Abstract
A real-time reverse-transcription PCR (RRT-PCR) was developed to detect avian paramyxovirus 1 (APMV-1) RNA, also referred to as Newcastle disease virus (NDV), in clinical samples from birds. The assay uses a single-tube protocol with fluorogenic hydrolysis probes. Oligonucleotide primers and probes were designed to detect sequences from a conserved region of the matrix protein (M) gene that recognized a diverse set (n = 44) of APMV-1 isolates. A second primer-probe set was targeted to sequences in the fusion protein (F) gene that code for the cleavage site and detect potentially virulent NDV isolates. A third set, also directed against the M gene, was specific for the North American (N.A.) pre-1960 genotype that includes the common vaccine strains used in commercial poultry in the United States. The APMV-1 M gene, N.A. pre-1960 M gene, and F gene probe sets were capable of detecting approximately 103, 102, and 104 genome copies, respectively, with in vitro-transcribed RNA. Both M gene assays could detect approximately 101 50% egg infective doses (EID50), and the F gene assay could detect approximately 103 EID50. The RRT-PCR test was used to examine clinical samples from chickens experimentally infected with the NDV strain responsible for a recent epizootic in the southwestern United States. Overall, a positive correlation was obtained between the RRT-PCR results and virus isolation for NDV from clinical samples.
Newcastle disease virus (NDV), formally recognized as avian paramyxovirus 1 (APMV-1), is the etiological agent of Newcastle disease, an affliction which can cause severe losses in domestic poultry production (3). The virus is enveloped and has a negative-sense, single-stranded RNA genome of approximately 15 kb (8) which codes for six proteins, including an RNA-directed RNA polymerase (L), hemagglutinin-neuraminidase protein (HN), fusion protein (F), matrix protein (M), phosphoprotein (P), and nucleoprotein (N) (16). Newcastle disease virus isolates are characterized by virulence in chickens and may be categorized into three main pathotypes depending on severity of disease (3). Lentogenic isolates are of low virulence and may cause mild respiratory or enteric infections. Viruses of intermediate virulence that cause primarily respiratory disease are termed mesogenic, while virulent viruses that cause high mortality are termed velogenic. Velogenic forms of NDV are further classified as neurotropic or viscerotropic based on pathological manifestation (3, 4). The highly virulent types are list A agents that require reporting to the Office of International Epizootes, and their occurrence results in quarantine and trade embargoes (25).
Presently, the virulence of new NDV isolates is assessed primarily on the basis of in vivo tests, including the intracerebral pathogenicity index in 1-day-old chickens, the inoculation of embryonated eggs to determine mean death time of the embryo, and the intravenous pathogenicity index in 6-week-old chickens (4). Additionally, the virulence of NDV isolates is known to be related to the amino acid sequence at the F protein cleavage site (9, 17, 20) and the ability of specific cellular proteases to cleave the protein of different pathotypes (11, 22). Fewer basic amino acids are present in the fusion protein cleavage site of lentogenic NDV isolates than either mesogenic or velogenic strains, which have similar cleavage site sequences (20). The Office of International Epizootes now accepts reporting of the F cleavage sequence as a primary virulence determinant (25).
The first reported cases of the disease in poultry came from Java, Indonesia, and Newcastle-upon-Tyne during 1926, but the disease currently has a worldwide distribution (3). In the United States, an epizootic of the disease in California during the 1970s threatened the entire U.S. poultry and egg industry and led authorities to destroy nearly 12 million chickens. That outbreak lasted 3 years and cost $56 million to control (36). In October 2002, California again reported an outbreak of velogenic Newcastle disease (26), also referred to as exotic Newcastle disease, when the strain is not endemic to the country of detection. At the time of this writing, officials are in the final phases of exotic Newcastle disease eradication, an effort that has involved the euthanization of approximately 3.9 million infected or exposed birds and the quarantining of over 19,000 premises in four southwestern states. The total cost of these control measures has been estimated to be approximately $188 million (Elizabeth Fitzsimons, The San Diego Union-Tribune, San Diego, Calif., 5 August 2003). In such a situation, the rapid and specific detection of NDV in suspected flocks would allow more efficient decision making and thus help limit these significant economic losses involved with containment of such an outbreak.
Presently, virus isolation in embryonated chicken eggs followed by hemagglutination inhibition assays with NDV-specific antibodies represent the reference standard for virus detection (24). Although virus isolation is a sensitive and specific method, it takes several days to make a routine diagnosis. More rapid pathotyping methods based on reverse transcription (RT)-PCR followed by agarose gel electrophoresis (10, 14), sequencing RT-PCR products (30), a heteroduplex mobility assay (6), restriction endonuclease analysis of amplification products (7), and the use of fluorogenic probes (2) have all been reported. Real-time PCR and real-time reverse-transcription PCR (RRT-PCR) offer the high sensitivity afforded by conventional RT-PCR with the advantage that a post-PCR processing step is avoided, which allows a savings in both time and material (for review, see reference 19). Here, we report development of an RRT-PCR assay with fluorogenic, hydrolysis-type probes that was used to detect NDV nucleic acid in clinical samples from experimentally infected chickens.
MATERIALS AND METHODS
Primer and probe design.
The M gene sequences from 30 diverse NDV isolates from around the world were aligned with the ClustalW method available as part of the MegAlign program in the LaserGene sequence analysis package, version 5 (DNAStar, Inc. Madison, Wis.). Specific oligonucleotide primers and fluorogenic probes were designed with the Primer3 program (27). One primer-probe combination, selected from a conserved region of the M gene, was designed to amplify as many NDV isolates as theoretically possible. A second primer-probe set was selected to specifically detect only one phylogenetic cluster of NDV isolates, the North America (N.A.) pre-1960 genotype, which includes common vaccine and challenge strains used in the United States (30). This group of related strains is known as genotype II in the system proposed by Ballagi-Pordany et al. (5), Lomniczi et al. (18), and Herczeg et al. (13).
