Abstract
Aromatase (CYP19A1) is an integral membrane enzyme that catalyzes the removal of the 19-methyl group and aromatization of the A-ring of androgens. All human estrogens are synthesized from their androgenic precursors by this unique cytochrome P450. The crystal structure of active aromatase purified from human placenta has recently been determined in complex with its natural substrate androstenedione in the high-spin ferric state of heme. Hydrogen bond forming interactions and tight packing hydrophobic side chains closely complement puckering of the steroid backbone, thereby providing the molecular basis for the androgenic specificity of aromatase. In the crystal, aromatase molecules are linked by a head-to-tail intermolecular interaction via a surface loop between helix D and helix E of one aromatase molecule that penetrates the heme-proximal cavity of the neighboring, crystallographically-related molecule, thus forming in tandem a polymeric aromatase chain. This intermolecular interaction is similar to the aromatase-Cytochrome P450 reductase coupling and is driven by electrostatics between the negative potential surface of the D-E loop region and the positively charged heme-proximal cavity. This loop-to-proximal site link in aromatase is rather unique - there are only a few of examples of somewhat similar intermolecular interactions in the entire P450 structure database. Furthermore, the amino acids involved in the intermolecular contact appear to be specific for aromatase. Higher order organization of aromatase monomers may have implications in lipid integration and catalysis.
1. Introduction
Cytochrome P450 aromatase (CYP19A1) uses with high specificity the androgenic substrates androstenedione, testosterone, and 16α-hydroxytestosterone as substrates and converts them to their respective estrogens, estrone, 17β-estradiol, and 17β,16α-estriol, respectively [1–3]. The reaction requires coupling with its redox partner cytochrome P450 reductase (CPR) for the transfer of electrons from NADPH. Aromatase is the only known enzyme in vertebrates capable of catalyzing the aromatization of a six-membered ring. The functional human enzyme is monomeric, comprised of a heme group and a single polypeptide chain of 503 amino-acid residues. It is an integral membrane protein of the endoplasmic reticulum, anchored to the membrane by an amino terminal transmembrane domain [4,5], in addition to other membrane-associating regions. Human microsomal P450s catalyze metabolism of a wide variety of endogenous and xenobiotic compounds and drugs with low substrate specificities. However, those in the sex-steroid biosynthesis pathways, such as aromatase and 17α-hydroxylase/17,2-lyase (CYP17A1) have high substrate specificity [6].
More than 70% of all breast cancers are estrogen-dependent [7]. Inhibition of estrogen biosynthesis by aromatase inhibitors (AIs) constitutes one of the modern therapies for postmenopausal estrogen-dependent breast cancer [8,9]. Aromatase has been the subject of biochemical and biophysical investigations for the past 40 years [10]. Until recently, lack of a crystal structure of aromatase had impeded understanding of the mechanism of action at the molecular level and development of rationally designed AIs with higher specificity and efficacy. Using aromatase purified from term human placenta, we have been able to elucidate the crystal structure of the enzyme, delineate the molecular determinants of androgenic specificity and propose a mechanism for the third step of the aromatization reaction [11].
In the crystal, interestingly, aromatase molecules are linked by a head-to-tail intermolecular interaction via a surface loop between helix D and helix E of one aromatase molecule that penetrates the heme-proximal cavity of the next molecule, thereby forming in tandem a polymeric aromatase chain. This intermolecular interaction, analogous to the aromatase-Cytochrome P450 reductase coupling, is driven by electrostatics between the negative potential surface of the D–E loop region and the positively charged heme-proximal cavity. There are only a few examples of similar head-to-tail oligomerization in the P450 structure database. Here, we analyze in molecular detail this higher order organization of human aromatase, and compare it with other P450 structures in the 3-D structure database of Protein Data Bank (PDB) [12].
