Abstract
Disulfide bonds are a form of posttranslational modification that often determines protein structure(s) and function(s). In this work, we report a mass spectrometry method for identification of disulfides in degradation products of proteins, and specifically endogenous peptides in the human blood plasma peptidome. LC-Fourier transform tandem mass spectrometry (FT MS/MS) was used for acquiring mass spectra that were de novo sequenced and then searched against the IPI human protein database. Through the use of unique sequence tags (UStags) we unambiguously correlated the spectra to specific database proteins. Examination of the UStags’ prefix and/or suffix sequences that contain cysteine(s) in conjunction with sequences of the UStags-specified database proteins is shown to enable the unambigious determination of disulfide bonds. Using this method, we identified the intermolecular and intramolecular disulfides in human blood plasma peptidome peptides that have molecular weights of up to ~10 kDa.
INTRODUCTION
Polypeptides often achieve their stable conformations through formation of disulfides, and the mechanisms for formation of disulfides have been recently reviewed (1,2). The physiological and pathological relevance of disulfides to diseases and protein functions has been increasingly recognized for e.g. tumor immunity (3), hemostasis (4), cell death and neurodegenerative diseases (5), growth factors (6), G-protein-receptors (7), etc.
Determination of disulfides can be accomplished using X-ray crystallography (8) and NMR (9) analyses, but typically requires purified proteins having known sequences. Mass spectrometry (MS) enables identification of disulfides (10) and advances have been reported for working with complex mixtures of proteins (11–14). For example, Nair et al. used de novo sequencing to find disulfides in peptide Mo1274 from FT MS/MS spectra (11), Xu et al. investigated using the MassMatrix search engine for identification of disulfides from LC-LTQ spectra (12), Tiptom et al. analyzed disulfides in the GM2 protein activator protein (GM2AP) using LC-FT MS/MS (13), and Wu et al. analyzed the disulfides in tryptic peptides of three proteins (14). Due to the limited options for disulfide analysis using widely applied database searching programs, such as SEQUEST and Mascot, Choi et al. developed their own algorithm “DBond” for database searching of disulfides using Q-TOF spectra (15). However, reported studies were limited to known or standard model peptides and proteins (11–15), and this approach has not yet been demonstrated for proteomic or peptidomic analysis. Thus, databases used were very small, and e.g., the custom databases only incorporated sequences of the model proteins (12,13), and “enzyme rules” were needed for effective identification of disulfides in peptides through the database search (15). All these factors affect the methods developed for practical proteomic and peptidomic analyses that require searching huge numbers of spectra against large databases. Proteomics analysis commonly applies reduction/alkylation of cysteine residues in peptides generated by exogenous enzymes (e.g., trypsin), while there are few reports for peptidomic analysis for which there are no enzyme rules that can be applied to reduce the number of analysis targets.
In this work, we describe a de novo sequencing-UStags approach for identification of disulfide bonded species in the blood plasma peptidome peptides, as they can be potential biomarker candidates. Additionally, the methodology developed for the human blood plasma peptidome peptides can be applied for various types of applications, as the blood plasma peptidome peptides can have a large MW range of 1,000–15,000 Da, carry various charges (e.g., 1–15), and have termini of any of the amino acids (16). We show that the use of UStags greatly simplifies identification of both intramolecular and intermolecular disulfide bond species by searching large FT MS/MS spectral datasets against the large IPI human protein database and without the need to define (or assume) the peptide termini.
