Abstract
During July 2000 and October 2001, a total of 595 clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) were collected from six medical centers distributed in northern, central, and southern Taiwan. Specimen sources included blood (n = 279), pus (n = 173), sputum (n = 94), body fluids (n = 21), catheter tips (n = 20), and urine (n = 8). Pulsed-field gel electrophoresis (PFGE) with SmaI digestion was used to fingerprint these isolates. A total of 31 genotypes with 97 type-subtypes were identified. Subtypes could be identified in 7 genotypes. While there were 6 to 15 genotypes in each hospital, 433 isolates (73%) were shown to belong to a major type (genotype A, with 29 subtypes). This genotype was not only the type prevailing in all six hospitals but also the predominant clone in each hospital, accounting for 46 to 89% of all isolates in each hospital. Genotype C (16 subtypes) was the second dominant genotype, accounting for 9% of all isolates, and was distributed in five hospitals. Genotypes D (11 subtypes), E (5 subtypes), and B (6 subtypes) were distributed in five, four, and three hospitals, respectively. The other 26 types (30 type-subtypes) were minor. We conclude that the majority of MRSA clinical isolates shared a common PFGE pattern, indicating the presence of a single, epidemic MRSA clone prevailing in major hospitals in Taiwan.
Methicillin-resistant Staphylococcus aureus (MRSA) was first reported in the United Kingdom in 1961 (17), soon after the introduction of methicillin. Over the next 10 years, increasing numbers of isolates and outbreaks were reported, mainly in European countries (3). After a decline in the 1970s, new epidemic strains that differed from the original MRSAs emerged in Australia (20), the United States (4), and the Irish Republic (5) in the late 1970s and early 1980s and have now reached global proportions (3).
In Taiwan, MRSA was first documented in the early 1980s (8). The incidence of nosocomial MRSA infections increased remarkably in the 1990s (9). In 2000, MRSA accounted for 53 to 83% of all S. aureus clinical isolates in 12 major hospitals (16), and we believe that this is also true in most large hospitals in Taiwan.
Geographic spread of one or several MRSA clones in a city (21, 22), in a country (2, 13, 24, 28, 31), and even between countries (25, 30) and continents (1, 3) has been reported and proven by molecular evidence. In Taiwan, the islandwide molecular epidemiology of MRSA isolates has not yet been studied extensively, although a study has been reported (33). Hence, we conducted this study of 595 clinical isolates from six major hospitals to delineate the relationship among clinical MRSA isolates from different hospitals in Taiwan.
MATERIALS AND METHODS
A total of 595 MRSA clinical isolates were collected from six major hospitals in Taiwan from July 2000 to October 2001. Hospitals I, II, and III were in northern Taiwan, hospitals IV and V were in central Taiwan, and Hospital VI was in southern Taiwan. At times evenly distributed during the study period, the MRSA isolates were randomly selected from the strains stocked in hospitals I, II, III, and VI. In hospital IV, consecutive MRSA isolates were collected during July and September 2001, and in hospital V, consecutive MRSA bloodstream isolates were collected from July to December 2000. No duplicate isolates from a single patient were included. The number and sources of all clinical isolates from each hospital are given in Table 1. Identification of MRSA was confirmed according to the recommendations of the National Committee for Clinical Laboratory Standards.
TABLE 1.
Distribution of the sources of 595 MRSA clinical isolates from six major hospitals in Taiwan, 2000-2001
| Hospital | No. of Isolates | No. of isolates from source
|
|||||
|---|---|---|---|---|---|---|---|
| Blood | Pus | Sputum | Body fluid | Urine | CVC tipa | ||
| I | 91 | 49 | 26 | 14 | 1 | 1 | 0 |
| II | 85 | 50 | 12 | 8 | 15 | 0 | 0 |
| III | 107 | 9 | 60 | 30 | 1 | 0 | 7 |
| IV | 100 | 9 | 42 | 32 | 3 | 4 | 10 |
| V | 112 | 112 | 0 | 0 | 0 | 0 | 0 |
| VI | 100 | 50 | 33 | 10 | 1 | 3 | 3 |
| Total | 595 | 279 | 173 | 94 | 21 | 8 | 20 |
CVC, central venous catheter.