2. Experimental
2.1. Determination of the crystal structure of human placental aromatase
Details of purification of aromatase from human placenta, crystallization and structure determination have already been published [11,13] (PDB ID code for the coordinates: 3EQM). Only a brief account is being provided here. An immunoaffinity chromatography column made with the highly specific monoclonal antibody mAb 3-2C2 was used for the purification of aromatase. Freshly purified aromatase was crystallized by the vapor diffusion technique. The purification and crystallization experiments were conducted at 4°C. Diffraction data sets were gathered at the beam lines A-1 of Cornell High Energy Synchrotron Source, Cornell University, Ithaca, NY and 19ID of Advanced Photon Source, Argonne National Laboratory, Argonne, IL. The crystals were cryo-cooled at the liquid nitrogen temperature using glycerol as the cryo-protectant. The diffraction data was complete to 2.90• resolution. The space group is P3221 and the unit cell parameters are a=b=140.2•, c=119.3•, α=β=90°, γ=120°. The structure was solved by molecular replacement method coupled with Bijvoet difference Fourier synthesis utilizing the Fe-absorption edge datasets. The final refined model contained 452 amino acid residues (Ser45 to Asn496), a heme group, one androstenedione molecule, 35 solvent waters, and 2 phosphate ions (3792 total atoms). A putative model for the 33 amino-terminal residues (Asn12 to Thr44), consisting of a trans-membrane helix with terminal residues Ile16 and Val38, was built through the weak electron density trace, but not included in the refinement.
2.2. Analysis of the oligomeric structures
Oligomeric structure was constructed by applying the crystallographic symmetries using Coot [14] and analyzed by Chimera [15]. Illustrations shown in Figs. 1 and 2 were drawn using Chimera. The coordinates all other P450 crystal structures were obtained from the Protein Data Bank [12].
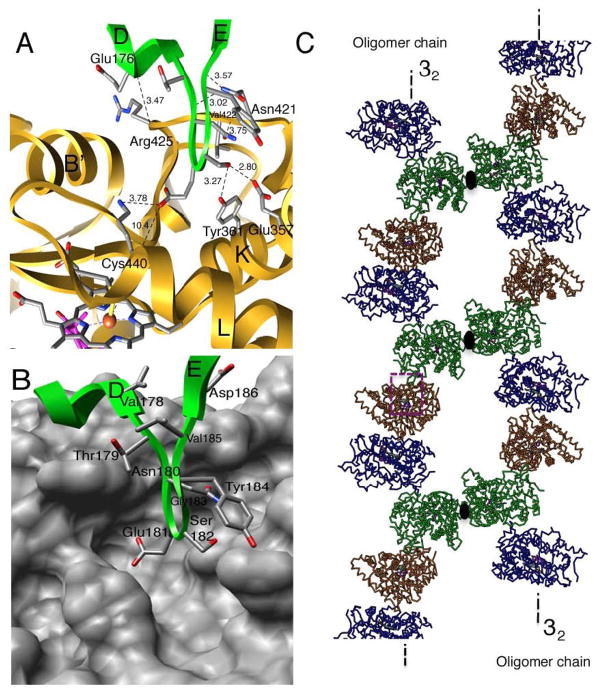
Fig. 1. Intermolecular interaction in human placental aromatase.
(A) A close-up view of the “head-to-tail” association of the D–E loop of the green molecule and the proximal cavity of the gold molecule. Intermolecular hydrogen bonds and salt bridges are indicated by dashed lines and the distances in angstrom are given. (B) The proximal site is shown by the van der Waals surface to illustrate the depth of the cavity. (C) Two polymeric aromatase chains formed by “head-to-tail” association in the crystal, each about a 32 crystallogarphic screw axis, are shown. The chains pack against each other about a 2-fold rotation axis shown. A magnified view of the boxed region of interaction is shown in (A) and (B). The protein backbone colors and viewing direction are same in all panels.