METHODS
Description of the FT MS/MS spectra datasets
The FT MS/MS spectral datasets used in this work have been submitted to the public repository pride (http://www.ebi.ac.uk/pride/). These datasets were obtained using LC-Orbitrap MS/MS analysis of the human blood plasma peptidome. The peptidome was isolated from early-stage breast cancer patients and healthy individuals using the method previously described (16). Briefly, the plasma sample was first depleted of its 12 high-abundance proteins using a 12.7 × 79.0 mm IgY12 LC10 AC column (Beckman Coulter, Fullerton, CA) according to the manufacturer’s recommendations. The AC flow through fraction (i.e., the depleted sample) was separated with use of a Superdex 200 10/300 GL SEC column (GE Healthcare, Piscataway, NJ). Based on the elution time calibrated with standard proteins beta-galactosidase, phosphorylase, bovine serum albumin, carbonic anhydrase, beta-lactoglobulin, and cytochrome c (Sigma-Aldrich, St. Louis, MO), the peptidome <20 kDa species were collected, and the contents of the resultant peptidome species were determined by BCA assay (Pierce, Rockford, IL). The isolated peptidome species (50 μg) were separated using HRLC equipped with a 100 cm × 100 μm i.d. capillary column that was manufactured in our lab with use of 3-μm porous (300 Å size) C4-bonded silica particles (Sepax Technologies, Inc. Newark, DE). The flow rate measured with the mobile phase A (acetonitrile/H2O/acetic acid, 10:90:0.2, v/v/v) was 0.5 μl/min. The separation was completed with a mobile phase gradient from mobile phase A to 70% B (acetonitrile/isopropyl alcohol/H2O/acetic acid/trifluoroacetic acid, 60:30:10:0.2:0.1, v/v/v/v/v) in 1600 min (the exponent gradient was generated in a static mixer under a constant pressure of 13.000 psi and room temperature). Two LC-MS/MS runs were performed for each sample, and all spectra collected were given in the public repository pride. The separated components were analyzed with an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific., San Jose, CA), and the spectra were collected at a 60K resolution for both survey scans and the following MS/MS. A survey scan with 400 ≤ m/z ≤ 2000 was followed by FT MS/MS of the 5 most intense ions from the survey scan (monoisotopic precursor selection not enabled). FT MS measurements with 3 micro scans were applied for all FT MS/MS analyses, and the minimum signal required for a selected precursor ion to trigger a subsequent MS/MS scan was set to 500. FT MS/MS employed 35% normalized collision energy for CID. Mass calibration was performed prior to analysis according to the method recommended by the instrument manufacturer. The same mass spectrometry conditions were used for analyses of various sizes of the peptidome components (isolated with a cutoff of 20 kDa), regardless of the presence of disulfide bonds.
The computational tools and methods
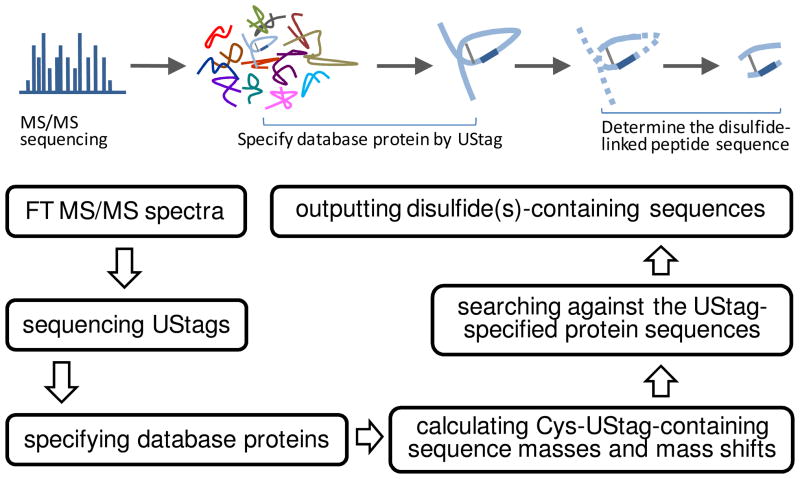
Figure 1 shows the flowchart for use of the de novo-UStags approach (17) to determine disulfides. Briefly, de novo sequencing was based on the monoisotopic masses de-isotoped from the Orbitrap FT MS/MS spectra using software ICR2LS (http://ncrr.pnl.gov/software/ICR2LS.stm). A mass error tolerance of 0.005 u was used for sequencing, and no gaps of >1 amino acid residues were allowed. The generated amino acid sequences were matched to the IPI human protein database (ipi.HUMAN.v3.39, ftp://ftp.ebi.ac.uk/pub/databases/IPI/, containing 69,731 sequence entries) to inspect for the sequence uniqueness (i.e., the criterion for establishing a UStag). If the sequences uniquely belong to specific proteins, they are referred as UStags; otherwise, the sequences were not accepted for