To detect exotic NDVs, particularly those from the 2002 California epizootic (26), the F gene sequence of several California isolates, related isolates from the Mexico 2000 outbreak (23), and other velogenic isolates were aligned as described above. Primers were selected to surround the fusion cleavage site, and the hydrolysis-type probe was designed to anneal directly to the fusion cleavage site. The probe selected, designated VFP-1 (virulent fusion probe 1), was a modified version of probe 1V reported by Aldous and Alexander (2). Primer and probe information is summarized in Table 1.
TABLE 1.
NDV real-time RT-PCR primer and hydrolysis probe sequences
| Specificity, target gene, and amplicon size | Primer-probe namea | Sequenceb |
|---|---|---|
| APMV-1 | M+4100 | 5′-AGTGATGTGCTCGGACCTTC-3′ |
| Matrix | M+4169 | 5′-[FAM]TTCTCTAGCAGTGGGACAGCCTGC[TAMRA]-3′ |
| 121 bp | M−4220 | 5′-CCTGAGGAGAGGCATTTGCTA-3′ |
| Velo- and mesogensc | F+4839 | 5′-TCCGGAGGATACAAGGGTCT-3′ |
| Fusion | F+4894 (VFP-1) | 5′-[FAM]AAGCGTTTCTGTCTCCTTCCTCCA[TAMRA]-3′ |
| 101 bp | F−4939 | 5′-AGCTGTTGCAACCCCAAG-3′ |
| N.A. pre-1960 genotyped | M+4213 | 5′-TCCTCAGGTGGCCAAGATAC-3′ |
| Matrix | M−4268 | 5′-[FAM]TTTTAACGCTCCGCAGGCAC[TAMRA]-3′ |
| 138 bp | M−4350 | 5′-TGCCCCTTCTCCAGCTTAGT-3′ |
Primer-probe designation refers to the nucleotide position where the 5′ end of the oligonucleotide anneals to the cDNA of the full-length NDV genome. +, sense; −, antisense.
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
This primer-probe set was designed specifically to detect isolates from the 2002 exotic NDV outbreak in the southwestern U.S. (26) and the closely related isolates of the Mexico 2000 (23) outbreak. It will also detect a wide range of other velo- and mesogenic strains (Table 2).
Includes U.S. vaccine strains chicken/US/B1/48 and chicken/US/LaSota/46.
Virus strains.
APMV-1 isolates examined in this study are listed in Table 2 with strain designation, year, and country of origin along with virulence-type designation. All isolates have been described previously (6, 29-32), except for the current game chicken/US(CA)/02 isolate.
TABLE 2.
Detection patterns of selected NDV isolates by RRT-PCR primer-probe sets
| Isolate | Patho- typea | Primer-probe set specificity (gene target)
|
||
|---|---|---|---|---|
| APMV-1 (matrix) | N.A. pre-1960 (matrix) | Velogen/ mesogen (fusion) | ||
| Chicken/US/B1/48 | L | + | + | − |
| Chicken/US/LaSota/46 | L | + | + | − |
| Chicken/Northen Ireland/ Ulster/64 | L | + | − | − |
| Chicken/Australia/QV4/66 | L | + | − | − |
| Turkey/US/VGGA/87 | L | + | + | − |
| Chicken/US(Nebraska)/54 | L | + | + | − |
| Chicken/US/23984/96 | L | + | + | − |
| Chicken/US/Kimber/47 | M | + | + | + |
| Chicken/US/Roakin/48 | M | + | + | + |
| Chicken/US(Texas)/ DK1155/59 | M | + | + | + |
| Chicken/US(Michigan)/ 46967/46 | M | + | + | + |
| Chicken/US (Mass.)/ MK107/45 | M | + | + | + |
| Anhinga/US/44083/93 | M | + | − | + |
| Mixed species/US/Largo/71 | V | + | − | + |
| Chicken/US/BeaudetteC/52 | V | + | + | + |
| Chicken/US(Texas)/GB/48 | V | + | + | + |
| Chicken/UK/Herts/33 | V | + | − | + |
| Chicken/Australia/Victoria/ 11176/32 | V | + | − | + |
| Cockatiel/US(Florida)/FL/80 | V | + | − | + |
| Cormorant/US(Minn.)/ 40068/92 | V | + | − | + |
| Turkey/US(N. Dak.)/43084/92 | V | + | − | + |
| Cockatoo/Indonesia/14698/90 | V | + | − | + |
| Parakeet/Tanz,Belg,China/ 28710/93 | V | + | − | + |
| Pigeon/US(Ga.)/21042/98 | V | + | − | + |
| Chicken/Italy/3286/00 | V | + | − | + |
| Dove/Italy/2736/00 | V | + | − | − |
| Pigeon/Italy/1166/00 | V | + | − | + |
| Chicken/US/CA1083 (Fontana)/72 | V | + | − | + |
| Game chicken/US(Ca.)/ 24255/98 | V | + | − | + |
| Parakeet/Myanmar/11592/91 | V | + | − | + |
| Chicken/Honduras/44813/00 | V | + | − | + |
| Pigeon/US(Tex.)/17498/98 | V | + | − | + |
| Chicken/Mexico/37821- 550-2/96 | V | + | − | + |
| Pheasant/US(Ca.)/F98- 1208/98 | V | + | − | + |
| Pigeon/CA/1307/75 | V | + | − | + |
| Chicken/Kenya/KRC139/90 | V | + | − | + |
| Pigeon/US(N.Y.)/84 | V | + | − | + |
| Game chicken/US(CA)/ 211472/02 | V | + | − | + |
| APMV-2/chicken/CA/ Yucaipa/56 | NA | − | − | − |
| APMV-3/turkey/Wisconsin/68 | NA | − | − | − |
| APMV-4/duck/Hong Kong/ D3/75 | NA | − | − | − |
| APMV-7/dove/Tennesse/4/75 | NA | − | − | − |
L, lentogen; M, mesogen; V, velogen. NA, not applicable.
RNA extraction and RRT-PCR.
