Abstract
The mechanisms by which bacteria resist killing by antibiotics and biocides are still poorly defined, although repeated exposure to sublethal concentrations of antibacterial agents undoubtedly contributes to their development. This study aimed both to investigate the potential of Salmonella enterica and Escherichia coli O157 for adaptive resistance to commonly used biocides and to determine any cross-resistance to antibiotics. Strains were repeatedly passaged in media containing increasing concentrations of a biocide or antibiotic until adaptive resistance was obtained. A wide panel of antimicrobial agents was then screened by using the adapted strain to determine cross-resistance, if any. Adaptive resistance was readily achieved for both S. enterica and E. coli O157. Cross-resistance in adaptively resistant S. enterica varied with the serotype; Salmonella enterica serovar Enteritidis expressed cross-resistance to chloramphenicol, whereas Salmonella enterica serovar Typhimurium expressed cross-resistance to chlorhexidine. Benzalkonium chloride-resistant Salmonella enterica serovar Virchow showed elevated resistance to chlorhexidine; however, chlorhexidine-resistant Salmonella serovar Virchow did not demonstrate reciprocal cross-resistance to benzalkonium chloride, suggesting specific rather than generic resistance mechanisms. E. coli O157 strains acquired high levels of resistance to triclosan after only two sublethal exposures and, when adapted, repeatedly demonstrated decreased susceptibilities to various antimicrobial agents, including chloramphenicol, erythromycin, imipenem, tetracycline, and trimethoprim, as well as to a number of biocides. These observations raise concern over the indiscriminate and often inappropriate use of biocides, especially triclosan, in situations where they are unnecessary, whereby they may contribute to the development of microbial resistance mechanisms.
Following the discovery and clinical application of antibacterial agents, the morbidity and mortality caused by bacterial infections were considerably reduced. Today, however, public health is facing a new challenge due to the alarming increase in bacterial resistance to most of the existing antibacterial agents as well as the emerging link between the resistance strategies employed by bacteria toward antibiotics and biocides. In bacteria there are four main sites—cell wall synthesis, protein synthesis, nucleic acid synthesis, and cell membrane function—which serve as targets for antibiotic action. The overall mechanism of action of biocides, however, suggests that unlike antibiotics, which act selectively against specific cell targets, biocides act at one or several other sites within the cell; the overall damage to these sites results in the bactericidal effect (2, 17). Cationic agents such as quaternary ammonium compounds, chlorhexidine (CHX), and triclosan (TLN) have been implicated as the possible causes for the selection and persistence of bacterial strains with low-level antibiotic resistance (25).
The continuous and uncontrolled usage of antibacterial agents has no doubt contributed significantly to bacterial resistance, and there are many similarities between the ways in which bacteria are able to resist antibiotics and biocides (24). These mechanisms include decreased drug accumulation, decreased permeability, modification of the target, and modification of the antibiotic itself (20). Active efflux systems are also increasingly implicated in bacterial drug resistance (15). It seems entirely feasible, therefore, that exposure and development of resistance to one antimicrobial agent could evoke mechanisms of resistance which were cross-protective against another. Some organisms are resistant to certain antimicrobial agents due to their innate metabolic characteristics (intrinsic resistance), whereas others may develop mechanisms to protect themselves (27). Exposure to a single drug may lead to cross-resistance to many other structurally and functionally unrelated drugs (7). It has been suggested that this might be due to the expression of efflux mechanisms; efflux pumps with a variety of structures have been described. A single protein may act alone to perform efflux. Traditional efflux pumps excrete only one drug or one class of drugs, whereas a multidrug efflux system is able to pump out a wide range of compounds (15, 23).
At present, there are comparatively few reports of cross-resistance between clinically used antibiotics and biocides; however, the inclusion of biocides such as benzalkonium chloride (BKC), CHX, and particularly TLN in home cleaning and hygiene products, where their contribution to product efficacy is debatable, has become worryingly commonplace. Microorganisms are infinitely adaptable and have already demonstrated mechanisms of resistance to these biocides; the concern is that these mechanisms may confer cross-resistance to clinically important antibiotics. In this study we investigated whether there is any potential link between adaptive resistance to biocides and cross-resistance to other antibacterial agents in Salmonella enterica and Escherichia coli O157.
MATERIALS AND METHODS
Bacterial strains.
