Abstract
Five fungal endophytes (K4, K5, K6, K9, K14) producing Cajaninstilbene acid (CSA, 3-hydroxy-4-prenyl-5-methoxystilbene-2-carboxylic acid) were isolated from the roots of pigeon pea [Cajanus cajan (L.) Millsp.]. CSA is responsible for the prominent pharmacological activities in pigeon pea. The amount of CSA in culture solution varied among the five fungal endophytes. K4 produced the highest levels of CSA (1037.13 µg/L) among the endophytes tested after incubation for five days. Both morphological characteristics and molecular methods were used for species identification of fungal endophytes. The five endophytic isolates were characterized by analyzing the internal transcribed spacer (ITS) rRNA and β-tubulin genes. The K4, K5, K9 and K14 strains isolated from pigeon pea roots were found to be closely related to the species Fusarium oxysporum. K6 was identified as Neonectria macrodidym. The present study is the first report on the isolation and identification of fungal endophytes producing CSA in pigeon pea. The study also provides a scientific base for large scale production of CSA.
Introduction
Pigeon pea [Cajanus cajan (L.) Millsp.], also called red gram, Congo pea, Gungo pea and no-eye pea, is a member of the legume family (Leguminosae). It is mainly distributed in tropical and subtropical countries as a substitute for expensive animal protein sources [1]. Pigeon pea has also been widely used as a traditional folk medicinal plant. The leaves of pigeon pea have been reported to arrest blood flow, relieve pain and kill worms [2]. They can be used to treat hepatitis, measles, dysentery, jaundice, diarrhea, sores, cough, bronchitis, bladder-stones, diabetes and many other illnesses [3]–[7].
In our previous work, the extracts of pigeon pea leaves showed significant antioxidant, antibacterial and antiherpetic activities [8]–[10]. The extracts also have hypoglycemic activity and potential in the treatment of postmenopausal osteoporosis [11]. Pigeon pea leaves are rich in flavonoids and stilbenes, which are known for their beneficial influence on human health [11]–[13].
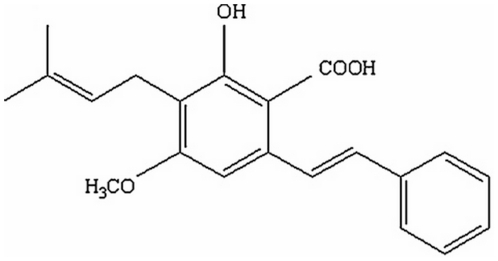
Cajaninstilbene acid (CSA), 3-hydroxy-4-prenyl-5-methoxystilbene-2-carboxylic acid, is the main active constituent in pigeon pea leaves and exists in few plants [14] other than pigeon pea. The chemical structure of CSA is shown in Fig. 1. Though the general biosynthetic pathway of CSA in pigeon pea has not been elucidated in detail, the biosynthetic pathway of plant stilbenes has been well studied [15]. A large body of evidence shows that stilbenes participate in both constitutive and inducible defense mechanisms in plants [15]. Certain stimuli such as UV light, NO, H2O2, jasmonic acid and fungal infections induce the accumulation of stilbenes.
Figure 1. Chemical structure of cajaninstilbene acid.
We hypothesized that endophytes in pigeon pea may contribute to CSA production. Endophytes infect living plants tissues without causing apparent disease symptoms in the host plant [16]. Endophytes are ubiquitous in a wide variety of plant species and most have a mutualistic relationship with their host [17]. Endophytes can improve plant adaptation to stress, increase biomass, or promote resistance to pathogen damage [18]–[20]. Some endophytes are thought to prevent their host from attacks by fungi, pests and mammals by producing secondary metabolites. Currently, endophytes are viewed as an outstanding source of bioactive products [21], [22]. Fungal endophytes possess a broad synthetic capability and synthesize abundant secondary metabolites with potential economic significance. Some endophytes produce biologically active constituents that are similar to or the same as those produced by their host, such as one fungal endophyte that produces taxol [23]. In addition, the production of secondary metabolites with pharmaceutical relevance is relatively common in endophytes, and thus they have great potential value as a source of drugs [24], [25]. Therefore, investigating endophytes represents a promising strategy to identify novel nature compounds. Endophytes can be cultured easily and quickly, and biomass can be accumulated by large scale fermentation, which is a unique advantage compared to plant resources [26].
The fungus Fusarium sp. has been isolated from a wide variety of sources, including soil, crops, plants and animals [27]. There are many non-pathogenic strains of Fusarium sp. including endophytes that utilize unique metabolic pathways to protect their host plants from invasion by pathogenic strains, pests and mammals [28]. Identification of Fusarium species has always been difficult due to ill-defined phenotypic classification systems [29], [30]. Fusarium taxonomy is primarily based on morphological characteristics of the anamorph, including the size and shape of macroconidia, the presence or absence of microconidia and chlamydospores, colony colour, and conidiophore structure [29], [30]. These characteristics can be highly variable depending on media and culturing conditions [31]. In addition, degeneration of cultures and growth of mutants may further complicate fungal identification [32]–[35]. Most teleomorphs of Cylindrocarpon sp. are currently classified as Neonectria [36], [37]. Neonectria sp. has also been reported to exhibit strong antimicrobial activity [38]. Neonectria taxonomy is based on anatomical characters of the perithecial wall and partly on characters of ascospores [39], [40].