identifications. Once UStags were obtained, the theoretical mass values of the UStag prefixes and suffixes were calculated according to the location of the UStags among the UStags-specified database protein sequences; the experimental mass values of the UStag prefixes were derived from masses of the smallest fragments in the UStags and the experimental mass values of the UStag suffixes were determined from the mass differences between the precursor and largest fragment. When the differences between the experimental and theoretical values were −2n×1.008 u and the UStag prefix and/or suffix contained 2n Cys (n: 1, 2, 3, …), disulfides were directly assigned for the sequences. When the UStag prefix or suffix contained (2n-1) Cys, the mass differences were calculated and searched against the database sequence specified by the UStag (the pseudo-algorithm is given in Table 1). The mass agreement (i.e., <10 ppm mass error) determines the two pieces of sequences linked by the disulfide bridge. All spectra that generated the UStags, but had the prefixes and suffixes not agreeing with the database predictions, were used for identification of peptides that contain disulfide bonds, and no a minimum number of counts required for individual MS/MS spectra.
Figure 1.
The flowchart of de novo-UStags approach for identification of disulfides in peptidome peptides. The dark blue segment of the blue loop-like diagram represents the UStag. Explanation of the flowchart is described in the text.
Table 1.
Pseudo code for searching disulfide(s) within the UStag-specified proteins with anchor sequences.
FUNCTION FindDisulfides (Pept, Prot, dM, Prec)
CONST dMCys := −1.007825 – mass modification on Cys-residue
Input
Pept – anchor peptide sequence (contains at least 1 Cys)
Prot - protein sequence containing Pept
dM – mass difference to explain (k * dMCys: k=1,2,3,…)
Prec – mass precision in amu (0.005)
Output
Disulfides – array of disulfides explaining dM in Pept-Prot
context
Main Loop
cntC – count of Cys residues in prefix and suffix part of Pept
Pept1 – Prot predicted peptide linked to Pept via disulfide bond
Pept1M – expected mass of Pept1
IF (cntC MOD 2 = 0) THEN
FOR i := 1 TO cntC \ 2
IF Abs(dM − i * 2 * dMCys) < Prec THEN
[Pept & “d-bond” & i] -> Disulfides
END IF
END FOR
ELSE
CPos := Find 1st Cys in (Prot-Pept); −1 if not found
DO WHILE CPos > 0
Pept1M := dM – dMCys
i := 0
DO WHILE Pept1M > (dM – (cntC – 1) * dMCys - Prec)
GENERATE all sequences Seq around CPos such that
Abs(Pept1M – SeqM) < Prec
IF i > 0 THEN
[[Pept & “d-bond” & i] & “d-bond” & Seq] ->
Disulfides
ELSE
[Pept & “d-bond” & Seq] -> Disulfides
END IF
Pept1M := dM – 2 * dMCys
i := i −1
LOOP
CPos := Find next Cys in (Prot-Pept); −1 if not found
LOOP
END IF
RETURN Disulfides
|
RESULTS
We identified 58 plasma peptidome peptides that contain the disulfide bonds among the ~990 in total modified and non-modified peptides identified (not shown). Table 2 shows examples of disulfides-containing peptidome peptides identified from the human blood plasma peptidome using the de novo-UStags approach. The disulfides identified can be classified into 3 categories: the intramolecular disulfides that are situated in the single peptide sequences, the intramolecular disulfides that link two pieces of separate sequences belonging to the same molecule of the protein, and the intermolecular disulfides that link two pieces of separate sequences belonging to the different molecules of the protein. Determinations of these disulfides are demonstrated below with examples, showing that identification of disulfides from complex mixtures by searching large (i.e., IPI human) protein database can be greatly simplified by use of the de novo-UStags approach.
Table 2.