Salmonella enterica serovar Enteritidis was a clinical isolate from Birmingham Heartlands Hospital, Birmingham, United Kingdom; Salmonella enterica serovar Virchow was a food isolate from Campden and Chorleywood Food Research Association, Gloucestershire, United Kingdom; and Salmonella enterica serovar Typhimurium was a reference isolate obtained from the National Collection of Type Cultures (NCTC 74). E. coli O157 was a verotoxin-negative strain obtained from the National Collection of Type Cultures (NCTC 12900). All strains were stored on Protect beads (Pro-Lab Diagnostics, Neston, United Kingdom) at −70°C and were cultured at 37°C on nutrient agar (Oxoid, Basingstoke, United Kingdom) and in nutrient broth (Lab M, Lancashire, United Kingdom) where appropriate.
Antimicrobial agents and biocides.
All antimicrobial agent disks were supplied by Oxoid unless otherwise stated. These included amoxicillin (AMX; 25 μg/ml), amoxicillin-clavulanic acid (AMC; 30 μg/ml), chloramphenicol (CHL; 30/ml), ciprofloxacin (CIP; 1 μg/ml), clindamycin (CLI; 2 μg/ml), colistin sulfate (CS; 25 μg/ml), gentamicin (GEN; 10 μg/ml), imipenem (IPM; 10 μg/ml), rifampin (RIF; 5 μg/ml), tetracycline (TET; 10 μg/ml), and trimethoprim (TMP; 1.25 μg/ml). Fusidic acid (FD; 10 μg/ml) and vancomycin (VAN; 5 μg/ml) were included as negative controls, because they have no activity against the Enterobacteriaceae. Erythromycin (ERY) was purchased from Sigma (Poole, United Kingdom). The biocides BKC (Sigma-Aldrich, Dorset, Buckinghamshire, United Kingdom) and CHX (Sigma) were supplied as laboratory standard powders of known potency, and TLN (Aquasept, Oldham, United Kingdom) was purchased as a laboratory standard solution. All solutions were filter sterilized by use of a 0.2-μm-pore-size cellulose syringe filter (Nalgene, Leicester, United Kingdom).
Strain continuity.
Pre- and postadapted strains were characterized biochemically and genotypically by use of API 20E (Biomerieux, Marcy L'Etoile, France) and random amplification of polymorphic DNA (RAPD) assays (9) to confirm strain continuity.
MICs.
MICs were determined by a standard broth dilution method carried out by using a twofold dilution of each antibacterial agent. One colony of each organism was taken from an agar plate and inoculated into 30 ml of nutrient broth. The liquid culture was incubated overnight with shaking at 37°C. Following a 24-h incubation, the A470 was measured and used to determine the population density by reference to a calibration plot. The antibiotic or biocide dilution range was prepared in Bijoux bottles to a final volume of 5 ml, and each dilution was inoculated with approximately 108 bacteria. The MIC was determined as the lowest concentration of the antibiotic or biocide that inhibited growth.
Adaptive resistance.
The first tube showing growth below the MIC was used to inoculate nutrient broth containing increasing concentrations of an antibiotic or biocide. This procedure took place daily until a significant increase in the MIC (>1 log unit) occurred. At each passage the sub-MIC culture was cultured onto nutrient agar containing the same concentration of the antibiotic or biocide and onto nutrient agar without antibiotics. Cultures were stored at 4°C and were subcultured onto fresh medium once per month.
Cross-resistance to antimicrobial agents and biocides.
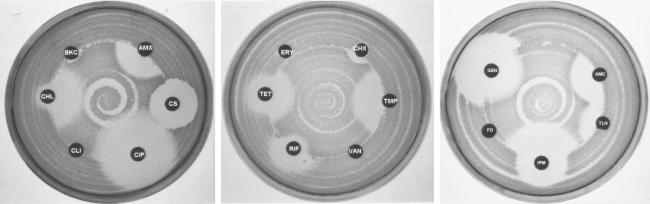
Cross-resistance toward various antibiotics and biocides was determined by Stoke's method (4). Suspensions of the parent E. coli O157 and Salmonella strains were inoculated over the central portions of the surfaces of separate Mueller-Hinton plates by using a rotary plater, leaving an outer ring of 1 cm (see Fig. 3). Adapted E. coli O157 and Salmonella strains were inoculated onto the remaining perimeter of the plate, and antibiotic disks were placed at the interface between the parent strain and the resistant strain. Plates were incubated overnight at 37°C and then examined for cross-resistance and antibiotic susceptibility by comparing zones of clearance around the disks. A >2-mm increase in the zone of clearance of the adapted strain over that of the parent strain was considered indicative of resistance, which was then confirmed by conducting a broth microdilution assay to determine the true MIC. Experiments were repeated a minimum of six times.