A number of molecular tools have been developed to support morphological identifications. One of the most powerful techniques used in fungal identification is nucleotide sequencing [41]. Analysis of 18S ribosomal RNA sequences, usually used in phylogenetic analysis at higher taxonomic levels, is generally not adequate to differentiate fungal species. However, the internal transcribed spacer (ITS) regions ITS1 and ITS2, flanking the 5.8S rRNA gene is very useful in distinguishing species in many eukaryotic organisms including fungi [42]. Nuclear protein-coding loci, namely β-tubulin, are also highly substituted and can be used for identification of fungi [43]. The close relationship between Fusarium species is supported by the ITS and β-tubulin sequences [44]. The accurate identification of five fungal endophytes should be correlated by both the description of morphological characters and phylogenetic analysis of ITS and β-tubulin gene sequences.
Although many fungal endophytes isolated from healthy plants have been described, there are no reports on fungal endophytes in pigeon pea. Pigeon pea endophytes are worth investigating because of their potential to produce CSA or other valuable bioactive compounds.
In this manuscript, fungal endophytes were isolated from pigeon pea and screened for CSA production. Endophytes producing CSA may provide a new valuable medicinal resource in addition to plant resources. Five CSA-producing fungal isolates were then identified based on morphological and molecular parameters, in particular phylogenetic relatedness in two genes: ITS and β-tubulin. The present study may also provide a basis for the study of CSA biosynthesis.
Results
1. Isolation of endophytes
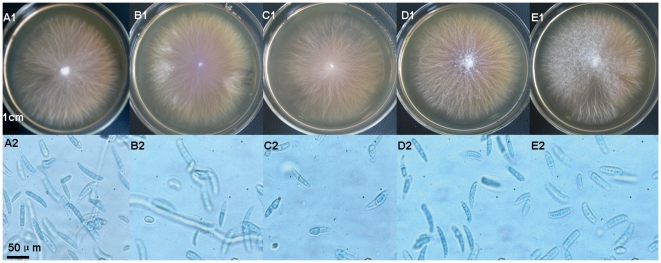
Endophytes were isolated from leaves, stems and roots of pigeon pea plants (72.2%, 76.2% and 67.4% of samples respectively). All endophytes grew 0.2–0.5 cm per day on PDA agar plate at 28°C. In the first 7 days, most fungal colonies on PDA were white and difficult to distinguish. After culturing for several more 7 days, different isolates were colored red, brown or black, and soft or rigid domes gradually formed in the center of colonies. Five CSA-producing isolates with similar biological characteristics were simultaneously found in roots of pigeon pea (Fig. 2). Fig. 2 shows the morphological characters of the CSA-producing endophytes K4, K5, K6, K9 and K14 on PDA agar plates. Fig. S1 shows the intercellular colonization of endphytes in the roots of pigeon pea.
Figure 2. Colonies on PDA plates and photomicrographs of endophytes.
A–E represent endophytes K4, K5, K6, K9 and K14. A1 to E1 show K4, K5, K6, K9 and K14 colonies on PDA medium after incubation for 14 days; A2 to E2 show photomicrographs of K4, K5, K6, K9 and K14 spores.
K4 and K5 isolates produced flat-growing colonies up to 7.0 cm diam. in two weeks. The colonies consisted of white aerial mycelium tinged with purple, which progressed to deep purple with aging and released purple pigment and droplets into the PDA. Spores were elliptical in shape, conidia were (30.0–60.8) µm×(7.8–12.0) µm. K9 and K14 isolates grew in flat colonies up to 6.5 cm diam. in two weeks. The colonies were white but turned pink with aging and released pink pigment into the PDA. Spore shape was falcate or elliptical and conidia were (40.3–80.4) µm×(8.0–13.6) µm. The K6 isolate produced flat-growing colonies up to 4.5 cm diam. in two weeks. Colonies were white with yellow in the center. Spore shape was cylindrical and conidia were (23.2–49.2) µm×(6.0–12.8) µm. Colonies and spore morphology of all isolates revealed them to be closest to strains of Fusarium sp. and Neonectria sp.
2. Quantitative determination of CSA
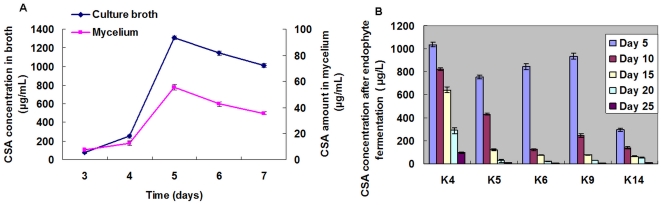
LC-MS/MS analysis showed that five of the pigeon pea endophytes tested produced CSA. The endophyte extracts shared the same LC-MS/MS retention time for CSA as the CSA standard (Fig. 3). Most of the endophytes producing a substantial amount of CSA were closely related to Fusarium sp. and Neonectria sp. CSA concentration in broth and mycelium of K4 endophyte is presented in Fig. 4A. CSA concentrations in broth and mycelium show a similar trend. The concentration of CSA increased from 3 to 5 days (77.91 µg/L to 1037.13 µg/L) and then declined. It is indicated that CSA is synthesized rapidly by endophyte's first growth stage in cultures, the accumulation of CSA in culture are both in total and relative to mycelial mass. Among the CSA-producing endophytes, the concentration of CSA varied with culture time. The K4 endophyte has the highest CSA concentration (1037.13 µg/L) after incubation for 5 days and the lowest concentration (97.33 µg/L) after 25 days. K5 showed a similar trend to K4. The highest CSA concentration (755.00 µg/L) was found after incubation 5 days and the lowest concentration (9.45 µg/L) after 25 days. The K6, K9 and K14 endophytes revealed a similar trend (Fig. 4A). Under the same culturing conditions used to measure CSA production, a compound with the same molecular weight and ion fragments as CSA appeared at 7.50 minutes. This compound's concentration gradually increased with incubation time, and it may be an isomer of CSA (Fig. 3).