Some of disulfides identified in the human blood plasma peptidome peptides.*
| Peptidome peptides |
Protein_ origin |
UStag | MW (Da) | CS | ME |
|---|---|---|---|---|---|
| R.KELFYKADGESC (–dehydro) SASMMYQEGKFR.Y; K.ANRPFLVFIREVPLNTIIFMGRVANPC (–dehydro)VK.- |
Antithrombin III | ELFYKAD | 6116.076 | 6 | 0.07 |
| R.KELFYKAD (–reduction) GESC(–dehydro) SASMMYQEGKFR.Y; L.NRVTFKANRPFLV FIREVPLNTIIFMGRVANPC (–dehydro)VK.- |
Antithrombin III | ELFYKA | 7072.595 | 7 | 0.54 |
| R.KELFYKADGESC (–dehydro) SASMMYQEGKFR.Y; R.SLNPNRVTFKANRPFLV FIREVPLNTIIFMGRVANPC (–dehydro)VK.- |
Antithrombin III | SASMMYQE | 7272.711 | 7 | 1.83 |
| R.TGKEKVTSGSTTTTRRSC (–dehydro) SKTVTKTVIGPDGHKE VTKEVVTSEDGSDC (–dehydro) PEAMDLGTLSGIGTLDGFRHRHP DEAAFFDTASTGKTFPGFF SPMLGEFVSETESRGSESGI FTNTKESSSHHPGIAEFPSRGK.S |
Fibrinogen α | LGEFVSE | 13994.71 | 13 | 0.28 |
| R.KELFYKADGESC (–dehydro) SASMMYQEGKFR.Y; R.SLNPNRVTFKANRPFL VFIREVPLNTIIFM (–hydroxylation) GRVANPC(dehydro)VK.- |
Antithrombin III | SASMMYQE | 7288.706 | 7 | 0.56 |
| R.SIAQYWLGC(–dehydro)PAPGHL.-; R.SIAQYWLGC(–dehydro)PAPGHL.- | Vitronectin | AQYWLG | 3221.558 | 3 | 1.52 |
| R.KELFYKADGESC (–dehydro) SASMMYQEGKFR.Y; R.VANPC(–dehydro)VK.- |
Antithrombin III | SMMYQE | 3531.630 | 4 | 4.79 |
| K.SGQSEDRQPVPGQQM TLKIEGDHGARVVLVAVDKGV FVLNKKNKLTQSKIWDVVE KADIGC(–dehydro) TPGSGKDYAGVFSDAG LTFTSSSGQQTAQRAELQC (–dehydro)PQPAA.R |
C3β | WDVVEKADIG | 10666.37 | 6 | 0.98 |
| R.SEGSSVNLSPPLEQC (–dehydro) VPDRGQQYQGRLAVTTHGLPC (–dehydro)LAWASAQAKALSKH QDFNSAVQLVENFC (–dehydro) RNPDGDEEGVWC (–dehydro) YVAGKPGDFGYC (–dehydro) DLNYC(–dehydro) EEAVEEETGDGLDEDSDRAIEGR.T |
Prothrombin | EEETGD | 12599.68 | 7 | 0.95 |
| R.SVPPSASHVAPTETFTYE WTVPKEVGPTNADPVC (–dehydro) LAKMYYSAVDPTKDIFTGLIGPM (–hydroxylation) KIC(–dehydro) KKGSLHANGRQKDVDK EFYLFPTVFDENESLLLEDNIR.M |