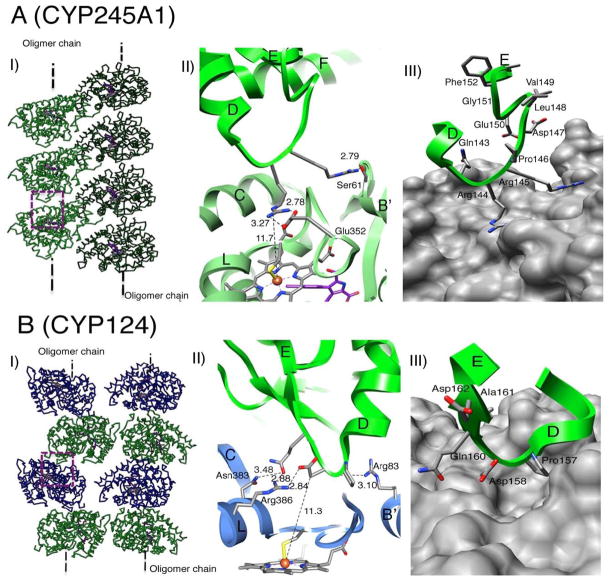
Fig. 2. Similar D-E loop to proximal site intermolecular association in other P450s.
(A) CYP245A1 and (B) CYP124: (I) Two oligomer chains in the crystal, (II) Close-up view of the boxed region in (I). The intermolecular polar interactions and the closest distance of approach to heme Fe are indicated. See text for more details. (III) Rendering of the proximal cavity by van der Waals surface illustrates their shallowness.
2.3. Structure-based sequence alignment
Owing to low homology, structure-based alignment of the sequences of D–E and K–L loops of aromatase and four other P450s (CYP101, CYP124, CYP245A1 and CYP3A4) was carried out. CYP3A4 and CYP101 were included as representative P450 structures. MOE [16] packages of software were used to least-square superimpose the structures in a region-specific manner. The 3-D alignment provided the information on the lengths of the peptides to compare.
3. Results
3.1. The D-E loop to proximal site interaction
The proximal site is the surface cavity behind the cysteine thiolate ligand of the heme moiety and the distal site comprises the substrate-binding pocket. The D–E loop consists of residues Val178-Thr179-Asn180-Glu181-Ser182Gly183-Tyr184-Val185-Asp186 between helices D and E (Fig. 1A). The nine-residue long loop inserts itself in the heme-proximal cavity of the neighboring molecule. The proximal cavity, shown by depicting the van der Waals surface in Fig. 1B, is comprised primarily of residues 354–361 from the K helix, 418–432 from the K–L loop and 440–444 from the L helix. The D–E loop makes several polar contacts, forming five hydrogen bonds and one salt bridge with residues within the cavity. The hydrogen bonds are: backbone O of Tyr 184 to ND2 of Asn421 (3.02 •), backbone N of Asn180 to ND2 of ASN 421 (3.58 •), OD of Asn180 to backbone N of Val422 (3.75 •), OG of Ser182 to OH of Tyr361 (3.27 •), and OG of Ser182 to OE2 of Glu357 (2.80 •). In addition, there is Coulombic interaction between two charged side chains OE1 of Glu181 to NZ of Lys440 (3.78 •), and a hydrogen bond between backbone O of Glu176 and backbone N of Arg425 (3.47 •). Together, these provide additional stability to the loop-proximal cavity association. Furthermore, calculation of electrostatic potentials at these two interacting regions shows that electrostatic attraction between the two oppositely charged surfaces, and their shape complementarity could actually be responsible for driving the head-to-tail dimer formation (D. Ghosh et al., unpublished data). The D–E loop is primarily negatively charged (Glu181, Asp186), whereas the heme-proximal site contains predominantly positively charged side chains (Lys108, Lys354, Lys420, Arg425, Lys440, Lys448).
In the aromatase crystal, this head-to-tail intermolecular interaction between the D–E loop of one monomer and the proximal cavity of the other through shape complementarity and electrostatic interactions generates a polymeric aromatase chain about the 32-screw axis, as illustrated in Fig. 1C. Additionally, two polymer chains pack about a 2fold rotational symmetry axis normal to the screw axis, forming the P3221 space group symmetry. The formation of an open proximal cavity is made possible by a long K–L loop and two helices K and L that line the cavity optimally in depth to accommodate the D–E loop (Fig. 1B). The innermost point of the loop, the OE1 atom of Glu181, is 10.4• from the heme Fe of the neighboring monomer (Fig. 1A).