Figure 3. Representative LC–MS/MS chromatograms of endophyte fermentation extracts of K4, K5, K6, K9 and K14.
A1–A5 show representative LC–MS/MS chromatograms of K4 after 5, 10, 15, 20 and 25 days of incubation; B1–B5 show representative LC–MS/MS chromatograms of K5 after 5, 10, 15, 20 and 25 days of incubation; C1–C5 show analogous representative LC–MS/MS chromatograms of K6; D1–D5 show analogous representative LC–MS/MS chromatograms of K9; E1–E5 showed representative LC–MS/MS chromatograms of K14.
Figure 4. CSA concentration in mycelium and culture broth of endophyte K4 (A) (day 3–7) and in culture broth of five endophyte strains (day 5–25) (B).
The fungal Mycelium extracts treating by 80% ethanol and culture liquid were extracted by ethyl acetate three times. The evaporated ethyl acetate extracts were subsequently dissolved in chromatogram methanol for determination of CSA production by LC-MS/MS.
3. Molecular identification of endophytes
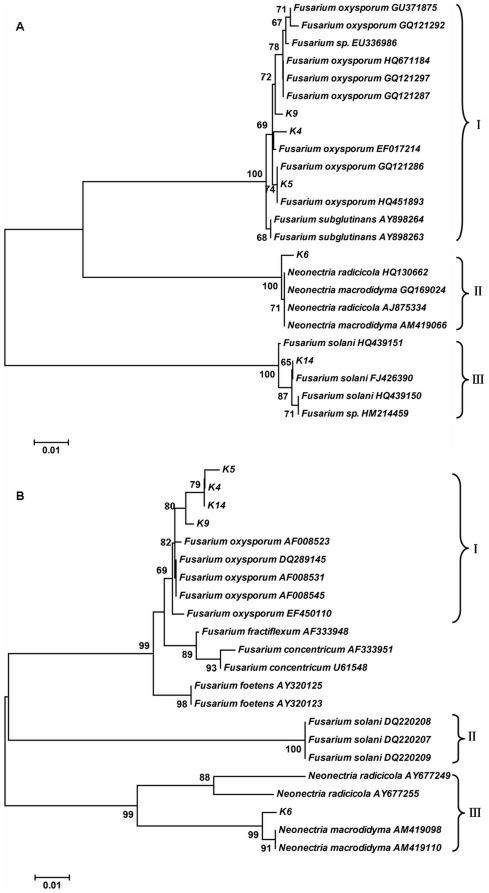
Neighbour-joining (NJ) trees inferred from the aligned ITS1–5.8S–ITS2 and β-tubulin genes suggested that the K4, K5, K9 and K14 isolates were most closely related to Fusarium oxysporum (Fig. 5). In the ITS1–5.8S–ITS2 sequences, K4, K9 and K14 were different from F. oxysporum at one out of 495, four out of 501 and two out of 518 base positions, respectively. K5 was a 100% match with F. oxysporum. In β-tubulin genes sequences, K4, K5, K9 and K14 were different from F. oxysporum at two out of 526, five out of 526, one out of 526 and one out of 521 base positions, respectively. ITS1–5.8S–ITS2 and β-tubulin gene sequences of K6 were most similar to those of N. radicicola and N. macrodidyma (Fig. 6), respectively. Alignment of the ITS1–5.8S–ITS2 sequences of K6 indicated that K6 was different from N. radicicola at two out of 499 base positions and from N. macrodidyma at four out of 498 base positions. In β-tubulin gene sequences, K6 was different from N. macrodidyma at two out of 554 base positions and from N. radicicola at 66 of 505 base positions.
Figure 5. Phylogenetic reconstruction based on two gene sequences of endohytes.
ITS1–5.8S–ITS2 sequences (A) and β-tubulin gene sequences (B) of endophytes were used for phylogenetic reconstruction. The trees were constructed using a neighbour-joining distance matrix. Evolutionary distances were computed using the Maximum Composite Likelihood method. Bootstrap values (1000 tree interactions) are indicated at the nodes.
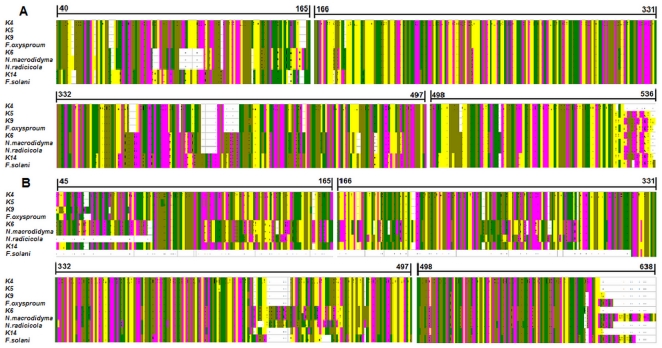
Figure 6. Alignment of ITS and β-tubulin gene sequences from five endophytes with reference sequences.
(A) The master alignment of ITS sequences from K4 to K14 with F. oxysporum, F. solani, N. radicicola and N. macrodidym. The position of the first and final nucleotide alignment are highlighted. (B) The master alignment of β-tubulin gene sequences from K4 to K14 with F. oxysporum, F. solani, N. radicicola, N. macrodidym. The position of the first and final nucleotide alignment are highlighted. Nucleotides are shown by color bars (A: yellow, G: purple, T: green, C: brown).
Morphological and biological characteristics of the endophytes also supported the conclusions reached through sequencing. In summary, the alignment of sequences of the five endophytes tested with known related species was used to distinguish differences between the two (Fig. 6).