Ceruloplasmin | SLLLEDNIR | 10929.41 | 9 | 0.59 |
| R.SEGSSVNLSPPLEQC (–dehydro) VPDRGQQYQGRLAVTTHGLPC (–dehydro) LAWASAQAKALSKHQDFNSAVQLVENFC (–dehydro) RNPDGDEEGVWC (–dehydro) YVAGKPGDFGYC(–dehydro) DLNYC(–dehydro) EEAVEEETGDGLDEDSDRAIEGR.T_(hydroxylation) |
Prothrombin | EAVEEETGD | 12615.68 | 7 | 2.64 |
| Q.KPRLLLFSPSVVHL GVPLSVGVQLQDVPRGQV VKGSVFLRNPSRNNVPC (–dehydro) SPKVDFTLSSERDFAL LSLQVPLKDAKSC (–dehydro) GLHQLLRGPEVQLVAHSPWL KDSLSR.T |
C4 | SVGVQL | 11393.22 | 10 | 0.35 |
| R.RSC(–dehydro) SKTVTKTVIGPDGHKEV TKEVVTSEDGSDC (–dehydro) PEAMDLGTLSGIGTLDGFR HRHPDEAAFFDTAS TGKTFPGFFSPM (–hydroxylation) LGEFVSETESRGSESGIFT NTKESSSHHPGIAEFPSRGK.S |
Fibrinogen α | LGEFVSE | 12475.91 | 12 | 3.31 |
| R.SVPPSASHVAPTETFT YEWTVPKEVGPTNADPVC (–dehydro) LAKMYYSAVDPTKDI FTGLIGPMKIC(–dehydro) KKGSLHANGRQKDVDKE FYLFPTVFDENESLLLEDNIR.M |
Ceruloplasmin | ENESLLLE | 10913.42 | 7 | 0.05 |
| R.TGKEKVTSGSTTTTRRSC (–dehydro) SKTVTKTVIGPDGHK EVTKEVVTSEDGSDC (–dehydro) PEAMDLGTLSGIGTLDGF RHRHPDEAAFFDTAST GKTFPGFFSPM(–hydroxylation) LGEFVSETESRGSES GIFTNTKESSSHHPGIAEFPSRGK.S |
Fibrinogen α | LGEFVSETE | 14010.70 | 13 | 0.66 |
| A.IQRTPKIQVYSR HPAENGKSNFLNC (–dehydro) YVSGFHPSDIEVDLLKN GERIEKVEHSDLSFSKDW SFYLLYYTEFTPTEKDEYAC (–dehydro) RVNHVTLSQPKIVKWDRDI (->M-oxidation).- |
β-microglobulin | IVKWDRD | 11737.77 | 7 | 0.89 |
| R.SEGSSVNLSPPLEQC (–dehydro) VPDRGQQYQGRLAVTTHGLPC (–dehydro) LAWASAQAKALSKH QDFNSAVQLVENFC (–dehydro) RNPDGDEEGVWC (–dehydro) YVAGKPGDFGYC (–dehydro) DLNYC(–dehydro) EEAVEEETGDGLDEDSDR.A |
Prothrombin | TGDGLD | 12073.40 | 7 | 1.28 |
| A.IQRTPKIQVYSRHP AENGKSNFLNC (–dehydro) YVSGFHPSDIEVDLLK NGERIEKVEHSDLSFSKD WSFYLLYYTEFTPTEKDEYAC (–dehydro) RVNHVTLSQPKIVKWDRDI (->M).- |
β-microglobulin | IVKWDRD | 11721.77 | 7 | 2.55 |
CS: charge state; ME: molecular mass error (ppm).