3.2. Intermolecular interaction via the proximal site in other P450’s
We have analyzed the intermolecular interaction and crystal packing for all 54 P450 structures available in the Protein Data Bank [12] (Table 1). Only in three cases the interactions involve the proximal sites. Furthermore, only two of those display a similar albeit less pronounced D–E loop to proximal site association, shown in Figs. 2A and B. Like aromatase, both CYP245A1 and CYP124 form oligomers in the crystals via crystallographic symmetry. In the CYP245A1 oligomer (Fig. 2A(I)) the D–E loop consisting of Gln143-Arg144-Arg145-Pro146-Asp147-Leu148Val149-Glu150-Gly151-Phe152-Ala153 forms a hydrogen bond, NH2 of Arg145 to OG of Ser 61 (2.79•) and two salt bridges, NH2 of Arg144 to OE2 of Glu352 (2.79•), and NH1 of Arg144 to OE1 of Glu352 (2.78•) (Fig. 2A(II)). The two interacting surfaces, therefore, have mixed positive and negative charges, unlike aromatase. In addition, access to the proximal cavity is partially blocked by the helices L and B′ and the K–L loop (Figs. 2A(II) and 2A(III)). Consequently, only a part of the D–E loop is inserted into the shallow proximal cavity, unlike aromatase. The closest approach to the heme iron is 11.7 • of NE of Arg144 side chain.
Table 1.
List of P450 crystal structures used in the analysis
| UniPort Entry | CYP | PDB ID | UniPort Entry | CYP | PDB ID |
|---|---|---|---|---|---|
| A9LLA5_DANRE | 8A1 | 3B98 | CP142_MYCTU | 142 | 2XKR |
| BIOI_BACSU | BIOI | 3EJB | CYPC_BACSU | BSBETA | 2ZQJ |
| C167_POLCB | EPOK | 1PKF | CYPX_BACSU | 134A1 | 3NC3 |
| C4B644_9PSEU | VDH | 3A4G | CPXA_PSEPU | CAM | 3L61 |
| C5B3_AMYOR | OXYB | 1LGF | CPXB_BACME | BM-3 | 1FAG |
| CP1A2_HUMAN | 1A2 | 2HI4 | CPXE_STRGO | 105A1 | 2ZBX, 3CV8 |
| CP2A6_HUMAN | 2A6 | 2FDU, 3EBS, 2PG5 | CPXJ_SACEN | ERYF | 1OXA |
| CP2A13_HUMAN | 2A13 | 2P85 | CPXL_PSESP | TERP | 1CPT |
| CP2B4_RABIT | 2B4 | 2Q6N, 3MVR | CPXQ_SACEN | ERYK | 2XFH |
| CP2B6_HUMAN | 2B6 | 3IBD | CPXW_SULSO | 119 | 1F4U |
| CP2C8_HUMAN | 2C8 | 1PQ2, 2NNI, 2VN0 | Q2G8A2_NOVAD | 101D2 | 3NV5 |
| CP2C9_HUMAN | 2C9 | 1OG2, 1R9O | Q2GB12_NOVAD | 101D1 | 3LXH |
| CP2D6_HUMAN | 2D6 | 2F9Q | Q2L6S8_9ACTO | 105 | 2Z36 |
| CP2E1_HUMAN | 2E1 | 3E4E | Q385E8_9TRYP | 51 | 2X2N |
| CP2R1_HUMAN | 2R1 | 3C6G | Q53W59_THET8 | HB8 | 1WIY |
| CP3A4_HUMAN | 3A4 | 2J0D | Q6KZ68_PICTO | 231A2 | 2RFB |
| CP46A_HUMAN | 46A1 | 2Q9F | Q6N8N2_RHOPA | 199A2 | 2FR7 |
| CP74A_ARATH | 74A | 2RCH | Q6ZZI7_ACTTI | 165D3 | 3O1A |
| CP7A1_HUMAN | 7A1 | 3DAX | Q83WG3_9ACTO | 245A1 | 3A1L |
| CP19A_HUMAN | 19A1 | 3EQM | Q939Y1_9PSEU | OXYD | 3MGX |
| CP11A_HUMAN | 11A1 | 3NA0 | Q93H80_STRAW | 105D6 | 3ABB |
| CP24A_RAT | 24A1 | 3K9V | Q93H81_STRAW | 105P1 | 3E5J |