FIG. 3.
Cross-resistance between TLN-resistant E. coli O157 strains and several antibacterial agents by Stoke's method. See Materials and Methods for antibiotic and biocide abbreviations and concentrations.
Stability of adaptive resistance.
The stabilities of the adaptive resistances of the most resistant passaged bacterial cells of Salmonella serovar Enteritidis, Salmonella serovar Typhimurium, Salmonella serovar Virchow, and E. coli O157 were determined by passage in antibiotic-free and biocide-free broth. The procedure was repeated every 24 h for 30 days. MICs were determined every 5 days.
RESULTS
Progress of resistance.
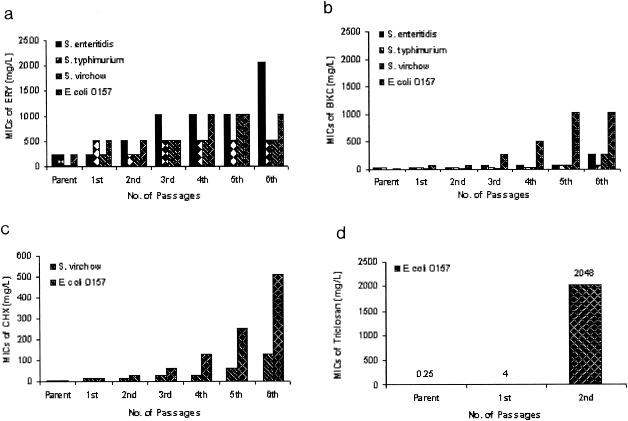
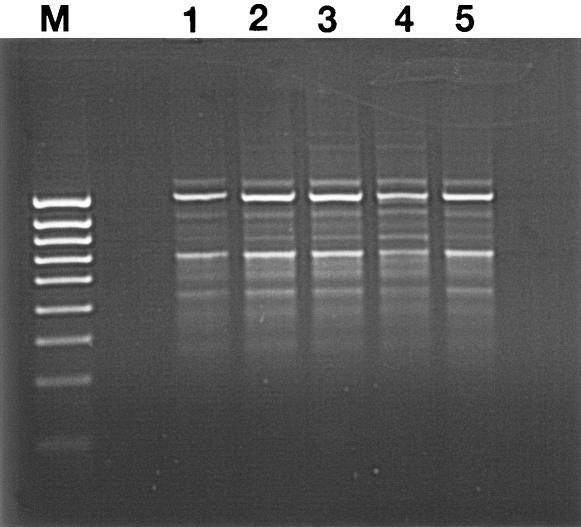
The profiles of adaptation of Salmonella serovar Enteritidis, Salmonella serovar Typhimurium, Salmonella serovar Virchow, and E. coli O157 to ERY and to the biocides tested are shown in Fig. 1. In each case the continuity of the adapted strain following repeated passages was confirmed by biochemical and RAPD profiling (Fig. 2). Similar results were obtained for the Salmonella isolates (data not shown). All the serotypes of Salmonella and E. coli O157 were adapted to high concentrations of ERY. The concentration of BKC that inhibited E. coli O157 was far below that for Salmonella serovar Enteritidis and Salmonella serovar Typhimurium; however, E. coli O157 acquired the highest resistance more rapidly than the rest of the strains investigated in this study. In addition, when Salmonella serovar Virchow and E. coli O157 were exposed to CHX, they repeatedly acquired resistance, even from their first exposure, a phenomenon which was most pronounced in E. coli O157. When E. coli O157 was adapted to TLN, it was initially sensitive to low levels; however, susceptibility was soon lost and very high concentrations were required to inhibit this bacterium. This rapid development of high-level resistance was highly reproducible upon repeat experimentation.
FIG. 1.
(a) Adaptation of Salmonella serovar Enteritidis, Salmonella serovar Typhimurium, Salmonella serovar Virchow, and E. coli O157 to ERY. (b) Adaptation of Salmonella serovar Enteritidis, Salmonella serovar Typhimurium, Salmonella serovar Virchow, and E. coli O157 to BKC. (c) Adaptation of Salmonella serovar Virchow and E. coli O157 to CHX. (d) Adaptation of E. coli O157 to TLN.
FIG. 2.
RAPD profiles obtained from preadapted E. coli O157 (lane 1) and from E. coli O157 strains postadapted to ERY (lane 2), BKC (lane 3), CHX (lane 4), and TLN (lane 5). Lane M, 1-kbp molecular weight marker.