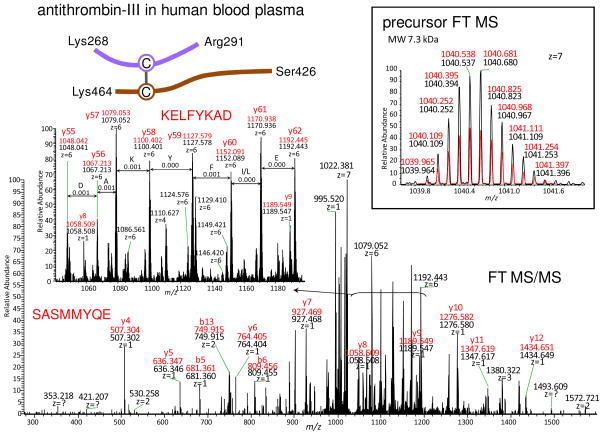
Figure 2 shows an identification of a disulfide bridge that links two separate sequences originating from the same molecule of the protein. The amino acid sequence SASMMYQE was de novo sequenced and this sequence is a UStag (Ser280–Glu287) of antithrombin-III (AT). The fragments used for sequencing of the UStag are located as the consecutive y4–y12 ions (labeled in the figure) counted from AT Arg291, and the UStag has a prefix sequence of GKFR. Extending the UStag suffix sequence from Ser280 towards the N-terminal direction generates a new sequence Cys279–Arg291 (CSASMMYQEGKFR) that contains one Cys residue (Cys279). This sequence has a mass value (mono isotopic, the same below except for the specified) of 1536.652 u, smaller than the precursor measurement (7272.697 u). The mass difference (5736.045 u) is searched against the masses that are generated by combination of the sequences extended from Cys279 towards the N-terminal direction with all pieces of the Cys-containing AT sequences. The searching outputs sequence Lys268–Ser278 (KELFYKADGES) extended from Cys279 plus another piece of sequence Ser426–Lys464 (SLNPNRVTFKANRPFLVFIREVPLNTIIFMGRVANPCVK) that contains Cys462. Therefore, the peptide is determined as to be constructed by two sequences Lys268-Arg291 and Ser426-Lys464 linked by a disulfide bridge Cys279–Cys462. The identified peptide has a molecular mass in agreement with the precursor measurement (e.g., ~1.0 ppm mass error, see the labels inserted in the figure) and explains additional abundant ions as fragments y56–y61 (counted from Arg291 and including sequence Ser426–Lys464; these fragments construct sequence KELFYKAD between Lys268 and Ser278, see those labeled in the figure). The disulfide bridge Cys279–Cys462 has been reported within AT (18), and therefore, the two different sequences linked by the identified disulfide bridge originate from the same AT molecules.
Figure 2.
The example showing the de novo-UStags approach for identification of an intramolecular disulfide that links two pieces of separate sequences within the same human blood plasma antithrombin (AT) molecule. Black peaks and numbers represent the experimental measurements and red peaks and numbers represent the predictions of the assigned-peptide (same below). The m/z values labeled for y66-y62 represent those of the most abundant isotopic peaks. Explanation of the identification is described in the text.
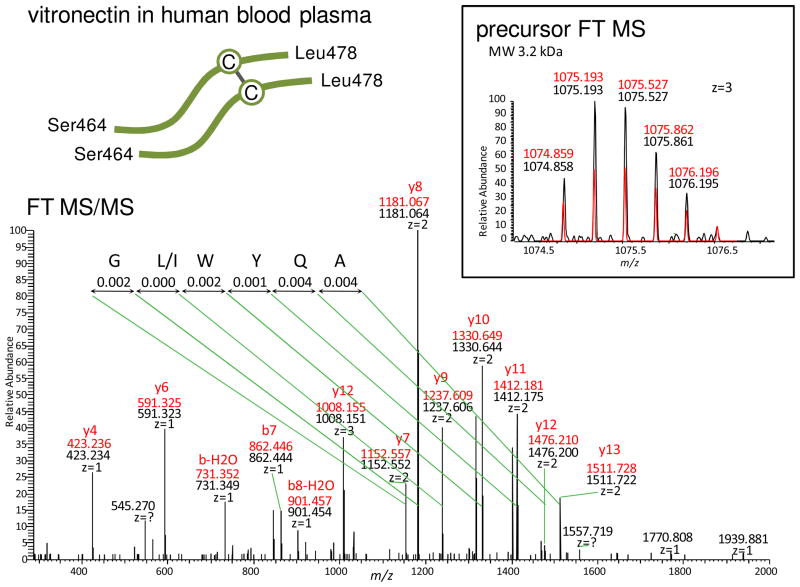
Figure 3 shows the identification of a disulfide bridge that apparently links two separate sequences originating from different protein molecules. The sequence AQYWLG was de novo sequenced, and it is a UStag (Ala466–Gly472) of vitronectin (VN). This UStag is sequenced from C- to N-terminal direction, but consecutive ions used for sequencing cannot be located in the VN database sequence. This means that the UStag prefix differs from VN database sequence. Extending the UStag prefix sequence from Gly472 toward to the C-terminal direction generates a new sequence that contains one Cys residue (Cys473). Further extending the sequence to the C terminus Leu478 of VN has a mass of 693.327 u, smaller than the neutral mass of the smallest ion (2303.089 u) used for sequencing of the UStag. The mass difference (1609.762 u) is searched against all pieces of Cys-containing VN sequences. The searching outputs a combination of the UStag suffix sequence Ser464–Ile465 (SI) and another VN sequence (Ser464–Leu478) SIAQYWLGCPAPGHL that contains Cys473. Therefore, the peptide is identified as to be constructed from two Ser464–Leu478 sequences linked by a disulfide Cys473–Cys473. This identified peptide has a molecular mass agreeing with the precursor measurement (e.g., ~0.9 ppm mass error, see measurements labeled in the figure), explains the UStag sequence as fragments y7–y13 (counted from the VN C terminus and including Ala466–Leu478), and explains additional low mass and other abundant ions in the MS/MS spectrum as y and b fragments (see those labeled in the figure).