For allantoic fluid and clinical swab samples, the Qiagen (Valencia, Calif.) RNeasy procedure was used to extract RNA following the manufacturer's recommended procedure for fluid samples with a vacuum manifold. For the RRT-PCR, the Qiagen one-step RT-PCR kit was used as previously described (34), except 25-μl reaction volumes were used.
Extensive optimization was performed on all three primer-probe sets on the following parameters: annealing temperature, MgCl2 concentration, primer concentration, probe concentration, and primer ratios. The assays were developed for use on a SmartCycler (Cepheid, Inc., Sunnyvale, Calif.). For the F gene primers, the following amounts per reaction were used: 1 μl of kit-supplied enzyme mix (including Hot Start Taq polymerase and RT), 5 μl of kit-supplied buffer (5×), 10 pmol of the reverse primer, 30 pmol of the forward primer, 6 pmol of probe, 0.8 μl of kit-supplied deoxynucleoside triphosphate s (final concentration: 320 μM each), 1.25 μl of 25 mM MgCl2 (combined with MgCl2 in kit-supplied buffer, final concentration = 3.75 mM) and 13 U of RNase inhibitor (Promega, Madison, Wis.). The reaction mixture for the APMV-1 and N.A. pre-1960 genotype M gene primers was the same as above except that the primers were used at 10 pmol each.
For each primer set, the RT step was 30 min at 50°C, followed by 15 min at 95°C. The cycling conditions for the APMV-1 matrix primers consisted of 40 cycles of 10 s of denaturation at 94°C, 30 s of annealing at 52°C, and extension at 72°C for 10 s. For the F gene primer set and N.A. pre-1960 M gene-specific set, the optimal annealing temperature was empirically determined to be 58°C.
Control RNA.
In vitro-transcribed RNA was used for positive controls and for the determination of the limit of detection for the assay. For the F gene and the APMV-1 M gene primer-probes, game chicken/US(CA)/02 cDNA was amplified with primers M629F (5′-TCGAGICTGTACAATCTTGC-3′) at positions 3884 to 3903 on the full-length NDV genome and F581R (5′-CTGCCACTGCTAGTTGIGATAATCC-3′) at positions 5054 to 5078 in the full-length NDV genome (30). This yielded a 1,195-bp product that spanned both the M and F gene target sites. The same primers were also used to amplify chicken/US/B1/48 cDNA. PCR products were cloned into TOPO-XL (Invitrogen, Carlsbad, Calif.) and linearized by endonuclease digestion at a unique site (either BamHI or SpeI) immediately downstream of the insertion. In vitro-transcribed RNA was generated from the T7 promoter following the manufacturer's recommendation (RiboMax kit, Promega Corp., Madison, Wis.). RNA was quantified spectrophotometrically following enzymatic removal of DNA.
Virus isolation and embryo titration.
Virus isolation procedures in embryonated chicken eggs followed standard protocols (4). For virus titration standards, NDV isolates with a predetermined 50% egg infective dose (EID50) were serially diluted in sterile brain heart infusion broth and used for RNA extraction.
Comparison with a previously published primer set.
The sensitivity of the F gene primer pair was compared to that of an APMV-1 F gene primer pair recently described (7). The Creelan et al. (7) primer set, NDV F and 4829/F-5031, was developed for standard RT-PCR, so it was necessary to optimize this set for the RRT-PCR procedure for the following parameters: annealing temperature, MgCl2 concentration, primer concentration, probe concentration, and primer ratios for use with the VFP-1 probe. Direct comparisons of sensitivity were made with the in vitro-transcribed game chicken/US(CA)/02 RNA and 10-fold serial dilutions of RNA extracted from clinical samples from experimentally infected chickens.
Phylogenetic analysis.
Nucleotide sequence editing, analysis, and alignments were conducted with IntelliGenetics GeneWorks 2.5 software (IntelliGenetics, Mountain View, Calif.) and the LaserGene package. Phylogenetic analysis was performed utilizing the neighbor-joining method (28) with Phylogenetic Analysis using Parsimony (PAUP) software (35). An avian paramyxovirus-2 sequence (APMV2/chicken/CA/Yucaipa/56; GenBank accession no. D13977) served as an outgroup (33) following 2,000 bootstrap replicates (12).
Detection of virus in samples from experimentally infected chickens.
For the first phase of the animal experiments, six groups of five 8-week-old Leghorn male chickens were inoculated by intraconjunctival instillation of 100 μl of phosphate-buffered saline or one of five doses of game chicken/US(CA)/2002 virus. The doses for the five virus-inoculated groups were 10-fold serial dilutions ranging from 101.9 to 105.9 EID50/bird (see Table 4). Oropharyngeal and cloacal swabs were collected into brain-heart infusion broth on day 2 and day 4 postinfection from each bird for comparison of virus isolation to RRT-PCR.
TABLE 4.