Stability of adaptive resistance.
The adaptive resistances of Salmonella serovar Enteritidis, Salmonella serovar Typhimurium, Salmonella serovar Virchow, and E. coli O157 to ERY, BKC, CHX, and TLN were not lost following more than 30 days of passage in antibiotic- or biocide-free medium. In the absence of a selective pressure, none of the strains returned to the preadaptation sensitivity.
Cross-resistance.
Resistance or sensitivity to an antibiotic or biocide was determined by the zone of inhibition (measured in millimeters) around the impregnated disk which was placed at the interface between the pre- and postadapted strains. The results for Salmonella serovar Enteritidis and Salmonella serovar Typhimurium are shown in Tables 1 and 2, respectively. Where a >2-mm difference in the zone of clearing was observed, a broth microdilution assay was performed to determine whether a true MIC change had taken place. In each case, a minimum 2-log-unit change in the MIC was found for 2-mm zone differences, and a MIC change of as much as 9 log units was found for 11-mm zone differences. Cross-resistance to antibiotics and biocides was demonstrated in a majority of cases, but not for Salmonella serovar Enteritidis. For Salmonella serovar Enteritidis, cross-resistance occurred only between ERY and CHL. Salmonella serovar Typhimurium demonstrated cross-resistance between antibiotics and biocides, as is apparent with ERY and CHX, as well as between CHX and other biocides.
TABLE 1.
Zones of inhibition of parent and adapted strains of Salmonella serovar Enteritidis
| Antibiotic or biocide in which strain was passaged | Zone of inhibition of parent strain/zone of inhibition of adapted straina with the following antibiotic or biocide:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMX | BKC | CHL | CHX | CIP | CLI | CS | ERY | FD | GEN | IPM | RIF | TET | TLN | TMP | VAN | |
| ERY | 15/15 | 15/15 | 4/4 | 14/11 | 10/10 | 16/16 | 0/0 | 10/10 | 10/3 | 0/0 | 14/14 | 16/16 | 5/5 | 10/10 | 13/13 | 13/13 | 0/0 |
| BKC | 14/14 | 14/14 | 11/6 | 14/14 | 6/6 | 16/16 | 0/0 | 10/10 | 0/0 | 0/0 | 16/16 | 16/16 | 4/4 | 10/11 | 17/17 | 10/10 | 0/0 |
Zones of inhibition are measured in millimeters. Boldfaced data indicate cross-resistance.
TABLE 2.
Zones of inhibition of parent and adapted strains of Salmonella serovar Typhimurium
| Antibiotic or biocide in which strain was passaged | Zone of inhibition of parent strain/zone of inhibition of adapted straina with the following antibiotic or biocide:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMX | BKC | CHL | CHX | CIP | CLI | CS | ERY | FD | GEN | IPM | RIF | TET | TLN | TMP | VAN | |
| ERY | 13/15 | 16/16 | 4/3 | 12/11 | 14/8 | 17/17 | 0/0 | 9/9 | 10/3 | 0/0 | 16/16 | 15/15 | 5/0 | 7/8 | 15/13 | 10/10 | 0/0 |
| BKC | 14/14 | 15/14 | 12/1 | 15/15 | 7/5 | 13/15 | 0/0 | 9/9 | 0/0 | 0/0 | 13/11 | 17/16 | 4/4 | 6/9 | 4/3 | 13/13 | 0/0 |
Zones of inhibition are measured in millimeters. Boldfaced data indicate cross-resistance.
When Salmonella serovar Virchow was tested, there was a high degree of cross-resistance between antibiotics and biocides (e.g., between ERY and TLN and between ERY and CHX) as well as among biocides (e.g., between BKC and TLN and between CHX and TLN). Generally, cross-resistance was observed between ERY and CHL, ERY and TMP, BKC and AMX, BKC and AMC, BKC and CHL, BKC and IPM, BKC and TMP, and CHX and TET. The results are summarized in Table 3.
TABLE 3.