Figure 3.
The example showing the de novo-UStags approach for identification of an intermolecular disulfide that links two pieces of separate sequences originated from two human blood plasma vitronectin (VN) molecules. Explanation of the identification is described in the text.
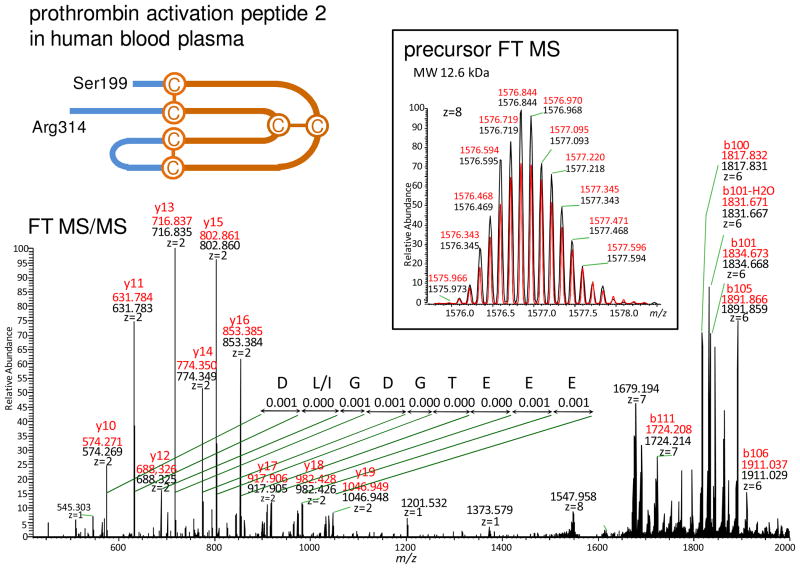
Determination of disulfide bridge(s) situated in the single-sequence peptides is a special situation of the de novo-UStags method, i.e., the determination is simply achieved with extension of the UStag prefix and/or suffix, as illustrated in Figure 4. The UStag DLGDGTEEE was de novo sequenced for prothrombin (FII) (Asp305–Glu297), and is constructed by fragments y10–y19 counted from FII Arg314. Extending the UStag suffix sequence to Ser199 directly leads to identification of three intramolecular disulfides in a single sequence peptide. The peptide (Ser199–Arg314) identified is a truncation of the intact FII activation peptide-2 (Ser199–Arg327) (19).
Figure 4.
The example showing the de novo-UStags approach for identification of an intramolecular disulfide within a single piece of sequence of prothrombin (FII). Explanation of the identification is described in the text.