Results of RRT-PCR and virus isolation from oral and cloacal swabs collected from chickens experimentally inoculated intraconjunctivally with game chicken/US(CA)/2002 at 8 weeks of age
| Group | Bird no. | Strain and dose | Result at 2 days postinfection
|
Result at 4 days postinfection
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RRT-PCR fusion
|
RRT-PCR APMV-1 matrix
|
Virus isolation
|
RRT-PCR fusion
|
RRT-PCR APMV-1 matrix
|
Virus isolation
|
|||||||||
| Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | |||
| 1 | 434 | PBS | − | − | − | − | − | − | − | − | − | − | − | − |
| 435 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 436 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 437 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 438 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 2 | 439 | CA/02 101.9 | − | − | − | − | − | − | − | − | − | − | − | − |
| 440 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 441 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 442 | − | − | − | − | − | − | − | − | − | − | − | + | ||
| 443 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 3 | 444 | CA/02 102.9 | − | − | − | − | − | − | − | − | − | − | − | + |
| 445 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 446 | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 447 | − | − | − | − | + | + | + | + | + | + | + | + | ||
| 448 | − | − | − | + | − | + | + | + | + | + | + | + | ||
| 4 | 449 | CA/02 103.9 | − | − | − | − | + | + | + | + | + | + | + | + |
| 450 | − | − | − | − | − | + | − | + | − | + | + | + | ||
| 451 | − | + | − | + | − | + | + | + | + | + | + | + | ||
| 452 | − | + | − | + | + | + | + | + | + | + | + | + | ||
| 453 | − | + | − | + | − | + | + | + | + | + | + | + | ||
| 5 | 454 | CA/02 104.9 | − | − | − | + | + | + | + | + | + | + | + | + |
| 455 | − | − | − | + | + | + | + | + | + | + | + | + | ||
| 456 | − | − | − | + | + | + | + | + | + | + | + | + | ||
| 457 | − | + | − | + | + | + | + | + | + | + | + | + | ||
| 458 | − | + | − | + | + | + | + | + | + | + | + | + | ||
| 6 | 459 | CA/02 105.9 | − | + | + | + | + | + | + | + | + | + | + | + |
| 460 | − | − | − | + | + | + | + | + | + | + | + | + | ||
| 461 | − | + | + | + | + | + | + | + | + | + | + | + | ||
| 462 | − | − | + | + | + | + | + | + | + | + | + | + | ||
| 463 | − | + | − | + | + | + | + | + | + | + | + | + | ||
The second group of animal experiments consisted of three sets of eight 8-week-old birds. Two of the groups received an initial intraocular vaccination of chicken/US/B1/48, at either a low (103.9 EID50) or high (106.9 EID50) dose (see Table 5). The third group received only phosphate-buffered saline. Seroconversion of vaccinated chickens was determined by a conventional microtiter hemagglutination inhibition method with 4 hemagglutination (HA) units of inactivated NDV-LaSota antigen (15). At 2 weeks postvaccination, all birds, including controls, were challenged by intraconjunctival instillation of 105.9 EID50 of game chicken/US(CA)/2002. Oral and cloacal swabs were taken at day 2, day 4, and day 6 postchallenge. Also, swab samples were obtained from all birds 3 days after vaccination.
TABLE 5.
Results of RRT-PCR and virus isolation from oral and cloacal swabs collected from chickens vaccinated with either a high or low dose of chicken/US/B1/48 and challenged 2 weeks postvaccination with 105.9 EID50 of game chicken/US(CA)/02
| Vaccine dose | Bird no. | Result at 3 days postvaccination
|
Result at 2 days postchallenge
|
Result at 4 days postchallenge
|
Result at 6 days postchallenge
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RRT-PCR N.A. pre-1960
|
RRT-PCR fusion
|
Virus isolation
|
RRT-PCR fusion
|
RRT-PCR APMV-1 matrix
|
Virus isolation
|
RRT-PCR Fusion
|
RRT-PCR APMV-1 Matrix
|
Virus isolation
|
RRT-PCR Fusion
|
RRT-PCR APMV-1 Matrix
|
Virus isolation
|
||||||||||||||
| Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | Cloacal | Oral | ||
| 106.9 (EID50) | 464 | NDa | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 465 | ND | + | ND | − | + | + | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | |
| 466 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 467 | ND | + | ND | − | − | + | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | |
| 468 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 469 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | |
| 470 | ND | + | ND | − | − | + | − | − | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | |
| 471 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 103.9 | 472 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| 473 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 474 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 475 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 476 | ND | + | ND | − | + | + | − | − | − | − | − | + | − | − | − | − | − | + | − | + | − | + | − | + | |
| 477 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | |
| 478 | ND | + | ND | − | − | + | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | |
| 479 | ND | + | ND | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| None | 480 | ND | − | ND | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 481 | ND | − | ND | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 482 | ND | − | ND | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | Dead | Dead | Dead | Dead | Dead | Dead | |
| 483 | ND | − | ND | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | Dead | Dead | Dead | Dead | Dead | Dead | |
| 484 | ND | − | ND | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | Dead | Dead | Dead | Dead | Dead | Dead | |
| 485 | ND | − | ND | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | Dead | Dead | Dead | Dead | Dead | Dead | |
| 486 | ND | − | ND | − | − | − | − | + | − | + | + | + | + | + | + | + | + | + | Dead | Dead | Dead | Dead | Dead | Dead | |
| 487 | ND | − | ND | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
ND, not determined
Nucleotide sequencing.
The full-length M gene and partial F gene sequence of the game chicken/US(CA)/02 virus were amplified with primers M26F (5′-TGTGCCAAGATGGACTCA-3′), corresponding to positions 3281 to 3298 on the full NDV genome, and F581R. The amplification product was cloned as described above, and double-stranded nucleotide sequencing reactions with fluorescently labeled dideoxynucleotide terminators were completed with an ABI 3700 automated sequencer (Applied Biosystems Inc., Foster City, Calif.). Nucleotide sequence editing and analysis were conducted with Lasergene software.
Nucleotide sequence accession number.
The sequence game chicken/US(CA)/211472/02) has been deposited in GenBank and assigned accession number AY246050.
RESULTS
Primer and probe specificity for NDV isolates.
The specificity of the primer-probe sets was examined with RNA extracted from a diverse array of NDV isolates collected worldwide (Table 2). The NDV primer-probe set directed to the M gene detected every APMV-1 specimen tested and did not amplify the other APMV serotypes examined. The N.A. pre-1960 genotype set was only positive with members of the target group. Although the F gene primers were originally created to specifically amplify isolates from the California 2002 outbreak, they appear to have much broader specificity, as they could detect the vast majority of velogenic and mesogenic isolates examined when used in combination with the VPF-1 probe.