| CP120_SYNY3 | 120A1 | 2VE3 | Q9EW92_9ACTO | PIMD | 2X9P |
| CP121_MYCTU | 121 | 3CXV, 3G5F | Q9FCA6_STRCO | 158A2 | 2D09 |
| CP124_MYCTU | 124 | 2WM4 | Q9K498_STRCO | 170A1 | 3DBG, 3EL3 |
| CP125_MYCTU | 125 | 3IW1 | Q9KZF5_STRCO | 158A1 | 2DKK |
| CP130_MYCTU | 130 | 2UUQ | O87605_9ACTO | PIKC | 2VZ7, 2CA0 |
The D–E loop, Pro157-Asp158-Arg159-Gln160-Ala161-Asp162, in the CYP124 oligomer (Fig. 2B(I)) is considerably shorter than those in aromatase and CYP245A1. It makes two hydrogen-bonding contacts, OE1 of Glu160 to ND2 of Asn383 (3.48 •), and backbone O of Pro157 to NH1 of Arg83 (3.10 •), and two salt bridges, OE1 of Glu 160 to NH1 of Arg386 (2.88 •), and OD2 of Asp158 to NH2 of Arg386 (2.84 •) to the proximal cavity. Again, the cavity itself is shallow, partially blocked by helices L and B′ and the K–L loop (Figs. 2B(II) and 2B(III)). The D–E loop penetrates the cavity minimally and residues Pro157 and Asp158 stay outside the cavity (Fig. 2B(III)). The closest approach of a D–E loop atom to heme iron is 11.3• by CB of Asp158.
3.3. Analysis of amino acid sequences
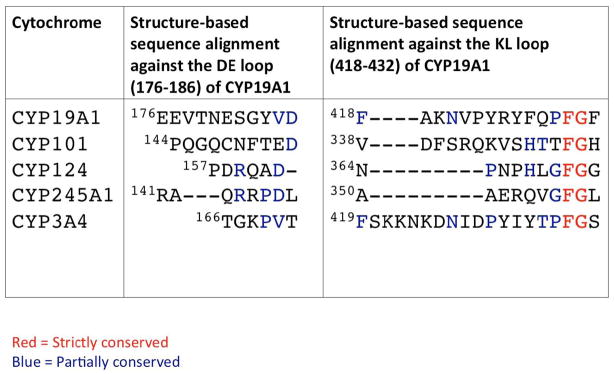
Alignment of the sequences of the D–E loop of the P450’s of interest and the most common region of the proximal site, the residues from the K–L loop, are shown in Fig. 3. It suggests very little, if any, conservation of the D–E loop residues among the P450’s. Based on the 3-D structure alignment, the loop also has a wide range of variability in terms of its length, and aromatase has one of the longest. Interestingly, while the other D–E loops in Fig. 3 have mixed charged residues, all charged residues in aromatase are Glu and Asp. Among the elements that make up the proximal cavity, the K–L loop is the longest and perhaps shows some homology (Fig. 3). The part of the loop associated with the proximal site is considerably longer in aromatase than in CYP124 and CYP245A1. The shortening of this section of the loop is partially responsible for limited opening to the proximal cavity in these two P450 structures. There is a propensity of Arg and Lys side chains in the positively charged proximal cavity of aromatase, which is reflected as well in the K–L loop. However, the same is not strictly true for other P450’s. The strictly conserved Phe-Gly pair of the loop is closest in space to the Cys thiolate ligand to heme as the Phe side chain packs against the porphyrin moiety and the Cys side chain, thus providing an explanation for the conservation. All in all, sequence comparison in the regions of intermolecular association suggests some uniqueness of aromatase, and rationale for the distinctive quaternary organization.