The genotyping method used in this study was pulsed-field gel electrophoresis (PFGE), which was performed according to the procedure described previously, with some modifications (27). Bacterial colonies grown overnight on blood agar plates were suspended in 10 mM Tris-0.1 mM EDTA and cast into gel plugs. The plugs were treated in lysis solution (6 mM Tris-HCl [pH 7.6], 1 M NaCl, 100 mM EDTA [pH 7.5], 0.5% Brij, 0.2% deoxycholate, 0.5% sodium lauroyl sarcosine, 30 μg of RNase [DNase free] per ml, 1 mg of lysozyme per ml) with 1 mg of lysostaphin/ml at 37°C for 24 h and were further incubated in ESP buffer (0.5 M EDTA [pH 9 to 9.5], 1% sodium lauroyl sarcosine, 500 μg of proteinase K per ml) at 50°C for 24 h. Plugs were thoroughly washed; then thin slices of the DNA plugs were cut and incubated overnight with 50 U of SmaI (New England Biolabs, Beverly, Mass.) at 25°C. Plugs were then loaded onto a 1% agarose gel, and PFGE was carried out with a CHEF Mapper XA system (Bio-Rad Laboratories) at 14°C. An autoalgorithm mode was chosen, with the running molecular sizes ranging from 30 to 500 kb. The gel was stained with ethidium bromide and photographed with UV illumination.
The criteria proposed by Tenover et al. were employed to analyze the DNA fingerprints generated by PFGE (29). Briefly, strains with banding patterns identical in the size and number of bands were considered genetically indistinguishable and assigned to the same type; strains with banding patterns that differed by only three or fewer bands were considered closely related and described as subtypes of a given clonal type; and strains with banding patterns that differed by four or more bands were considered different and assigned to separate types.
RESULTS
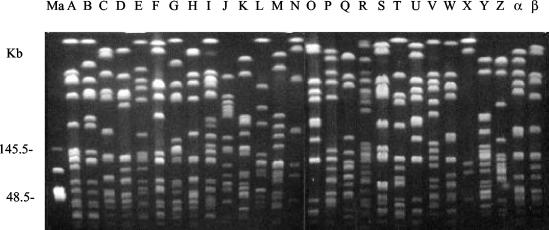
Among the 595 MRSA isolates, a total of 31 genotypes with 97 type-subtypes were identified by PFGE. Subtypes could be identified in seven genotypes (A, B, C, D, E, F, and H). There were a total of 29 subtypes in genotype A, 6 subtypes in genotype B, 16 subtypes in genotype C, 11 subtypes in genotype D, 5 subtypes in genotype E, 4 subtypes in genotype F, and 2 subtypes in genotype H. The banding patterns of most genotypes are shown in Fig. 1. The 31 genotypes found comprised 15 each from hospitals I and VI and 6 each from hospitals II, III, IV, and V. The distribution of PFGE patterns of MRSA isolates in each hospital is shown in Table 2. Among the 595 MRSA isolates, 433 isolates (73%) were shown to belong to a major genotype (type A, with 29 subtypes). This MRSA clone was the predominant clone in each hospital, accounting for 46 to 89% of isolates in each hospital. Genotype C (16 subtypes) was the second dominant clone; it was prevalent in five hospitals, particularly in hospital VI (22%), and accounted for 9% of all isolates. Genotypes D (11 subtypes) and B (6 subtypes) were distributed in five and three hospitals, respectively, and accounted for 6 and 5% of all isolates, respectively. Genotype E (5 subtypes) was distributed in four hospitals, but only eight strains belonged to this type. The other 35 isolates belonged to 26 other, minor genotypes.
FIG. 1.
PFGE patterns of SmaI-digested genomic DNA from MRSA isolates. Ma, lambda DNA concatemer standard; A to β, 28 various genotypes.
TABLE 2.