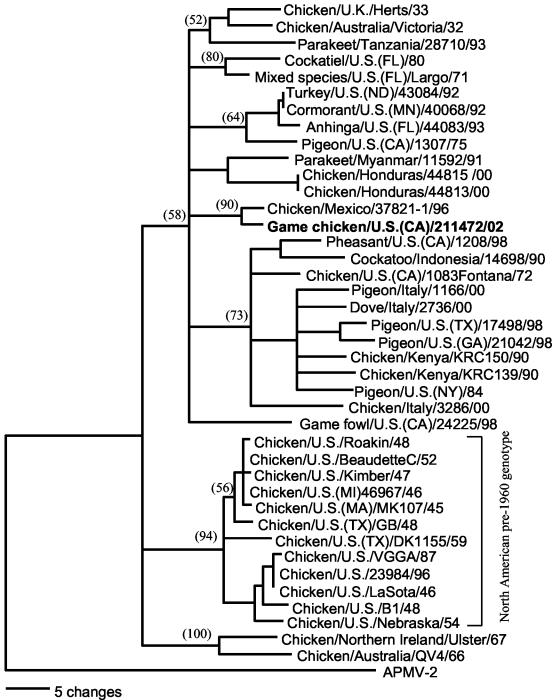
The phylogenetic relationship of the NDV strains used in this study is depicted in Fig. 1. All the virulent, velogenic NDV isolates obtained after 1960 to the present are related to chicken/Australia/Victoria/32 as the earliest possible progenitor-type virus. Recent NDV isolates highly virulent for chickens are all related to viruses considered exotic to North America. This included chicken/U.S./CA1083(Fontana)/72 and cormorant/U.S.(MN)/40068/92, isolated during previous outbreaks of Newcastle disease in the United States, as well as viruses isolated from poultry in Mexico during 1996 and the recent California isolate from the outbreak that began during the autumn of 2002. Representative virulent NDV isolates recovered in the United States prior to 1970 contain only neurotropic viruses and are not phylogenetically related to isolates obtained since the chicken/U.S./CA1083(Fontana)/72 strain.
FIG. 1.
Phylogenetic analysis of nucleotide sequences from Newcastle disease virus fusion protein genes. Isolates include those examined by RRT-PCR as listed in Table 2. The phylogenetic tree was generated with the neighbor-joining method (28) following alignment of sequences. Numbers represent bootstrap confidence limits following 2,000 replications (12) with the avian paramyxovirus type 2 (APMV-2) sequence as an outgroup.
Sensitivity with in vitro-transcribed RNA and detection during virus titration.
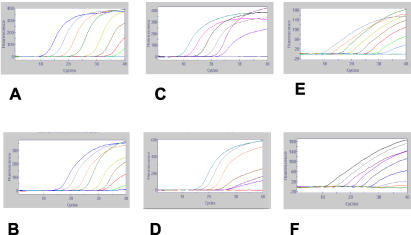
Representative SmartCycler fluorographs illustrating sensitivity measurements are presented in Fig. 2. Overall, the RRT-PCR primer-probe sets were capable of detecting from 102 to 104 copies of NDV RNA and as few as 10 infectious particles. For the APMV-1 M gene set, the procedure could consistently detect in the range of 103 copies of the target gene (Fig. 2A) and approximately 10 EID50 (Fig. 2B). The minimum copy number that could be detected consistently with the F gene set was 104 (Fig. 2C), and the procedure could detect an EID50 of 103 (Fig. 2D). For the N.A. pre-1960 genotype set, the limit of detection was in the range of 102 copies (Fig. 2E) and 10 EID50 (Fig. 2F).
FIG. 2.
SmartCycler fluorographs illustrating 10-fold serial dilutions used to determine limits of detection of the RRT-PCR assays for viral nucleic acid. For the APMV-1 matrix gene with in vitro-transcribed game chicken/US(CA)/02 RNA (A), the limit of detection was approximately 103 copies of the target gene, and the assay could detect approximately 10 EID50 (B). For the fusion gene assay, the limit of detection as measured with in vitro-transcribed game chicken/CA/02 RNA (C) was approximately 104 target copies, and the assay could detect approximately 103 EID50 (D). The N.A. pre-1960 genotype specific assay could detect approximately 102 target copies of chicken/US/B1/48 RNA (E) and approximately 10 EID50 (F).
Comparison with a previously published F gene primer pair.
With the in vitro-transcribed game chicken/US(CA)/02 RNA, the limit of detection for the Creelan et al. (7) primer set was approximately 105 copies of the target gene (data not shown). We also directly compared 10-fold serial dilutions of RNA extracted from clinical samples from experimentally infected chickens (Table 3). Generally, the F gene primers described in this investigation detected more samples and lower concentrations of RNA than the Creelan et al. (7) primer pair.
TABLE 3.
RRT-PCR cycle threshold values of two F gene primer sets with diluted clinical samples
| Sample no. | Dilution | Primer set cycle thresholds (Ct)a
|
Ct difference | |
|---|---|---|---|---|
| F+4829/F−5031b | F+4839/F−4939c | |||
| 12 | 100 | 26.39 | 25.45 | 0.94 |
| 10−1 | 0 | 29.44 | NAd | |
| 10−2 | 0 | 0 | 0 | |
| 14 | 100 | 22.53 | 21.39 | 1.14 |
| 10−1 | 26.58 | 24.46 | 2.12 | |
| 10−2 | 30.53 | 27.95 | 2.58 | |
| 10−3 | 0 | 32.16 | NA | |
| 10−4 | 0 | 0 | 0 | |
| 15 | 100 | 0 | 30.69 | NA |
| 10−1 | 0 | 0 | 0 | |
| 18 | 100 | 22.90 | 22.14 | 0.76 |
| 10−1 | 26.36 | 25.42 | 0.94 | |
| 10−2 | 29.95 | 28.87 | 1.08 | |
| 10−3 | 0 | 35.83 | NA | |
| 10−4 | 0 | 0 | 0 | |
| 19 | 100 | 0 | 29.86 | NA |
| 10−1 | 0 | 35.41 | NA | |
| 10−2 | 0 | 0 | 0 | |
| 29 | 100 | 0 | 29.19 | NA |
| 10−1 | 0 | 35.55 | NA | |
| 10−2 | 0 | 0 | 0 | |
| 35 | 100 | 21.30 | 21.31 | −0.01 |
| 10−1 | 24.83 | 24.26 | 0.57 | |
| 10−2 | 28.91 | 27.45 | 1.46 | |
| 10−3 | 0 | 36.98 | NA | |
| 10−4 | 0 | 0 | 0 | |
| 36 | 100 | 27.15 | 23.73 | 3.42 |
| 10−1 | 0 | 26.19 | NA | |
| 10−2 | 0 | 29.78 | NA | |
| 10−3 | 0 | 0 | 0 | |
| 38 | 100 | 21.09 | 21.19 | −0.1 |
| 10−1 | 25.09 | 23.67 | 1.42 | |
| 10−2 | 29.25 | 27.06 | 2.19 | |
| 10−3 | 30.74 | 31.65 | −0.91 | |
| 10−4 | 0 | 34.66 | NA | |
Cycle threshold (Ct) was defined as the first cycle where detected fluorescence was 30 units over background.