Fig. 3. Structure-based alignment of the sequences of the D–E and K–L loops of aromatase (CYP19A1), CYP124, CYP245A1 and two other representative P450’s.
The alignment shows variability of the lengths and low homology among the sequences.
4. Discussion
Analysis of the crystal structure of aromatase elucidates that Coulombic forces between the two oppositely charged surfaces and their shape complementarity could actually be responsible for driving the head-to-tail dimer formation, which could be the basis for higher order organization of aromatase. This is consistent with a recent report providing evidence for aromatase dimer, and possibly higher order oligomer formation within the lipid membrane for both full length and N-terminal modified enzyme [17]. This association is akin to the results from docking of CPR with aromatase demonstrating that the highly negatively charged FMN-binding domain of CPR binds to the positively charged proximal surface of aromatase through electrostatic attractions [18]. It is possible that in fact an aromatase oligomer, and not a monomer, associates with the electron donor through electrostatic interactions. The association between the loop and the proximal residues by polar and electrostatic interactions, as well as complementarity of the conjoining interfacial surfaces are probably sufficient for maintaining the specific quaternary organization.
Additionally, the negatively charged D-E loop-to-proximal site interaction could also influence the electronics of the neighboring active site heme only 11• away. This interaction may be important for the maintenance of the integrity of heme oxidation state, and for molecule-to-molecule transfer of electrons that drives the hydroxylation reaction. In the interaction between P450cam and its electron donor putidaredoxin, a proximal site Gln residue was shown to control electron transfer rate by modulating the redox potential of the heme iron and an Arg residue to regulate the formation of electron transfer pathway [19, 20].
However, CPR couples to a P450 with the FMN-binding domain near the same proximal cavity [21,22]. The current working hypothesis is that CPR-binding and loop-to-proximal cavity association are mutually exclusive. One possible way to achieve this would be if CPR binds not to a monomeric aromatase, but to a higher order organization with an open proximal end, or by displacing a monomer in a closed or cyclized oligomeric state. The oligomerization could also be a means of protection of aromatase in its resting low spin or substrate-bound high spin ferric state, until CPR couples to the proximal site by displacing the neighboring aromatase molecule.
A recent report suggests increased aromatase activity due to phosphorylation of Tyr361 by nongenomic signaling of 17β-estradiol in breast cancer cell lines [23]. Tyr361 at the proximal site is involved in a hydrogen-bonding interaction with the D–E loop. Phosphorylation of this residue would significantly alter the electrostatic potential of the proximal cavity. Furthermore, a phosphorylated Tyr361 would interfere with the D–E loop insertion and oligomer formation. Oligomeric aromatase could thus be self-protective by preventing a kinase from constitutively binding and phophorylating at the proximal site. More in-depth analysis of the molecular basis for the coupling and functional consequences thereof is underway and will be published elsewhere.
