many different host immune responses and environmental pressures (4).
Phage typing is used as an extension to serotyping to allow a further level of discrimination between strains. Comparison of C. coli FAFLP genotype and phage type data showed that PT2, which accounts for more than half of the strains (9), was the most closely related FAFLP-defined group of strains and is therefore a good epidemiological marker. By FAFLP analysis, PT6 and PT33 were the most closely related groups of C. jejuni strains. However, the PT1 group (the most prevalent phage type in C. jejuni, accounting for 19.6% of strains) was only 57.3% genetically similar to other PT1 strains. The PT1 phage type is defined on the basis of sensitivity to two related phages, but this reaction is difficult to reproduce and so may result in misidentification (9).
Combining phage typing with serotyping allows identification of 6 to 29 phage types within each of the predominant serotypes, although within some serotypes only one or two phage types predominate, possibly indicating a closer epidemiological relationship (9). With 66 different HS serotypes and 76 phage types, a total possible 5,016 serophage types could be identified if the two characteristics are unrelated (10). In this study, as expected, a serophage type comprised of a HS serotype and a phage type that gave good differentiation between strains was a good marker of genetic relatedness. However, there were exceptions, such as C. jejuni HS50/PT33, which was a poor marker of genetic relatedness. These data reinforce the need for a highly discriminatory genotypic method or combination of methods, as well as phenotypic typing to be available for epidemiological studies to allow the recognition of unrelated strains in an investigation, for example, of outbreaks or sources of infection. Tracing the source of infection is complicated by the carriage of multiple types and the high genetic diversity of strains within numerous hosts (25). This study, along with previous studies, has demonstrated that genotyping of C. jejuni by FAFLP, as with other methods, may not be fully effective in tracing sources of infection. However, FAFLP has revealed that pig and poultry C. coli strains may be genetically distinct, and this requires confirmation.
Between 18.9 and 36.6% of C. jejuni and C. coli isolates are untypeable using the current antiserum panel for serotyping (10, 16). At present new serotypes are derived from clusters of untypeable strains that have common profiles by one or more fingerprinting techniques, and therefore, the data produced in this study may prove useful in generating new serotypes and lowering the percentage of untypeable strains. Alternatively, AFs that are specific to strains of particular epidemiological importance could be used as markers in the form of PCR assays or DNA arrays. Identification and meaningful subtyping of Campylobacter have proved challenging. The data presented here show that FAFLP genotyping is capable of identifying Campylobacter strains to the species level and of concurrently subtyping strains in an information-rich genotyping scheme.
Acknowledgments
This work was supported by a grant (B03014) from the Food Standards Agency, London, United Kingdom.
We thank D. Newell and staff at the Veterinary Laboratory Agency, Weybridge, and D. Wareing and staff at the Campylobacter Collaborating Unit, Preston Microbiology Laboratory, for providing strains and epidemiological data. We thank Philip Mortimer for critical reading of the manuscript.