Creelan et al. (7).
This report.
NA, not applicable.
Detection of virus in samples from experimentally infected chickens.
During the first phase of the animal experiment, five groups of five birds each were inoculated with a serially increasing dose of game chicken/US(CA)/02. Virus isolation results and RRT-PCR results with both the F gene and APMV-1 M gene assays are presented in Table 4. At 2 days postinfection, three cloacal swabs, all at the 105.9 EID50 dose, were positive with the M gene RRT-PCR. Among the oral swabs at 2 days postinfection, 8 of the 17 (47.1%) samples positive by virus isolation were also detectable with the F gene assay, and 14 of the 17 (82.4%) were positive with the APMV-1 M gene procedure. By 4 days postinfection, 88.2% of the cloacal swabs that were eventually positive by virus isolation were also positive with the F gene and the APMV-1 M gene RRT-PCR. With oral swabs the number of virus isolation positives that were also positive by both these assays increased to 89.5%.
Over the course of the second phase of the animal study, groups of chickens were vaccinated with either a high (106.9 EID50) or low (103.9 EID50) dose of chicken/US/B1/48. Birds were subsequently challenged 2 weeks later with 105.9 EID50 of game chicken/US(CA)/02. All chickens in the vaccinated groups seroconverted, and all controls were uniformly negative prior to challenge. Both the low- and high-dose vaccinated groups had identical geometric mean hemagglutination inhibition titers of 256, although there was slight variation among individual titers.
The birds were swabbed 3 days postvaccination, and all oral swabs were positive for virus isolation and RRT-PCR with the N.A. pre-1960 genotype specific primer-probe set (Table 5). As expected, these samples were all negative with the velogen-specific F gene primer-probe set. Two days after challenge with virulent NDV, five of the oral swabs from vaccinated birds were virus isolation positive. RNA extracted from the oral swab from vaccinated bird 470 was positive with APMV-1 M gene RRT-PCR, but the remaining samples were all negative. At this point, all the oral swab samples from the challenge controls were positive by both virus isolation and APMV-1 M gene RRT-PCR, and 62.5% of these were also positive with the F gene assay.
Swab samples from vaccinated birds at 4 days postchallenge were negative by virus isolation except for the oral sample from one bird (no. 476) that received the low dose of vaccine. Three of the combined 32 oral and cloacal swabs from this group were weakly positive by APMV-1 M gene RRT-PCR, but all were negative with the F gene RRT-PCR assay. At this time point (4 days postinfection), both cloacal and oral swab specimens from all the unvaccinated challenge controls were positive by all three methods.
Swabs taken 6 days after challenge showed that all the vaccinated birds remained negative by virus isolation except for the oral swab from bird 476. This specimen was the only one of the group that was also positive by both the F gene and APMV-1 M gene RRT-PCR.
DISCUSSION
The objective of this study was to develop a rapid real-time RT-PCR for detection and differentiation of Newcastle disease viruses in avian clinical samples. To accomplish this aim, we first focused on an assay designed to detect all APMV-1 isolates. Due to the known high sequence heterogeneity among APMV-1 genomes (18, 30, 31), the design of PCR primers that can amplify every APMV-1 isolate has proven to be somewhat problematic (1). The majority of such efforts have focused on the F gene (1), but an examination of the M gene sequence from a diverse array of isolates revealed a relatively conserved region near the 5′ end of the gene (32). Although the oligonucleotides used in this study are not an exact match to all APMV-1 isolates whose sequence is available in the databases, the most heterologous sequence examined, that of broiler chicken/Italy/3286/00, was still positive with the APMV-1 primer-probe combination. Evidently the RT-PCR conditions used were not too stringent to exclude isolates with a small number of primer-probe nucleotide mismatches, yet the assay was specific enough to prevent a positive reaction with the other APMV serotypes tested.
A similar phenomenon was observed with F gene primer-probe set. The original intention was to design primer-probe combinations useful for the rapid detection of RNA from the current game chicken/US(CA)/02 isolate for use in controlling the 2002-2003 outbreak of this virus in the southwestern United States. However, specificity testing of this set on a panel of isolates demonstrated that we could detect a diverse array of velogenic and mesogenic APMV-1 strains. The only member of the panel that was negative with this set, dove/Italy/2736/00, is an isolate known to have an unusual fusion cleavage amino acid sequence with five basic amino acids in series that includes a lysine residue at position 114 (39). This is in contrast to most other APMV-1 strains, which have a glutamine at this position (30, 31).
The N.A. pre-1960 primer-probe set was devised for use to detect vaccine strains, such as chicken/US/B1/48 and chicken/US/LaSota/46, which may be present in clinical swabs from recently vaccinated birds. It bears mentioning that this test is not specific for all lentogenic strains; vaccine strains used outside the United States, such as chicken/Northern Ireland/Ulster/64 and chicken/Australia/QV4/66, form a separate phylogenetic cluster, genotype I (5, 13, 18, 30, 31), that is not detected with this primer-probe set. The N.A. pre-1960 primer-probe set will also react positively with some virulent strains that are also members of genotype II originally found in North America pre-1960, such as chicken/US/BeaudetteC/52 and chicken/US(Texas)/GB/48. These isolates are not known to presently circulate among free-living birds or commercial poultry and are used currently only as experimental challenge strains (30, 31).