Discussion
By means of a highly sensitive LC-MS/MS based method, the present study verified that several fungal endophytes isolated from pigeon pea produce CSA. CSA production is higher in the leaves than in the roots of pigeon pea [3]. These results are based on the assumption that CSA may be synthesized in both roots and leaves. CSA is unstable and is transferred to other structures within the roots under the alkaline conditions created by the nitrogen fixation of leguminous pigeon pea roots. However, it can be detected in the leaves even in an acidic environment. However, the culturing medium used provides a stable pH that is suitable for the endophytes' CSA production. By this method, CSA can be detected in root endophytes. The isolation of endophytes from leaves is still in progress. Preliminary evidence suggests leaf endophytes can produce a limited amount of CSA (unpublished data).
Among the endophytes isolated from roots, the CSA generation rate varied with culture time. The results (Fig. 4A) confirmed that CSA was biosynthesized by the endophyte and biosynthesis initially proceeded quickly. The production of CSA occurs only during rapid mycelial growth as after incubate 5 days, the total concentration in the medium and the concentration relative to mycelial mass declined at a similar rate. Fig. 4B demonstrated differences between different strains in the rate of decline from maximum accumulation 5 to 25 days. Among these endophytes, K4 showed the maximum accumulation and slow decline rate. Meanwhile, an isomer of CSA was also detected and its amount gradually increased during the incubation time. Hence, we suppose that CSA maybe converted into this isomer during incubation.
Certain stimuli such as UV light, NO, H2O2, jasmonic acid and fungal infections induce the accumulation of stilbenes [15], all environment factors maybe contribute to the CSA biosynthesis. Meanwhile, the genes for CSA biosynthesis may be transferred between endophytes and plant cells by horizontal gene transfer, leading to the production of CSA by endophyte itself. As a result of these long-held associations, it is plausible that some of the endophytes may have developed genetic systems promoting the transfer of information between themselves and the host plant [24]. Further studies are warranted to test this hypothesis. The present study may facilitate the elucidation of the biosynthetic pathway for CSA. The presence of CSA-producing endophytes in pigeon pea supports the hypothesis that during the long co-evolution of endophytes and their host plants, one way in which endophytes adapted to their particular microenvironments was through the uptake of host plant DNA into their own genomes. This could have led to endophytes with the ability to synthesize phytochemicals originally associated with the host plant [45], [46].
Fusarium sp. and Neonectria sp. are very difficult to distinguish by their morphological characteristics. The close relationship between these species is supported by the ITS and β-tubulin sequences [44].
The morphology of the five isolated endophytes suggested a close relationship with Fusarium sp. and Neonectria sp.. Phylogenetic analysis of ITS and β-tubulin sequences supported the identification of the K4, K5, K9 and K14 as F. oxysporum. Identification of K6 was not as simple as that of K4, K5, K9 and K14, since analyzing different sequences provided inconclusive results. As the ITS region is generally considered to be less conserved than either small or large subunits of rRNA genes [47], the ITS analysis may be more informative in distinguishing closely related species. A single two base substitutions out of 499 positions in ITS1–5.8S–ITS2 between K6 and Neonectria radicicola compared to four base substitutions out of 498 positions for K6 and N. macrodidyma suggests that K6 may be more closely related to N. radicicola. However, in the β-tubulin gene sequences, K6 differs from N. macrodidyma at two out of 554 base positions and from N. radicicola at 66 out of 505 base positions, indicating that K6 is more different form N. radicicola. Considering both sequences, we conclude that K6 is more closely related to N. macrodidyma.
In the ITS1–5.8S–ITS2 tree generated with neighbour-joining analyses using the Maximum Composite Likelihood method, isolate K6 formed a cluster clade II supported by 100% bootstrap value with N. radicicola and N. macrodidyma and showed 99% similarity with both in BLAST analysis. Clade I composed of the isolates K4, K5 and K9 showed 99%, 100% and 99% similarity to F. oxysporum in BLAST analysis, and the bootstrap of clade I was 100%. Isolate K14 showed 99% similarity to F. solani in BLAST analysis, but the bootstrap analysis did not support the conclusion well (<70%).
According to the Neighbor-Joining (NJ) tree based on the β-tubulin gene sequences, isolates of K6 form a well-defined clade III with N. macrodidyma (bootstrap support value of 99%). Isolates in the “F. oxysporum” complex clade I (82% bootstrap support) are mainly in basal positions with a small number of defined clades with high bootstrap support values. K4, K5 and K14 both showed 99%, similarity to F. oxysporum in BLAST analysis, and the bootstrap value was 79%. K9 showed 99% similarity to F. oxysporum in BLAST analysis, and the bootstrap value was 80%.
Firstly, our results indicate that the endophytes produce CSA. Endophytes producing valuable products could not only reduce the use of slow-growing and rare plants in industrial production but also preserve the world's ever-diminishing biodiversity. Furthermore, using a microbial source to produce a certain valued product such as CSA may be easier and more economical, effectively reducing the product's market price [48]. Endophytes can be easily cultured in vitro and accumulate biomass through large-scale fermentation in a process distinctive from that for plants [26]. In the future, CSA may be more quickly and efficiently obtained from endophytes than from plants. Therefore, CSA-producing endophytes from pigeon pea may provide valuable natural medicinal resources for large-scale production of CSA. Further optimization of submerged cultivation conditions and molecular studies of K4 are merit future investigation.