Zones of inhibition of parent and adapted strains of Salmonella serovar Virchow
| Antibiotic or biocide in which strain was passaged | Zone of inhibition of parent strain/zone of inhibition of adapted straina with the following antibiotic or biocide:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMX | BKC | CHL | CHX | CIP | CLI | CS | ERY | FD | GEN | IPM | RIF | TET | TLN | TMP | VAN | |
| ERY | 16/16 | 16/15 | 0/0 | 16/13 | 0/0 | 15/15 | 0/0 | 13/13 | 8/0 | 0/0 | 14/16 | 17/17 | 5/5 | 11/10 | 21/0 | 12/6 | 0/0 |
| BKC | 16/0 | 16/1 | 5/0 | 14/2 | 9/4 | 0/0 | 0/0 | 9/11 | 4/4 | 0/0 | 16/15 | 16/12 | 5/5 | 8/8 | 17/0 | 14/0 | 0/0 |
| CHX | 15/15 | 15/15 | 0/0 | 0/0 | 10/5 | 15/15 | 0/0 | 10/10 | 5/5 | 0/0 | 16/15 | 16/16 | 5/5 | 8/10 | 14/0 | 16/16 | 0/0 |
Zones of inhibition are measured in millimeters. Boldfaced data indicate cross-resistance.
In E. coli O157, cross-resistance between antibiotics and biocides occurred frequently. The results are shown in Table 4 and Fig. 3. Strains that were selected for adaptation to BKC exhibited increased resistance to AMC, AMX, CHL, IPM, TET, and TMP. Those that were selected for adaptation to TLN showed signs of a higher degree of resistance to the same antibiotics plus ERY and CIP (Fig. 3). Cross-resistance between biocides was observed only in one circumstance: when bacteria were adapted to CHX, they consequently became cross-resistant to TLN. However, cross-resistance between biocides and other antibiotics was demonstrated for resistance to both BKC and TLN. Adaptation of E. coli O157 to ERY also conferred cross-resistance to several other antibiotics, including CHL, CIP, TET, and TMP as well as to the biocide TLN. As expected, no resistance to VAN or FD was observed.
TABLE 4.
Zones of inhibition of parent and adapted strains of E. coli O157
| Antibiotic or biocide in which strain was passaged | Zone of inhibition of parent strain/zone of inhibition of adapted straina with the following antibiotic or biocide:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMX | BKC | CHL | CHX | CIP | CLI | CS | ERY | FD | GEN | IPM | RIF | TET | TLN | TMP | VAN | |
| ERY | 12/12 | 11/11 | 5/5 | 14/7 | 4/4 | 17/9 | 0/0 | 9/9 | 6/2 | 0/0 | 12/12 | 15/15 | 4/4 | 10/7 | 20/5 | 14/12 | 0/0 |
| BKC | 12/0 | 12/0 | 6/0 | 19/0 | 8/0 | 14/14 | 0/0 | 10/10 | 4/4 | 0/0 | 13/13 | 15/10 | 5/5 | 10/4 | 18/0 | 14/0 | 0/0 |
| CHX | 12/14 | 12/15 | 4/3 | 14/15 | 8/3 | 14/14 | 0/0 | 9/10 | 11/11 | 0/0 | 13/14 | 13/15 | 5/5 | 10/10 | 19/15 | 14/13 | 0/0 |
| TLN | 11/0 | 13/0 | 6/0 | 13/5 | 6/0 | 14/14 | 0/0 | 9/10 | 7/0 | 0/0 | 12/12 | 15/11 | 5/5 | 17/14 | 11/4 | 13/0 | 0/0 |
Zones of inhibition are measured in millimeters. Boldfaced data indicate cross-resistance.
DISCUSSION
Three functional classes of macrolide resistance mechanisms exist in pathogenic bacteria: those that modify the ribosome, those that modify the antibiotic itself, and those that affect the rate of transport of the antibiotic across the cell membrane (5). With ERY it was possible through passage to reach high levels of resistance in all strains tested. At present, it is unknown which mechanisms are contributing to the adaptive resistance observed in the strains under study; however, this resistance is likely due to the presence of active efflux (14).
Reduced susceptibility to biocides is apparently increasing (18, 25), and the ability of strains to rapidly develop enhanced resistance to BKC in the present investigation is in agreement with the findings of other studies (10). The MIC of BKC changed from 4 to 256 μg/ml as adaptation progressed in Salmonella serovar Virchow. The MIC of CHX for E. coli O157 began at 4 μg/ml and increased by 7 log units to 512 μg/ml. When E. coli O157 was adapted to TLN, the MIC changed from 0.25 to 4 μg/ml. It is of interest that while the E. coli O157 parent strain was sensitive to extremely low concentrations (0.25 μg/ml) of TLN at the first exposure, this bacterium acquired resistance rapidly: after the 1st-passage, growth at extremely high concentrations (1,024 μg/ml) was observed. The speed and extent to which E. coli O157 becomes resistant to BKC and TLN are of particular concern. Both BKC and TLN are common biocide components in a range of domestic disinfectant products, which are often used inappropriately and at subinhibitory concentrations (13). For instance, the TLN concentration usually found in soap is approximately 2,500 μg/ml (13); however, it has been proposed that soap reduces the efficacy of TLN, thereby reducing its effective concentration. In this study E. coli O157 was found to grow in 1,024 μg of pure TLN/ml. It appears possible that the domestic kitchen may provide a selective environment for adaptive resistance to biocides, which may eventually lead to the undesirable situation of resident strains resistant to disinfection.