The assays that targeted the M gene, both the APMV-1 and N.A. pre-1960 genotype, were substantially more sensitive than the F gene assay. It is unclear why this is the case, but it may be related to sequence-specific phenomenon such as secondary structure, primer-dimers, etc., that may occur with F gene primers or target. The one-step RT-PCR method used here, although providing ease of use and reduced chance for cross-contamination, is potentially less sensitive than two-step RT-PCR (21). However, since this assay was designed to be used during an outbreak of exotic Newcastle disease where high throughput is important, the decrease in sensitivity was determined to be a necessary compromise.
During the first phase of the animal experiment, the correlation of M gene APMV-1 RRT-PCR-positive specimens with virus isolation positives over the two sampling days was 88.6% with oral swab samples but only 57.6% with cloacal swabs. However, the correlation of virus isolation positives from cloacal swabs to APMV-1 RRT-PCR positives increased to 94.1% (16 of 17) when the comparison was limited to samples collected at 4 days postinfection Fewer RRT-PCR-positive cloacal swabs among the early samples may be due to the presence of PCR inhibitors in these samples (38), but it is more likely virus had not yet circulated systemically at this stage (1 to 2 days postinfection), and extensive shedding in the cloaca was therefore minimal.
As may be expected by the sensitivity difference between the APMV-1 M gene assay and the velogenic F gene RRT-PCR, the APMV-1 primer-probe set consistently detected viral RNA earlier in the infection than the F gene set. However, by 4 days postinfection both sets were essentially equivalent in their ability to detect the game chicken/US(CA)/02 virus in both oral and cloacal sample types, with an overall positive correlation with virus isolation of >90%. The RRT-PCR assay is intended to be used to screen on a per flock basis, and the stage of infection will vary with individual birds. Therefore, negative results for the birds in the earliest stages of infection would be acceptable if other birds at later stages (>2 days postinfection) are sampled concurrently.
The second phase of the animal experiment demonstrated the efficacy of vaccination with chicken/US/B1/48. Among vaccinated chickens, virus could be isolated in only 5 of the 32 swab samples taken 2 days postchallenge. No virulent virus could be isolated from the vaccinated birds 4 days postchallenge, with the exception of the oral swab from bird 476, which was virus isolation positive as well as M and F gene RRT-PCR positive at 4 and 6 days postchallenge. Four swab samples were weakly positive with the APMV-1 M gene RRT-PCR but negative by virus isolation. Previous investigations have suggested that NDV isolation frequencies decrease following increased NDV antibody titers (10, 37), so these results may be due to the effect of neutralizing antibodies.
In summary, we report a RRT-PCR assay for the detection of APMV-1 that correlates well with virus isolation yet offers the advantage of being much faster. Although the fact that viral nucleic acids were not detected in all samples that were virus isolation positive suggests that RRT-PCR cannot replace virus isolation completely on an individual sample basis, this technique should be useful for rapid screening and surveillance of at-risk poultry flocks.
Acknowledgments
Appreciation is extended to Melissa Scott, Joyce Bennett, Phillip Curry, Suzanne DeBlois, and Tracy Smith-Faulkner for technical assistance.
These studies were supported by ARS, USDA, CRIS project number 6612-32000-015-00D-085, and Homeland Security Funds 346-6612-480.
REFERENCES
- 1.Aldous, E. W., and D. J. Alexander. 2001. Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol. 30:117-128. [DOI] [PubMed] [Google Scholar]
- 2.Aldous, E. W., M. S. Collins, A. McGoldrick, and D. J. Alexander. 2001. Rapid pathotyping of Newcastle disease virus (NDV) with fluorogenic probes in a PCR assay. Vet. Microbiol. 80:201-212. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, D. J. 1997. Newcastle disease and other Paramyxoviridae infections, p. 541-569. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. McDougald, and J. Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University, Ames, Iowa.
- 4.Alexander, D. J. 1998. Newcastle disease virus and other avian paramyxoviruses, p. 156-168. In D. E. Swayne, J. R. Glisson, M. W. Jackwood, J. E. Pearson, and W. M. Reed (ed.), A laboratory manual for the isolation and identification of avain pathogens, 4th ed. American Association of Avian Pathologists, Kennett Square, Pa.
- 5.Ballagi-Pordany, A., E. Wehmann, J. Herczeg, S. Belak, and B. Lomniczi. 1996. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch. Virol. 141:243-261. [DOI] [PubMed] [Google Scholar]
- 6.Berinstein, A., H. S. Sellers, D. J. King, and B. S. Seal. 2001. Use of a heteroduplex mobility assay to detect differences in the fusion protein cleavage site coding sequence among Newcastle disease virus isolates. J. Clin. Microbiol. 39:3171-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creelan, J. L., D. A. Graham, and S. J. McCullough. 2002. Detection and differentiation of pathogenicity of avian paramyxovirus serotype 1 from field cases with one-step reverse transcriptase-polymerase chain reaction. Avian Pathol. 31:493-499. [DOI] [PubMed] [Google Scholar]
- 8.de Leeuw, O., and B. Peeters. 1999. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J. Gen. Virol. 80:131-136. [DOI] [PubMed] [Google Scholar]
- 9.Glickman, R. L., R. J. Syddall, R. M. Iorio, J. P. Sheehan, and M. A. Bratt. 1988. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J. Virol. 62:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gohm, D. S., B. Thuer, and M. A. Hofmann. 2000. Detection of Newcastle disease virus in organs and faeces of experimentally infected chickens with RT-PCR. Avian Pathol. 29:143-152. [DOI] [PubMed] [Google Scholar]
- 11.Gotoh, B., Y. Ohnishi, N. M. Inocencio, E. Esaki, K. Nakayama, P. J. Barr, G. Thomas, and Y. Nagai. 1992. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J. Virol. 66:6391-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedges, S. B. 1992. The number of replications needed for accurate estimation of the bootstrap P value in phylogenetic studies. Mol. Biol. Evol. 9:366-369. [DOI] [PubMed] [Google Scholar]
- 13.Herczeg, J., E. Wehmann, R. R. Bragg, P. M. Travassos Dias, G. Hadjiev, O. Werner, and B. Lomniczi. 1999. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch. Virol. 144:2087-2099. [DOI] [PubMed] [Google Scholar]
- 14.Kant, A., G. Koch, D. J. Van Roozelaar, F. Balk, and A. Ter Huurne. 1997. Differentiation of virulent and non-virulent strains of Newcastle disease virus within 24 hours by polymerase chain reaction. Avian Pathol. 26:837-849. [DOI] [PubMed] [Google Scholar]
- 15.King, D. J. 1999. A comparison of the onset of protection induced by Newcastle disease virus strain B1 and a fowl poxvirus recombinant Newcastle disease vaccine to a viscerotropic velogenic Newcastle disease virus challenge. Avian Dis. 43:745-755. [PubMed] [Google Scholar]
- 16.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: The viruses and their replication, p. 1177-1204. In B. N. Fields, D. M. Knipe, and P. M. e. a. Howley (ed.), Fields Virology, 3rd ed. Lippincott, Philadelphia, Pa.