Materials and Methods
1. Isolation of fungal endophytes
Fungal endophytes were isolated from healthy roots, stems and leaves of pigeon pea [Cajanus cajan (L.) Millsp.]. Fresh tissue of pigeon pea were collected from the arboretum of the Key Laboratory of Forest Plant Ecology, Ministry of Education, Northeast Forestry University, Harbin, PR China, Voucher specimens (No. 052056001001001) were deposited in the herbarium of this Key Laboratory. The plant materials were subjected to endophyte isolation within 3 hours of harvest. Specifically, the roots, stems and leaves of pigeon pea were washed with running tap water, sterilized with 75% ethanol for 60, 60 and 30 seconds, rinsed in sterile water and 5% sodium hypochlorite for 7, 5, and 3 minutes, rinsed in sterile water three times respectively, and finally cut into 1 cm long segments. Both edges of sterilized segments were cut off, and the segments were incubated at 28±1°C on potato dextrose agar (PDA) medium for 14–21 days until the colonies originating from the newly formed surface of the segments appeared. Penicillin (20 µg/mL) was added to culture medium to suppress bacterial growth. The effectiveness of the surface sterilization procedure was verified by plating uncut plant tissue and samples of the final rinse water. Furthermore, the purified fungal endophytes were numbered and transferred to fresh PDA slants separately, and these slants were incubated for 14–21 days and then kept at 4°C.
Isolates of five endophytes were identified according to their cultural and morphological characteristics as described by Nelson et al. (1983) [30]. The diameter and colour of colonies was measured and determined. The microscopic features such as size and shape of hyphae and conidia were examined and measured under a light microscope after maturation.
2. Preparation the endophytes samples and standard solutions
For quantitative determination of CSA, 15 conical flasks of 500 mL capacity were used, with each endophyte isolate cultured in three parallel samples containing 200 mL Potato Dextrose Broth (PDB) medium. Fungal endophytes were grown in PDB medium and cultured for 5, 10, 15, 20 and 25 days at 28°C with shaking (180 rev/min) on a rotary shaker. The fungal culture liquid was then filtered and then endophytes extracted by ethyl acetate three times. Mycelium was rinsed in sterile water three times, oven for 48 h at 40°C, then ultrasonic processing 30 min in 80% ethanol and extracted by ethyl acetate three times. The evaporated ethyl acetate extracts were subsequently dissolved in chromatogram methanol and filtered with a 0.22 µm member filter, before being used for determination of CSA production.
A stock solution of reference sample CSA was prepared in methanol at a final concentration of 100 µg/mL. The final concentrations of the standard curve samples were 10, 20, 50, 100, 200 500, 1000, 3000 and 6000 ng/mL. The standard stock solution and working standard solutions were all stored at 4°C. Standard calibration samples were stored at −20°C until analysis.
3. Quantitative determination of CSA by LC-MS/MS
The determination of CSA based on LC-MS/MS methods described by Hua et al. (2010) [49]. An Agilent 1100 series HPLC system (Agilent Technologies, San Jose, CA, USA) consisting of a G1312A binary pump, a 7725i manual injector, and a G1379A degasser were used for chromatographic analysis. Chromatographic separations were achieved on a HIQ Sil C18 column (4.6 mm×250 mm, KYA TEACH, made in Japan) maintained at 20°C. The column effluent was monitored by an API3000 triple-stage quadrupole mass spectrometer (Applied Biosystems, Concord, Canada) equipped with an electrospray ionization (ESI) source. The mobile phase consisted of water and methanol (9∶91, v/v) containing 0.1% formic acid. The flow rate was 1.0 mL/min, and the sample injection volume was 10 µL.
The ion spray voltage was set at −4500 V. Compound parameters viz., declustering potential (DP), collision energy (CE), entrance potential (EP) and collision exit potential (CXP) were set at −60, −25, −10, −5 V respectively for analysis of CSA. The mass spectrometer was operated in ESI negative ion mode, and the detection of the ions was performed in the multiple reaction monitoring (MRM) mode, monitoring the transition of m/z 337.1 precursor ion [M–H]− to the m/z 293.0 product ion for CSA. The compound of second peak were detected by LC-MS/MS in the same conditions as used to monitor the transition of m/z 337.1 precursor ion [M–H]− to the m/z 293.0 product ion.
4. Fungal genomic DNA extraction
The fungal genomic DNA extraction procedure was followed by extraction using a universal genomic DNA extraction kit (Tiangen, Beijing, China). After incubation for two weeks on PDA, fungal mycelia was harvested, frozen in liquid nitrogen, and ground to powder with a mortar. Subsequently, fungal genomic DNA was extracted and purified following the manufacturer's instructions. The genomic DNA was determined by electrophoresis through 1% agarose gel stained with ethidium bromide. The concentration of DNA was determined by an RNA/DNA Calculator Gene Quant II (Pharmarcia Biotech Amersham, UK).
5. ITS and β-tublin amplification and sequencing
The ITS region of the ribosomal DNA was amplified using the primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATG-C-3′) [50]. The β-tubulin gene was amplified using the primers T1 (5′-ATGCGTGAGATTGTAAGT-3′) and T22 (5′-TGACCGAAAACGAAGTTGTC-3′) [51]. The PCR reaction mixtures used to amplify the fragments consisted of final concentrations of 1×PCR buffer and contained Pfu DNA Polymerase (Sangon, Shanghai, China), 0.5 µM forward and reverse primers, and approximately 30 ng of fungal genomic DNA made up to a total reaction volume of 50 µL with sterile deionised water. Amplification was performed in an ST1000™ thermo cycler (BioRad, California, USA). The amplification program was as follows: 1 cycle of 3 minutes at 95°C, 30 cycles of 30 seconds at 95°C, 30 seconds at 55°C, 1 minute at 72°C, and finally 1 period of 5 minutes at 72°C. Amplified fragments were separated by electrophoresis through 1% agarose gels stained with ethidium bromide and purified using Gel Product Purification Kit (Tiangen, Beijing, China). DNA fragments were sequenced by the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) and an ABI 3730XL sequencer (Applied Biosystems) at the Sangon Biotech Co., Ltd (Shanghai, China).