The stability of adaptive resistance to ERY, BKC, CHX, and TLN was investigated by passage of the adapted cells in nonselective broth. The MICs of biocides were tracked as the passages continued over 30 days. When bacterial cells were passaged in an ERY-free medium, in no case did the MIC return to the wild-type level. Thus, it can be concluded that adaptive tolerance to ERY is subject to minor variability but remains stable. The stability of adaptation to BKC was also studied for Salmonella serovar Enteritidis, Salmonella serovar Typhimurium, Salmonella serovar Virchow, and E. coli O157. Acceptable variability was observed in all experiments that were performed; however, since none of the strains returned to their preadaptation sensitivities, it can be concluded that adaptive resistance to BKC was stable. Loughlin et al. (16) demonstrated that resistance to BKC was readily acquired when Pseudomonas aeruginosa strains were tested for 30 days and was retained in the absence of the disinfectant. Information in the literature regarding the stability of adaptive resistance of Salmonella or E. coli O157 to BKC is sparse; however, the results obtained here support the observations with Pseudomonas. The results suggest a degree of stability in adaptation to both CHX and TLN, but to a lesser extent for TLN. Adaptive resistance to TLN has been shown to decrease in Staphylococcus aureus with subculture in the absence of TLN (28), a result which does not entirely correlate with the findings for Salmonella and E. coli O157.
Concern about a possible linkage between antibiotic- and biocide-resistant strains has been expressed, and in some instances it has been claimed that biocides select for resistant gram-negative bacteria (A. D. Russell, U. Tattawasart, J. Y. Maillard, and J. R. Furr, Letter, Antimicrob. Agents Chemother. 42:2151, 1998). Cross-resistance between different classes of antibacterial agents, including quinolones, nalidixic acid, CHL, TMP, and in some cases β-lactam antibiotics, is a common phenomenon in gram-negative bacteria (8, 26). In the present study, broad-spectrum antimicrobial agents (CLI, TET, RIF, GEN, CHL, IPM, AMX, and AMC), including those specifically active against E. coli and Salmonella (TMP and CIP), were employed. There was good evidence that some types of biocide resistance could provide cross-protection in certain organisms. For Salmonella serovar Enteritidis, this was not the case; however, for Salmonella serovar Typhimurium, Salmonella serovar Virchow, and E. coli O157, cross-resistance between antibacterial agents and biocides was observed. In a majority of cases, the cross-resistance expressed moved strains from the category of “sensitive” to that of “resistant” according to the NCCLS guidelines on antimicrobial susceptibility testing (22). This was particularly the case when E. coli was adapted to BKC: it showed a reduction in the zone of inhibition from 19 to 0 mm. In other cases, the level of adaptive resistance was not as pronounced; when E. coli was adapted to TLN, it moved from “intermediately sensitive” to “resistant” to CHL. Although not observed in this study, even a level of cross-resistance below the criteria set by the NCCLS would still be important, since even a modest change in susceptibility may ultimately confer a growth advantage on a strain.
Cross-resistance between antibacterial agents or biocides and TLN was readily achieved for Salmonella serovar Virchow, for E. coli O157, and, in one instance, for Salmonella serovar Typhimurium. The widespread use of antimicrobial products containing TLN has been suggested as a possible cause of cross-resistance to other antibacterial agents (3). In Salmonella serovar Virchow, biocide resistance is most probably associated with exposure to CHX, since when adapted to BKC this organism showed elevated resistance to CHX but when adapted to CHX it did not display increased resistance to BKC. Russell et al. (letter) also reported that CHX-resistant strains of Pseudomonas stutzeri showed variable increases in resistance to quarternary ammonium compounds and to TLN. They also suggested that CHX-resistant strains showed elevated resistance to many antibiotics, including ERY; however, this was not observed for the Salmonella and E. coli O157 strains investigated in this study.