- 17.Le, L., R. Brasseur, C. Wemers, G. Meulemans, and A. Burny. 1988. Fusion (F) protein gene of Newcastle disease virus: sequence and hydrophobicity comparative analysis between virulent and avirulent strains. Virus Genes 1:333-350. [DOI] [PubMed] [Google Scholar]
- 18.Lomniczi, B., E. Wehmann, J. Herczeg, A. Ballagi-Pordany, E. F. Kaleta, O. Werner, G. Meulemans, P. H. Jorgensen, A. P. Mante, A. L. Gielkens, I. Capua, and J. Damoser. 1998. Newcastle disease outbreaks in recent years in western Europe were caused by an old (virus isolation) and a novel genotype (VII). Arch Virol. 143:49-64. [DOI] [PubMed] [Google Scholar]
- 19.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai, Y., H. D. Klenk, and R. Rott. 1976. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology 72:494-508. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, S., S. Katamine, T. Yamamoto, S. K. Foung, T. Kurata, Y. Hirabayashi, K. Shimada, S. Hino, and T. Miyamoto. 1993. Amplification and detection of a single molecule of human immunodeficiency virus RNA. Virus Genes 7:325-338. [DOI] [PubMed] [Google Scholar]
- 22.Ogasawara, T., B. Gotoh, H. Suzuki, J. Asaka, K. Shimokata, R. Rott, and Y. Nagai. 1992. Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo. EMBO J. 11:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Office of International Epizootes. 2000. Disease outbreaks reported to Office of International Epizootes in May-June 2000: Newcastle disease. OIE Bull. 112:250-253. [Google Scholar]
- 24.Office of International Epizootes. 2000. Manual of standards for diagnostic tests and vaccines, 4th ed. Office of International Epizootes, Paris, France.
- 25.Office of International Epizootes. 2000. Newcastle disease. International Health Code, 9th ed. Office of International Epizootes, Paris, France.
- 26.Office of International Epizootes. 2002. Newcastle disease in the United States of America. OIE Bull. 15. [Online.] http://www.oie.int.
- 27.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In M. S. Krawetz S (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 28.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 29.Seal, B. S., J. M. Crawford, H. S. Sellers, D. P. Locke, and D. J. King. 2002. Nucleotide sequence analysis of the Newcastle disease virus nucleocapsid protein gene and phylogenetic relationships among the Paramyxoviridae. Virus Res. 83:119-129. [DOI] [PubMed] [Google Scholar]
- 30.Seal, B. S., D. J. King, and J. D. Bennett. 1995. Characterization of Newcastle disease virus isolates by reverse transcription PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J. Clin. Microbiol. 33:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seal, B. S., D. J. King, D. P. Locke, D. A. Senne, and M. W. Jackwood. 1998. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J. Clin. Microbiol. 36:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seal, B. S., D. J. King, and R. J. Meinersmann. 2000. Molecular evolution of the Newcastle disease virus matrix protein gene and phylogenetic relationships among the paramyxoviridae. Virus Res. 66:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Smith, A. B. 1994. Rooting molecular trees: problems and strategies. J. Linn. Soc. 51:279-292. [Google Scholar]
- 34.Spackman, E., D. A. Senne, T. J. Myers, L. L. Bulaga, L. P. Garber, M. L. Perdue, K. Lohman, L. T. Daum, and D. L. Suarez. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swofford, D. 1998. PAUP*4.0: phylogenetic analysis with parsimony. Smithsonian Institution. Sinauer Associates, Inc. Sunderland, Mass.
- 36.Utterback, W. W., and J. H. Schwartz. 1973. Epizootiology of velogenic viscerotropic Newcastle disease in southern California, 1971-1973. J. Am. Vet. Med. Assoc. 163:1080-1088. [PubMed] [Google Scholar]
- 37.Westbury, H. A., G. Parsons, and W. H. Allan. 1984. Duration of excretion of virulent Newcastle disease virus following challenge of chickens with different titres of serum antibody to the virus. Aust. Vet. J. 61:44-46. [DOI] [PubMed] [Google Scholar]
- 38.Wilde, J., J. Eiden, and R. Yolken. 1990. Removal of inhibitory substances from human faecal specimens for detection of group A rotaviruses by reverse transcriptase and PCR. J. Clin. Microbiol. 28:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanetti, F., M. Rodriguez, D. J. King, I. Capua, E. Carrillo, B. S. Seal, and A. Berinstein. 2003. Matrix protein gene seauence analysis of avian paramyxovirus 1 isolates obtained from pigeons. Virus Genes 26:199-206. [DOI] [PubMed] [Google Scholar]