Resolution and distribution of PFGE patterns of 595 MRSA isolates from six major hospitals in Taiwan, 2000-2001
| Hospital | No. of isolates | No. of genotypes | No. of type-subtypes | Type A
|
Type B
|
Type C
|
Type D
|
No. (%) of isolates of other types | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) of isolates | No. of subtypes | No. (%) of isolates | No. of subtypes | No. (%) of isolates | No. of subtypes | No. (%) of isolates | No. of subtypes | |||||
| I | 91 | 15 | 31 | 59 (65) | 32 | 0 | 0 | 7 (8) | 7 | 12 (13) | 8 | 13 (14) |
| II | 85 | 6 | 18 | 76 (89) | 35 | 0 | 0 | 0 | 0 | 3 (4) | 3 | 6 (7) |
| III | 107 | 6 | 29 | 85 (79) | 23 | 0 | 0 | 11 (10) | 9 | 8 (8) | 5 | 3 (3) |
| IV | 100 | 6 | 24 | 89 (89) | 52 | 4 (4) | 2 | 2 (2) | 1 | 0 | 0 | 5 (5) |
| V | 112 | 6 | 22 | 78 (70) | 43 | 16 (14) | 7 | 13 (12) | 11 | 3 (3) | 3 | 2 (2) |
| VI | 100 | 15 | 39 | 46 (46) | 36 | 9 (9) | 9 | 22 (22) | 17 | 9 (9) | 6 | 14 (14) |
| Total | 595 | 31 | 97 | 433 (73) | 221 | 29 (5) | 18 | 55 (9) | 45 | 35 (6) | 25 | 43 (7) |
DISCUSSION
Results from this study demonstrate that there was a major PFGE genotype (genotype A, with 29 subtypes) of MRSA isolates prevailing in the six major hospitals in Taiwan, though a total of 97 subtypes could be identified among all 595 isolates. This clone was the predominant clone in each hospital and accounted for three-fourths of all isolates. In two of the six hospitals, this clone even accounted for nearly 90% of MRSA clinical isolates. Though a total of 31 genotypes could be identified, relatively few were found in four of the six hospitals, at each of which only 6 types were identified. A recent study from Taiwan (33) also documented that there was a major clone spreading all over Taiwan and accounting for 54% of 208 MRSA isolates collected from 22 hospitals distributed islandwide during a 3-month period in 1998. Comparison of the resolution of PFGE patterns shows that the major clone in that report was the same as genotype A in the present study. All these results suggested that this clone had prevailed and spread all over Taiwan at least for several years before 1998. A 5-year longitudinal analysis (1992 to 1996) of 140 MRSA isolates in hospital II (12) showed that this clone (genotype A in the present study) appeared in 1994 and became the predominant clone in 1996. However, we could not find any pattern among the major types in the present study that was identical or similar to the PFGE patterns of epidemic MRSA strains from European countries (19).
To monitor and investigate the epidemiology of MRSA isolates, a precise typing method is important. For typing of MRSA, genotyping methods are preferred for their high discriminatory power and reproducibility (15, 32). In the present study, although only one genotyping method was used, we have demonstrated a vast diversity of genotypes with an apparent epidemic clone among MRSA clinical isolates, again suggesting that PFGE is well suited for typing MRSA (32).
Previous studies (1, 2, 13, 21, 22, 24, 25, 28, 30, 31, 33) had documented that MRSA clones may spread in and between hospitals, cities, and countries and that even intercontinental spread may occur. The present study, including a large number of MRSA isolates, again demonstrated that a MRSA clone may spread all over the country. How the islandwide dissemination of this major clone occurred is an important issue. The principal island of Taiwan is about 390 km long and 145 km wide. The traffic system is well established. As much as 99% of the population of 23 million is included in the National Health Insurance System now, and it is very convenient for patients to seek medical care. Transfer of patients between hospitals and patient “shopping” among hospitals are not infrequently seen, and both conditions may facilitate the spread of particular MRSA clones. In many hospitals, not every effort is expended to prevent the spread of MRSA, and health care workers' adherence to infection control precautions is not always strict, which may facilitate the spread of MRSA. In addition, control of antibiotic use is neither well established nor strictly followed by clinicians (10, 11, 18), leading to the selection of resistant strains. All these phenomena may contribute to the current high prevalence of MRSA infections and the spread of a major MRSA clone in Taiwan. However, spread by healthy carriers cannot be ignored. Further studies should be conducted.