6. Analysis of DNA sequences
ITS1–5.8S–ITS2 and β-tubulin gene sequences were successfully amplified from each CSA-producing endophyte by means of PCR. The obtained sequences were screened for homology using the MEGA version 4.1 [52] (http://www.ncbi.nlm.nih.gov/BLAST). Sequence data has been submitted to Genbank under the accession numbers JF807393 to JF807397 and JF807403 to JF807407. When sequence identity was greater than 97% homologous, genus and species of the database results were accepted; when identity was 97 to 95% homologous, only genus was accepted [53]. To determine the taxonomic identity of the endophytes, the ITS sequences and β-tubulin gene sequences from both the CSA-producing endophytes and species thought to be of close homology (Table 1) within the Fusarium family were aligned using the Clustal of alignment in MEGA version 4.1 [52]. Phylogenetic analysis was then conducted with phylogeny in MEGA version 4.1. A neighbour-joining tree [54] was created from the alignment file using the Maximum Composite Likelihood method and bootstrapping of 1000 replicates [55].
Table 1. Sequences and GenBank accession numbers by DNA locus.
| Fungal taxon | Source and authors | Accession No. |
| Fusarium sp. | Sun, H. and Song, R. 2007 | EU338986 (ITS) |
| Fusarium sp. | Zhao, X. et al. 2010 | HM214459 (ITS) |
| Fusarium oxysporum | Yang, X. M. 2009 | GU371875 (ITS) |
| Fusarium oxysporum | Manici, L. M. and Caputo, F. 2009 | EF017214 (ITS) |
| Fusarium oxysporum | Shanmugam, V. 2009 | GQ121286 (ITS) |
| Fusarium oxysporum | Shanmugam, V. 2009 | GQ121297 (ITS) |
| Fusarium oxysporum | Shanmugam, V. 2009 | GQ121292 (ITS) |
| Fusarium oxysporum | Cheng, J. et al. 2010 | HQ671184 (ITS) |
| Fusarium oxysporum | Lang, C. Y. et al. 2010 | HQ451893 (ITS) |
| Neonectria sp. | Mitter, B. et al. 2010 | HQ130662 (ITS) |
| Neonectria radicicola | Oliveira, H. and Nascimento, T. C. 2008 | AJ875334 (ITS) |
| Neonectria macrodidyma | Liu, Y. et al. 2009 | GQ169024 (ITS) |
| Neonectria macrodidyma | Oliveira, H. et al. 2006 | AM419066 (ITS) |
| Fusarium solani | Sun, H. and Song, R. 2009 | FJ426390 (ITS) |
| Fusarium solani | Lee, H. B. 2010 | HQ439150 (ITS) |
| Fusarium subglutinans | Liu, X. and Wang, X. 2005 | AY898264 (ITS) |
| Fusarium subglutinans | Liu, X. and Wang, X. 2005 | AY898263 (ITS) |
| Fusarium oxysporum | Cigelnik, E. and O'Donnell, K.1998 | AF008545 (β-tubulin) |
| Fusarium oxysporum | Cigelnik, E. and O'Donnell, K.1998 | AF008531 (β-tubulin) |
| Fusarium oxysporum | Cigelnik, E. and O'Donnell, K.1998 | AF008523 (β-tubulin) |
| Fusarium oxysporum | Cigelnik, E. and O'Donnell, K.1998 | AF008522 (β-tubulin) |
| Fusarium oxysporum | Zhang, C. L. et al. 2006 | DQ289145 (β-tubulin) |
| Fusarium oxysporum | Garcia-Sanchez. M. A. et al. 2007 | EF450110 (β-tubulin) |
| Neonectria macrodidyma | Oliveira, H. et al. 2009 | AM419098 (β-tubulin) |
| Neonectria macrodidyma | Oliveira, H. et al. 2009 | AM419110 (β-tubulin) |
| Neonectria radicicola | Halleen, F. et al. 2004 | AY677249 (β-tubulin) |
| Neonectria radicicola | Halleen, F. et al. 2004 | AY677255 (β-tubulin) |
| Fusarium foetens | Schroers, H. J. et al. 2004 | AY320123 (β-tubulin) |
| Fusarium foetens | O' Donnell, K. 2003 | AY320125 (β-tubulin) |
| Fusarium concentricum | O' Donnell, K. et al. 1996 | U61548 (β-tubulin) |
| Fusarium concentricum | O' Donnell, K. 2001 | AF333951 (β-tubulin) |
| Fusarium solani | Bogale, M. et al. 2006 | DQ220207 (β-tubulin) |
| Fusarium solani | Bogale, M. et al. 2006 | DQ220208 (β-tubulin) |
| Fusarium solani | Bogale, M. et al. 2006 | DQ220209 (β-tubulin) |
Supporting Information
Intercellular colonization of endphytes (arrows) in the roots of pigeon pea.