TLN-resistant E. coli O157 strains often showed decreased susceptibility to a range of antimicrobial agents, including CHL, ERY, IPM, TET, and TMP, as well as to various biocides. The latter observation has been reported previously for P. aeruginosa by Chuanchuen et al. (6); however, Suller and Russell (28) demonstrated that TLN-resistant S. aureus strains exhibited no increase in resistance to ERY or TET. The fact that a link between TLN and TMP was shown is of particular concern, since TMP is an antimicrobial agent active against enteric pathogens such as E. coli (1, 12). Proposed mechanisms for TLN resistance and cross-resistance in E. coli are the efflux pump AcrAB and mutations in FabI active-site residues (19). Chuanchuen et al. (6) also proposed that resistance to both antibacterials and antibiotics occurs through efflux systems in P. aeruginosa.
Cross-resistance between antibiotics and biocides and between different biocides has been reported for P. aeruginosa by Lambert et al. (11) and Murtough et al. (21); however, no reports were found for E. coli O157 and Salmonella. It has been suggested that the possible linkage between resistance to antibiotics and resistance to biocides might be due to common resistance mechanisms (28); however, this hypothesis has never been proven conclusively. The linkage might be also due to a nonspecific reduction in cell permeability, which does not allow chemically unrelated molecules into the resistant cells. This, of course, does not exclude the possibility of the presence of an active efflux (29). No obvious correlation could be drawn between the Salmonella serotype and resistance to a particular class of antibiotics or group of biocides; however, for particular strain-antibiotic-biocide combinations, strong evidence of cross-resistance was observed.
Conclusion.
Development of resistance to antimicrobial agents and biocides is a particularly worrying problem which is compounded by cross-resistance mechanisms that may exist in certain pathogenic strains. In this study a high degree of cross-resistance to a range of biocides and antibiotics was observed in Salmonella serovar Virchow and E. coli O157, and a lower degree was observed in Salmonella serovar Typhimurium and Salmonella serovar Enteritidis, when strains were repeatedly exposed to subinhibitory concentrations of antimicrobial agents. The increasing popularity of biocide-containing domestic cleaning products which, when used inappropriately, may provide sublethal exposure represents a real risk for the development of resistance and the promotion of cross-resistance to a range of antimicrobial agents.
REFERENCES
- 1.Adrian, P. V., M. Plessis, K. P. Klugman, and S. G. B. Amyes. 1998. New trimethoprim-resistant dihydrofolate reductase cassette dfrXV, inserted in a class 1 integron. J. Antimicrob. Agents 42:2221-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beumer, R., S. F. Bloomfield, M. Exner, G. M. Fara, K. J. Nath, and E. Scott. September 2000. Microbial resistance and biocides. IFH review. [Online.] International Scientific Forum on Home Hygiene, Geneva, Switzerland. http://www.ifh-homehygiene.org/forum/antresFINAL.pdf.
- 3.Braid, J. J., and M. C. J. Wale. 2002. The antibacterial activity of triclosan-impregnated storage boxes against Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Bacillus cereus and Shewanella putrefaciens in conditions simulating domestic use. J. Antimicrob. Chemother. 49:87-94. [DOI] [PubMed] [Google Scholar]
- 4.British Society for Antimicrobial Chemotherapy. 1991. Journal of Antimicrobial Chemotherapy, vol. 27, suppl. D. Guide to sensitivity testing. Academic Press, London, United Kingdom. [PubMed]
- 5.Carsenti-Etesse, H., P.-M. Roger, B. Dunais, S. Durgeat, G. Mancini, M. Bensoussan, and P. Dellamonica. 1999. Gradient plate method to induce Streptococcus pyogenes resistance. J. Antimicrob. Chemother. 44:439-443. [DOI] [PubMed] [Google Scholar]
- 6.Chuanchuen, R., K. Beinlich, T. T. Hoand, A. Becher, R. R. Karkhoff-Schweizer, and H. Schweizer. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfx mutants overexpressing mexCD-oprJ. Antimicrob. Agents Chemother. 45:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.George, A. M. 1996. Multidrug resistance in enteric and other Gram-negative bacteria. FEMS Microbiol. Lett. 139:1-10. [DOI] [PubMed] [Google Scholar]