MICs of vancomycin were measured for all 595 isolates and were no more than 2 mg/liter for all but one strain, for which the vancomycin MIC was 4 mg/liter. Antibiograms were not collected and analyzed for each isolate. However, results from hospital III revealed that all 107 strains were resistant to oxacillin, penicillin, and erythromycin, while they were susceptible to vancomycin and teicoplanin. Eighty-eight percent of the strains were resistant to clindamycin, and resistance rates were not significantly different among strains of different genotypes. However, the resistance rate for trimethoprim-sulfamethoxazole was significantly higher (93 versus 5%; P < 0.001), and the resistance rate for chloramphenicol was significantly lower (13 versus 81%; P < 0.001), respectively, in genotype-A strains than in non-genotype-A strains.
All 595 MRSA isolates in the present study were clinical isolates from individual patients, and half were isolated from blood cultures. The clinical significance and impact cannot be overemphasized. Recently, vancomycin-intermediate and -resistant S. aureus (VISA and VRSA, respectively) strains have been identified and reported in several countries (6, 7, 14, 23, 26, 34). Alhough neither VISA nor VRSA has yet been documented in Taiwan, the emergence of these strains can be predicted. We are concerned that if no effective control measures are implemented in Taiwan, the dissemination of either VISA or VRSA could be as rapid as that of MRSA.
In conclusion, the majority of MRSA clinical isolates from six major hospitals in Taiwan shared a common PFGE pattern, indicating the presence of a single, epidemic MRSA clone prevailing in large hospitals in Taiwan.
Acknowledgments
This work was supported by a grant from the National Science Council of Taiwan (NSC90-2314-B-182A-08).
REFERENCES
- 1.Aires de Sousa, M., I. Santos Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aucken, H. M., M. Ganner, S. Murchan, B. D. Cookson, and A. P. Johnson. 2002. A new UK strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-17) resistant to multiple antibiotics. J. Antimicrob. Chemother. 50:171-175. [DOI] [PubMed] [Google Scholar]
- 3.Ayliffe, G. A. J. 1997. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24(Suppl. 1):S74-S79. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, J. M., and W. A. Causey. 1982. Increasing occurrence of methicillin-resistant Staphylococcus aureus in the United States. Infect. Control 3:377-378. [DOI] [PubMed] [Google Scholar]
- 5.Cafferkey, M. T., R. Hone, D. Coleman, H. Pomeroy, B. McGrath, R. Ruddy, and C. T. Keane. 1985. Methicillin-resistant Staphylococcus aureus in Dublin, 1971-1984. Lancet ii:705-708. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—-Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902-904. [PubMed] [Google Scholar]
- 8.Chang, S. C., L. Y. Hsu, K. T. Luh, and W. C. Hsieh. 1988. Methicillin-resistant Staphylococcus aureus infection. J. Formos. Med. Assoc. 87:157-163. [PubMed] [Google Scholar]
- 9.Chang, S. C., C. C. Sun, L. S. Yang, K. T. Luh, and W. C. Hsieh. 1997. Increasing nosocomial infections of methicillin-resistant Staphylococcus aureus at a teaching hospital in Taiwan. Int. J. Antimicrob. Agents 8:109-114. [DOI] [PubMed] [Google Scholar]
- 10.Chang, S. C., Y. C. Chen, and O. Y. P. Hu. 2001. Antibiotic use in public hospitals in Taiwan after the implementation of national health insurance. J. Formos. Med. Assoc. 100:155-161. [PubMed] [Google Scholar]
- 11.Chang, S. C., M. N. Shiu, and T. J. Chen. 2001. Antibiotic usage in primary care units in Taiwan after the institution of national health insurance. Diagn. Microbiol. Infect. Dis. 40:137-143. [DOI] [PubMed] [Google Scholar]
- 12.Chen, M. L., S. C. Chang, H. J. Pan, P. R. Hsueh, L. S. Yang, S. W. Ho, and K. T. Luh. 1999. Longitudinal analysis of methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J. Formos. Med. Assoc. 98:426-432. [PubMed] [Google Scholar]