(DOC)
Acknowledgments
The authors gratefully acknowledge Prof. Nie Shaoquan (Northeast Forestry University, Harbin, PR China) for plant authenticated. The authors thank Ms. Karen Duffy (Cornell University, Ithaca, NY, USA) for critically reading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Agricultural Science and Technology Achievements Transformation Fund Program (2009GB23600514), Special Fund of Forestry Industrial Research for Public Welfare of China (201004040), Heilongjiang Province Science Foundation for Excellent Youths (JC200704), Project for Distinguished Teacher Abroad, Chinese Ministry of Education (MS2010DBLY031) and the Key Program for Science and Technology Development of Harbin (2009AA3BS083). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fujita K, Kai Y, Takayanagi M. Genotypic variability of pigeonpea in distribution of photosynthetic carbon at low phosphorus level. Plant Sci. 2004;166:641–649. [Google Scholar]
- 2.Tang Y, Wang B, Zhou XJ. Effect of external application of herbal cajani preparation on the fibronection content during healing process of open wound. J Guangzhou U Tradit Chin Med. 1999;16:302–304. [Google Scholar]
- 3.Kong Y, Fu YJ, Zu YG, Liu W, Wang W, et al. Ethanol modified supercritical fluid extraction and antioxidant activity of cajaninstilbene acid and pinostrobin from pigeonpea [Cajanus cajan (L.) Millsp.] leaves. Food Chem. 2009;117:152–159. [Google Scholar]
- 4.Liu W, Zu YG, Fu YJ, Kong Y, Ma W, et al. Variation in contents of phenolic compounds during growth and post-harvest storage of pigeon pea seedlings. Food Chem. 2010;121:732–739. [Google Scholar]
- 5.Grover JK, Yadav S, Vats VJ. Medicinal plants of India with anti-diabetic potential. J Ethnopharmacol. 2002;81:81–100. doi: 10.1016/s0378-8741(02)00059-4. [DOI] [PubMed] [Google Scholar]
- 6.Morton JF. The pigeon pea (Cajanus cajan Millsp.), a high protein, tropicalbush legume. HortScience. 1976;11(1):11–19. [Google Scholar]
- 7.Sun SM, Song YM, Liu J. Studies on the pharmacology of Cajanin preparation. Chinese Traditional and Herbal Drugs. 1995;26(3):147–148. [Google Scholar]
- 8.Fu YJ, Zu YG, Liu W, Efferth T, Zhang NJ, et al. Optimization of luteolin separation from pigeonpea [Cajanus cajan (L.) Millsp.] leaves by macroporous resins. J Chromatogr A. 2006;1137(2):145–152. doi: 10.1016/j.chroma.2006.08.067. [DOI] [PubMed] [Google Scholar]
- 9.Fu YJ, Zu YG, Liu W, Zhang L, Tong MH, et al. Determination of vitexin and isovitexin in pigeonpea using ultrasonic extraction followed by LC–MS. J Sep Sci. 2008;31(2):268–275. doi: 10.1002/jssc.200700312. [DOI] [PubMed] [Google Scholar]
- 10.Wu N, Fu K, Fu YJ, Zu YG, Chang FR, et al. Antioxidant activities of extracts and main components of pigeonpea [Cajanus cajan (L.) Millsp.] leaves. Molecules. 2009;14:1032–1043. doi: 10.3390/molecules14031032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duker-Eshun G, Jaroszewski JW, Asomaning WA, Oppong-Boachie F, Christensen SB. Antiplasmodial constituents of Cajanus cajan. Phytother Res. 2004;18:128–130. doi: 10.1002/ptr.1375. [DOI] [PubMed] [Google Scholar]
- 12.Zu YG, Fu YJ, Liu W, Hou CL, Kong Y. Simultaneous determination of four flavonoids in pigeonpea [Cajanus cajan (L.) Millsp.] leaves using RP-LC-DAD. Chromatographia. 2006;63:499–505. [Google Scholar]
- 13.Zheng YY, Yang J, Chen DH, Sun L. Effects of the stilbene extracts from Cajanus cajan L. on ovariectomy-induced bone loss in rats. Acta Pharm Sinica. 2007;42:562–565. [PubMed] [Google Scholar]
- 14.Cooksey CJ, Dahiya JS, Garratt PJ, Strange RN. Two novel stilbene-2-carboxylic acid phytoalexins from Cajanus cajan. Phytochemistry. 1982;21(12):2935–2938. [Google Scholar]
- 15.Chong JL, Poutaraud A, Hugueney P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009;177:143–155. [Google Scholar]
- 16.Petrini O. Fungal endophytes of tree leaves. In: Andrews JH, Hirano SS, editors. Microbial Ecology of Leaves. Springer, New York; 1991. pp. 179–197. [Google Scholar]
- 17.Herre EA, Knowlton N, Mueller UG, Rehne SA. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol Evol. 1999;14(2):49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- 18.Redman RS, Sheehan KB, Scout RG, Rodríguez RJ, Henson JM. Thermotolerance generated by plant/fungal symbiosis. Science. 2002;298:1581. doi: 10.1126/science.1072191. [DOI] [PubMed] [Google Scholar]
- 19.Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, et al. Fungal endophytes limit pathogen damage in a tropical tree. PNAS. 2003;100:15649–15654. doi: 10.1073/pnas.2533483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. PNAS. 2005;102:13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li WK, Hu ZB. Endophytes and natural medicines. Chin J Nat Med. 2005;4:193–199. [Google Scholar]
- 22.Poland WSW, Reinhard K, Thomas P, Eva M, Heidrum A. Anti-Candida metabolites from endophytic fungi. Phytochemistry. 2007;68:886–892. doi: 10.1016/j.phytochem.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Stierle A, Strobel G, Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science. 1993;5105:214–216. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- 24.Strobel GA. Rainforest endophytes and bioactive products. Crit Rev Biotechnol. 2002;22:315–333. doi: 10.1080/07388550290789531. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Guo LD, Hyde KD. Taxonomic placement of sterile morphotypes of endophytic fungi from Pinus tabulaeformis (Pinaceae) in northeast China based on rDNA sequences. Fungal Divers. 2005;20:235–260. [Google Scholar]