Acknowledgments
The research is supported in part by grant GM086893 from the National Institutes of Health, U.S.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thompson EA, Siiteri PK. Utilization of oxygen and reduced nicotinamide adenine dinucleotide phosphate by human placental microsomes during aromatization of androstenedione. J Biol Chem. 1974;249:5364–5372. [PubMed] [Google Scholar]
- 2.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD, Meldenson CR, Bulun SE. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 3.O’Neal Johnston J. Aromatase inhibitors. Critical Rev Biochem Mol Biol. 1998;33:375–405. [PubMed] [Google Scholar]
- 4.Shimozawa O, Sakaguchi M, Ogawa H, Harada N, Mihara K, Omura T. Core glycosylation of cytochrome P450(arom). Evidence for localization of N terminus of microsomal cytochrome P-450 in the lumen. J Biol Chem. 1993;268:21399–21402. [PubMed] [Google Scholar]
- 5.Amarneh B, Corbin CJ, Peterson JA, Simpson ER, Graham-Lorence S. Functional domains of human aromatase cytochrome P450 characterized by linear alignment and site-directed mutagenesis. Mol Endocrinol. 1993;7:1617–1624. doi: 10.1210/mend.7.12.8145767. [DOI] [PubMed] [Google Scholar]
- 6.Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32:0000–0000. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montemurro F, Aglietta M. Hormone receptor-positive early breast cancer: controversies in the use of adjuvant chemotherapy. Endocrine-Related Cancer. 2009;16:1091–1102. doi: 10.1677/ERC-09-0033. [DOI] [PubMed] [Google Scholar]
- 8.Brueggemeier RW. Update on the use of aromatase inhibitors in breast cancer. Expert Opin Pharmacother. 2006;7:1919–1930. doi: 10.1517/14656566.7.14.1919. [DOI] [PubMed] [Google Scholar]
- 9.Eisen A, Trudeau M, Shelley W, Messersmith H, Pritchard KI. Aromatase inhibitors in adjuvant therapy for hormone receptor positive breast cancer: A systematic review. Cancer Treat Rev. 2008;34:157–174. doi: 10.1016/j.ctrv.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Santen RJ, Brodie H, Simpson ER, Siiteri PK, Brodie A. History of aromatase: saga of an important biological mediator and therapeutic target. Endocr Rev. 2009;30:343–375. doi: 10.1210/er.2008-0016. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh D, Griswold J, Erman M, Pangborn W. Structural basis for androgen specificity and oestrogen biosynthesis in human aromatase. Nature. 2009;457:219–223. doi: 10.1038/nature07614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lala P, Higashiyama T, Erman M, Griswold J, Wagner T, Osawa Y, Ghosh D. Suppression of human cytochrome P450 aromatase activity by monoclonal and recombinant antibody fragments and identification of their stable antigenic complex. J Steroid Biochem Mol Biol. 2004;88:235–245. doi: 10.1016/j.jsbmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 15.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 16.Molecular Operating Environment (MOE), version 2009.10. Chemical Computing Group, Inc; Montreal, Quebec, Canada: 2009. [Google Scholar]
- 17.Praporski S, Ng SM, Nguyen AD, Corbin CJ, Mechler A, Zheng J, Conley AJ, Martin LL. Organization of Cytochrome P450 enzymes involved in sex steroid synthesis: protein-protein interaction in lipid membranes. J Bio Chem. 2009;284:33224–33232. doi: 10.1074/jbc.M109.006064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong Y, Li H, Yuan Y, Chen S. Molecular characterization of aromatase. Ann NY Acad Sci. 2009;1155:112–120. doi: 10.1111/j.1749-6632.2009.03703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tosha T, Yoshioka S, Hori H, Takahashi S, Ishimori K, Morishima I. Molecular mechanism of the electron transfer reaction in cytochrome P450(cam)--putidaredoxin: roles of glutamine 360 at the heme proximal site. Biochemistry. 2002;41:13883–13893. doi: 10.1021/bi0261037. [DOI] [PubMed] [Google Scholar]
- 20.Unno M, Shimada H, Toba Y, Makino R, Ishimura Y. Role of Arg112 of Cytochrome P450cam in the Electron Transfer from Reduced Putidaredoxin: Analyses with site-directed mutants. J Biol Chem. 1996;271:17869–17874. doi: 10.1074/jbc.271.30.17869. [DOI] [PubMed] [Google Scholar]
- 21.Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci USA. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong Y, Li H, Ye J, Miki Y, Yuan YC, Sasano H, Evans DB, Chen S. Epitope characterization of an aromatase monoclonal antibody suitable for the assessment of intratumoral aromatase activity. PLoS One. 2009;30:e8050. doi: 10.1371/journal.pone.0008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catalano S, Barone I, Giordano C, Rizza P, Qi H, Gu G, Malivindi R, Bonofiglio D, Ando S. Rapid estradiol/ER signaling enhances aromatase enzymatic activity in breast cancer cells. Mol Endocrinol. 2009;23:1634–1645. doi: 10.1210/me.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]