REFERENCES
- 1.de Boer, P., B. Duim, A. Rigter, J. Der Plas, W. F. Jacobs-Reitsma, and J. A. Wagenaar. 2000. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 38:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desai, M., J. M. Logan, J. A. Frost, and J. Stanley. 2001. Genome sequence-based fluorescent amplified fragment length polymorphism of Campylobacter jejuni, its relationship to serotyping, and its implications for epidemiological analysis. J. Clin. Microbiol. 39:3823-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai, M., A. Tanna, R. Wall, A. Efstratiou, R. George, and J. Stanley. 1998. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J. Clin. Microbiol. 36:3133-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duim, B., C. W. Ang, A. Van Belkum, A. Rigter, N. W. van Leeuwen, H. P. Endtz, and J. A. Wagenaar. 2000. Amplified fragment length polymorphism analysis of Campylobacter jejuni strains isolated from chickens and from patients with gastroenteritis or Guillain-Barre or Miller Fisher syndrome. Appl. Environ. Microbiol. 66:3917-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duim, B., P. A. Vandamme, A. Rigter, S. Laevens, J. R. Dijkstra, and J. A. Wagenaar. 2001. Differentiation of Campylobacter species by AFLP fingerprinting. Microbiology 147:2729-2737. [DOI] [PubMed] [Google Scholar]
- 8.Duim, B., T. M. Wassenaar, A. Rigter, and J. Wagenaar. 1999. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 65:2369-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frost, J. A., J. M. Kramer, and S. A. Gillanders. 1999. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol. Infect. 123:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost, J. A., A. N. Oza, R. T. Thwaites, and B. Rowe. 1998. Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J. Clin. Microbiol. 36:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, and K. R. Neal. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer, J. M., J. A. Frost, F. J. Bolton, and D. R. Wareing. 2000. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J. Food Prot. 63:1654-1659. [DOI] [PubMed] [Google Scholar]
- 13.Lastovica, A. J., and M. B. Skirrow. 2000. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli, p. 89-120, In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 14.Lindstedt, B. A., E. Heir, T. Vardund, K. K. Melby, and G. Kapperud. 2000. Comparative fingerprinting analysis of Campylobacter jejuni subsp. jejuni strains by amplified-fragment length polymorphism genotyping. J. Clin. Microbiol. 38:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lior, H., D. L. Woodward, J. A. Edgar, L. J. Laroche, and P. Gill. 1982. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J. Clin. Microbiol. 15:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKay, D., J. Fletcher, P. Cooper, and F. M. Thomson-Carter. 2001. Comparison of two methods for serotyping Campylobacter spp. J. Clin. Microbiol. 39:1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.On, S. L. 1996. Identification methods for campylobacters, helicobacters, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.On, S. L., and C. S. Harrington. 2000. Identification of taxonomic and epidemiological relationships among campylobacter species by numerical analysis of AFLP profiles. FEMS Microbiol. Lett. 193:161-169. [DOI] [PubMed] [Google Scholar]
- 19.On, S. L., E. M. Nielsen, J. Engberg, and M. Madsen. 1998. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol. Infect. 120:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen, R. J., and S. Leeton. 1999. Restriction fragment length polymorphism analysis of the flaA gene of Campylobacter jejuni for subtyping human, animal and poultry isolates. FEMS Microbiol. Lett. 176:345-350. [DOI] [PubMed] [Google Scholar]
- 21.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 22.Patton, C. M., I. K. Wachsmuth, G. M. Evins, J. A. Kiehlbauch, B. D. Plikaytis, N. Troup, L. Tompkins, and H. Lior. 1991. Evaluation of 10 methods to distinguish epidemic-associated Campylobacter strains. J. Clin. Microbiol. 29:680-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penner, J. L., and J. N. Hennessy. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson, J. F., J. A. Frost, J. M. Kramer, R. T. Thwaites, F. J. Bolton, D. R. Wareing, and J. A. Gordon. 2001. Coinfection with Campylobacter species: an epidemiological problem? J. Appl. Microbiol. 91:206-211. [DOI] [PubMed] [Google Scholar]
- 25.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. Willems, K. E. Dingle, F. M. Colles, and J. D. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sopwith, W., M. Ashton, J. A. Frost, K. Tocque, S. O'Brien, M. Regan, and Q. Syed. 2003. Enhanced surveillance of campylobacter infection in the north west of England 1997-1999. J. Infect. 46:35-45. [DOI] [PubMed] [Google Scholar]
- 27.Tam, C. C., S. J. O'Brien, G. K. Adak, S. M. Meakins, and J. A. Frost. 2003. Campylobacter coli—an important foodborne pathogen. J. Infect. 47:28-32. [DOI] [PubMed] [Google Scholar]
- 28.Tamada, Y., Y. Nakaoka, K. Nishimori, A. Doi, T. Kumaki, N. Uemura, K. Tanaka, S. I. Makino, T. Sameshima, M. Akiba, M. Nakazawa, and I. Uchida. 2001. Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]