- 26.Xu LL, Han T, Wu JZ, Zhang QY, Zhang H, et al. Comparative research of chemical constituents, antifungal and antitumor properties of ether extracts of Panax ginseng and its endophytic fungus. Phytomedicine. 2009;16:609–616. doi: 10.1016/j.phymed.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Nel B, Steinberg C, Labuschagne N, Viljoen A. Isolation and characterization of nonpathogenic Fusarium oxysporum isolates from the rhizosphere of healthy banana plants. Plant Pathol. 2006;55:207–216. [Google Scholar]
- 28.Sébastien A, Christelle C, Claude A, Chantal O. Comparative analysis of PR gene expression in tomato inoculated with virulent Fusarium oxysporum f. sp. lycopersici and the biocontrol strain F. oxysporum Fo47. Physiol Mol Plant P. 2008;73(1–3):9–15. [Google Scholar]
- 29.Booth C. The Genus Fusarium. Commonwealth Agricultural Bureau: Kew, Surrey, England; 1971. [Google Scholar]
- 30.Nelson PE, Toussoun TA, Mararas WFO. Fusarium species: An Illustrated Manual for Identification. University Park, PA: The Pennsylvania State University Press; 1983. [Google Scholar]
- 31.Windels CE. Fusarium. In: Singleton LL, Mihail JD, Rush CM, editors. Methods for Research on Soilborne Phytopathogenic Fungi. American Phytopathological Society Press. St Paul; 1992. pp. 115–128. [Google Scholar]
- 32.Ouellette GB, Bard G. A perennial canker of balsam fir on Anticosti Island. Plant Dis Rep. 1966;50:722–724. [Google Scholar]
- 33.Schultz ME, Parmeter JR. A canker disease of Abies concolor caused by Nectria fuckeliana. Plant Dis. 1990;74:178–180. [Google Scholar]
- 34.Brayford D, Honda BM, Mantiri FR, Samuels GJ. Neonectria and Cylindrocarpon: the Nectria mammoidea group and species lacking microconidia. Mycologia. 2004;96:572–597. [PubMed] [Google Scholar]
- 35.Vasiliauskas R, Stenlid J, Johansson M. Fungi in bark peeling wounds of Picea abies in central Sweden. Eur J Forest Pathol. 1996;26:285–296. [Google Scholar]
- 36.Rossman AY, Samuels GJ, Rogerson CT, Lowen R. Genera of Bionectria-ceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol. 1999;42:1–260. [Google Scholar]
- 37.Seifert KA, McMullen CR, Yee D, Reeleder RD, Dobinson KF. Molecular differentiation and detection of ginseng-adapted isolates of the root rot fungus Cylindrocarpon destructans. Phytopathology. 2003;93:1533–1542. doi: 10.1094/PHYTO.2003.93.12.1533. [DOI] [PubMed] [Google Scholar]
- 38.Bigelis R, He HY, Yang HY, Chang LP, Greenstein M. Production of fungal antibiotics using polymeric solid supports in solid-state and liquid fermentation. J Ind Microbiol Biot. 2006;33:815–826. doi: 10.1007/s10295-006-0126-z. [DOI] [PubMed] [Google Scholar]
- 39.Booth C. Studies of pyrenomycetes. IV. Nectria (part 1). Mycological Papers. 1959;73:1–115. [Google Scholar]
- 40.Samuels GJ, Brayford D. Phragmosporous Nectria species with Cylindrocarpon anamorphs. Sydowia. 1993;45:55–80. [Google Scholar]
- 41.Vasiliauskas R, Lygis V, Thor M, Stenlid J. Impact of biological (Rotstop) and chemical (urea) treatments on fungal community structure in freshly cut Picea abies stumps. Biol Control. 2004;31:405–413. [Google Scholar]
- 42.Xin G, Glawe D, Doty SL. Characterization of three endophytic, indole-3-acetic acid producing yeasts occurring in Populus trees. Mycol Res. 2009;113:973–980. doi: 10.1016/j.mycres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Anderson IC, Cairney JWG. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol. 2004;6:769–779. doi: 10.1111/j.1462-2920.2004.00675.x. [DOI] [PubMed] [Google Scholar]
- 44.O'Donnell K, Kistler HC, Tacke BK, Casper HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. PNAS. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Germaine K, Keogh E, Garcia-Cabellos G, Borremans B, Lelie D, et al. Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol Ecol. 2004;48:109. doi: 10.1016/j.femsec.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Tan RX, Zou WX. Endophytes: a rich source of functional metabolites. Nat Prod Rep. 2001;18:448–459. doi: 10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- 47.Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. Systematics of Basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 48.Strobel G, Daisy B. Bioprospecting for Microbial Endophytes and Their Natural Products. Microbiol Mol Biol R. 2003;67(4):491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hua X, Fu YJ, Zu YG, Wu N, Kong Y, et al. Plasma pharmacokinetics and tissue distribution study of cajaninstilbene acid in rats by liquid chromatography with tandem mass spectrometry. J Pharmaceut Biomed. 2010;52:273–279. doi: 10.1016/j.jpba.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 50.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, PCR Protocols: a guide to methods and applications. Academic Press, SanDiego; 1990. pp. 315–322. [Google Scholar]
- 51.O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7:103–106. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 52.Sánchez Márquez S, Bills GF, Zabalgogeazcoa I. The endophytic mycobiota of the grass Dactylis glomerata. Fungal Divers. 2007;27:171–195. [Google Scholar]
- 53.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 54.Saitou N, Nei M. The neighbor-joining method–a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 55.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intercellular colonization of endphytes (arrows) in the roots of pigeon pea.
(DOC)