- 13.de Lencastre, H., E. Severina, H. Milch, T. M. Konkoly, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 3:289-296. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 15.Hryniewicz, W. 1999. Epidemiology of MRSA. Infection 27(Suppl. 2):S13-S16. [DOI] [PubMed] [Google Scholar]
- 16.Hsueh, P. R., C. Y. Liu, and K. T. Luh. 2002. Current status of antimicrobial resistance in Taiwan. Emerg. Infect. Dis. 8:132-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jevons, M. P. 1961. “Celbenin”-resistant staphylococci. BMJ 1:124-125. [Google Scholar]
- 18.McDonald, L. C., H. T. Yu, H. C. Yin, A. C. Hsiung, M. Ho, et al. 2001. Use and abuse of surgical antibiotic prophylaxis in hospitals in Taiwan. J. Formos. Med. Assoc. 100:5-13. [PubMed] [Google Scholar]
- 19.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk diffusion susceptibility tests, 7th ed. Approved standard. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Pavillard, R., K. Harvey, D. Douglas, A. Hewstone, J. Andrew, B. Collopy, V. Asche, P. Carson, A. Davidson, G. Gilbert, J. Spicer, and F. Tosolini. 1982. Epidemic of hospital-acquired infections due to methicillin-resistant Staphylococcus aureus in major Victorian hospitals. Med. J. Aust. 69:451-455. [PubMed] [Google Scholar]
- 21.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, and the MRSA Collaborative Study Group. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 22.Roman, R. S., J. Smith, M. Walker, S. Byrne, K. Ramotar, B. Dyck, A. Cabani, and L. E. Nicolle. 1997. Rapid geographic spread of a methicillin-resistant Staphylococcus aureus strain. Clin. Infect. Dis. 25:698-705. [DOI] [PubMed] [Google Scholar]
- 23.Rotun, S. S., V. McMath, D. J. Schoonmaker, P. S. Maupin, F. C. Tenover, B. C. Hill, and D. M. Ackman. 1999. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from a patient with fatal bacteremia. Emerg. Infect. Dis. 5:147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmenlinna, S., O. Lyytikainen, P. Kotilainen, R. Scotford, E. Siren, and J. Vuopio-Varkila. 2000. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Finland. Eur. J. Clin. Microbiol. Infect. Dis. 19:101-107. [DOI] [PubMed] [Google Scholar]
- 25.Santos Sanches, I., M. Ramirez, H. Troni, M. Abecassis, M. Padua, A. Tomasz, and H. de Lencastre. 1995. Evidence of the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, and the Glycopeptide-Intermediate Staphylococcus aureus Working Group. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 27.Su, L. H., H. S. Leu, Y. P. Chiu, J. H. Chia, A. J. Kuo, C. F. Sun, and T. L. Wu. 2000. Molecular investigation of two clusters of nosocomial bacteraemia caused by multiresistant Klebsiella pneumoniae using pulsed-field gel electrophoresis and infrequent-restriction-site PCR. J. Hosp. Infect. 46:110-117. [DOI] [PubMed] [Google Scholar]
- 28.Teixeira, L., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Townsend, D. E., N. Ashdown, S. Bolton, J. Bradley, G. Duckworth, E. C. Moorhouse, and W. B. Grubb. 1987. The international spread of methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 9:60-71. [DOI] [PubMed] [Google Scholar]
- 31.van Belkum, A., W. van Leeuwen, R. Verkooyen, S. C. Sacilik, C. Cokmus, and H. Verbrugh. 1997. Dissemination of a single methicillin-resistant Staphylococcus aureus clone among Turkish hospitals. J. Clin. Microbiol. 35:978-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waller, T. M. 2000. Methicillin-resistant Staphylococcus aureus typing methods: which should be the international standard? J. Hosp. Infect. 44:160-172. [DOI] [PubMed] [Google Scholar]
- 33.Wang, J. T., Y. C. Chen, T. L. Yang, and S. C. Chang. 2002. Molecular epidemiology and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus in Taiwan. Diagn. Microbiol. Infect. Dis. 42:199-203. [DOI] [PubMed] [Google Scholar]
- 34.Wong, S. S. Y., P. L. Ho, P. C. Y. Woo, and K. Y. Yuen. 1999. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin. Infect. Dis. 29:760-767. [DOI] [PubMed] [Google Scholar]