- 8.Gutmann, L., R. Williamson, R. Moreau, M. D. Kinzis, E. Collatz, and J. F. Acar. 1995. Cross-resistance to nalidixic acid, trimethoprim and chloramphenicol associated with alterations in outer membrane proteins of Klebsiella, Enterobacter, and Serratia. J. Infect. Dis. 151:501-507. [DOI] [PubMed] [Google Scholar]
- 9.Hopkins, K. L., and A. C. Hilton. 2001. Optimisation of ramdom amplification of polymorphic DNA analysis for molecular subtyping of Escherichia coli O157. Lett. Appl. Microbiol. 32:126-130. [DOI] [PubMed] [Google Scholar]
- 10.Joynson, J. A., B. Forbes, and R. J. W. Lambert. 2002. Adaptive resistance to benzalkonium chloride, amikacin and tobramycin: the effect on susceptibility to other antimicrobials. J. Appl. Microbiol. 93:96-107. [DOI] [PubMed] [Google Scholar]
- 11.Lambert, R. J. W., J. Joynson, and B. Forbes. 2001. The relationships and susceptibilities of some industrial, laboratory and clinical isolates of Pseudomonas aeruginosa to some antibiotics and biocides. J. Appl. Microbiol. 91:972-984. [DOI] [PubMed] [Google Scholar]
- 12.Lee, J. C., J. Y. Oh, J. W. C. Cho, J. C. Park, J. M. Kim., S. Y. Seol, and D. T. Cho. 2001. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 47:599-604. [DOI] [PubMed] [Google Scholar]
- 13.Levy, S. B. 2001. Antibacterial household products: cause for concern. Emerg. Infect. Dis. 7:512-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy, S. B. 2002. Factors impacting on the problem of antibiotic resistance. J. Antimicrob. Chemother. 49:25-30. [DOI] [PubMed] [Google Scholar]
- 15.Levy, S. B. 2002. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. Symp. Suppl. 92:65S-71S. [PubMed] [Google Scholar]
- 16.Loughlin, M. F., M. V. Jones, and P. A. Lambert. 2002. Pseudomonas aeruginosa cells adapted to benzalkonium chloride shoe resistance to other membrane-active agents but not to clinically relevant antibiotics. J. Antimicrob. Chemother. 49:631-639. [DOI] [PubMed] [Google Scholar]
- 17.Maillard, J.-Y. 2002. Bacterial target sites for biocide action. J. Appl. Microbiol. Symp. Suppl. 92:16S-27S. [PubMed] [Google Scholar]
- 18.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Overexpression of marA, soxS or acrAB produces resistance to triclosan in Escherichia coli. FEMS Microbiol. Lett. 166:305-309. [DOI] [PubMed] [Google Scholar]
- 20.Moreillon, P. 2000. Means of bacterial resistance. Rev. Med. Suisse Romande 120:641-650. [PubMed] [Google Scholar]
- 21.Murtough, S. M., S. J. Hiom, M. Palmer, and A. D. Russell. 2001. Biocide rotation in the healthcare setting: Is there a case for policy implementation? J. Hosp. Infect. 48:1-6. [DOI] [PubMed] [Google Scholar]
- 22.NCCLS. 2002. Performance standards for antimicrobial susceptibility testing: supplemental tables. M100-S-12. NCCLS, Wayne, Pa.
- 23.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell, A. D. 2000. Do biocides select for antibiotic resistance? J. Pharm. Pharmacol. 52:227-233. [DOI] [PubMed] [Google Scholar]
- 25.Russell, A. D. 2002. Introduction of biocides into clinical practice and the impact on antibiotic-resistant bacteria. J. Appl. Microbiol. Symp. Suppl. 92:121S-135S. [PubMed] [Google Scholar]
- 26.Sanders, C. C., W. E. Sanders, R. V. Goering, and V. Werner. 1984. Selection of multiple antibiotic resistance by quinolones, β-lactams, and aminoglycosides with special reference to cross-resistance between unrelated drug classes. Antimicrob. Agents Chemother. 26:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, T. L., and W. R. Jarvis. 1999. Antimicrobial resistance in Staphylococcus aureus. Microbiol. Infect. 1:795-805. [DOI] [PubMed] [Google Scholar]
- 28.Suller, M. T. E., and A. D. Russell. 2000. Triclosan and antibiotic resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 46:11-18. [DOI] [PubMed] [Google Scholar]
- 29.Tattawasart, U., J.-Y. Maillard., J. R. Furr, and A. D. Russell. 1999. Development of resistance to chlorhexidine diacetate and cetylpyridinium chloride in Pseudomonas stutzeri and changes in antibiotic susceptibility. J. Hosp. Infect. 42:219-229. [DOI] [PubMed] [Google Scholar]