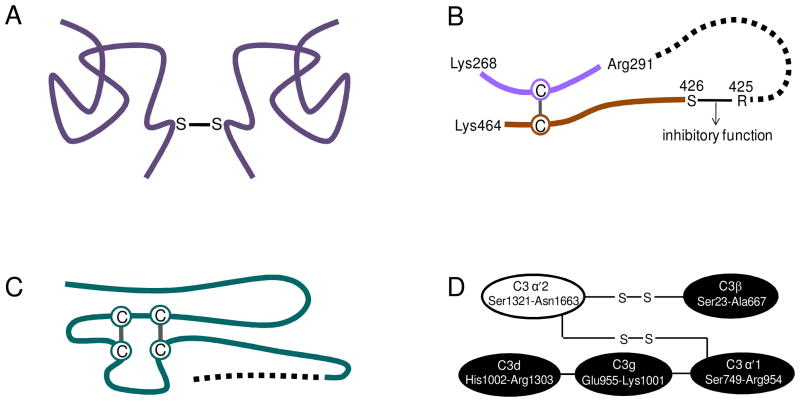
Identification of disulfide bonds in the peptidome peptides can reveal the protein structures arising from degradation, as shown in Figure 5. VN has been reported to exist as monomer and dimer in human blood plasma through formation of a disulfide bond (20). Using the de novo-UStag FT MS/MS analysis, we first find evidence that the disulfide bond responsible for formation of the VN dimer is the C terminal Cys473–Cys473 (Figure 4) between two VN molecules (Figure 5A). AT is a protease inhibitor to inhibit the protease (e.g., thrombin) proteolytic activities, and its inhibitory reaction site is at Arg425–Ser426 (21). Cleaving the inhibitory Arg425–Ser426 bond by degradation will lead to loss of the AT inhibitory function. Figure 5B shows an identified disulfide bond that links the sequence (Ser426–Lys464) containing the broken AT inhibitory bond to another piece of AT sequence (Lys268–Arg291). This reveals that AT should retain its structure during degradation of the sequence loop defined by the disulfide bond. A different situation is shown in Figure 5C for plasminogen preactivation peptide. This preactivation peptide has sequence loops that are formed by two disulfide bonds (22). Degradation cleavage occurs not to the loop sequence, but to the C terminal truncation of the preactivation peptide. Disulfide bonds also have functions to link different protein sequence domains. As shown in Figure 5D, C3α and C3β in C3b of the third complement component are linked by disulfide bonds (23). We detected the disulfide bonds within C3β (Table 2), but not that linking C3α and C3β, which suggests that C3b degradation occurs after disruption of the interchain disulfide bonds.
Figure 5.
Human plasma protein structures through identification of disulfide bonds in the degradation products. (A) Vitronectin dimer is formed by a Cys-Cys disulfide bond that linked the C terminal regions of the two protein molecules. (B) Antithrombin inhibitor inhibitory site degrades together with other sequence. (C) Plasminogen activation peptide degrades on the outside sequence of the disulfide bonds-linked sequence loop. (D) The protein state during degradation can be uncovered through identification of disulfide bond(s). Descriptions are detailed in the text.
DISCUSSION
The present results show that use of the UStags enables improving identification of disulfides through assignment of spectra to specific proteins from a large protein sequence database (e.g., IPI human protein database). Once spectra and proteins are specifically related by UStags, the protein sequences suggested by the database can be examined for disulfide bonds (and also other modifications) without the need for searching of all spectra against the large proteome database in conjunction with consideration of a range of other modifications for these species. The UStags identification is founded upon the accurate measurements provided by FT MS/MS; we did not find any incorrect UStags for the datasets tested (>100,000 FT MS/MS spectra) based upon examination with a decoy database (i.e., re-constructed from the IPI human protein database used for identification by reversing each protein sequence). The peptidome peptides containing the intermolecular and intramolecular disulfides identified by the de novo-UStags approach have molecular masses up to ~14,000 u and can incorporate termini of various amino acids. Arg and Lys are the residues mainly found for the P1 site of the disulfide-containing peptides identified, which is similar to that observed for overall peptides identified from the blood plasma peptidome (16). Plasmin may be one of the major proteases responsible for observation of the trypsin-like cleavage feature. In this work the longest Cys-containing sequences obtained from the de novo sequencing had 6 residues, and they are not the UStags (i.e., they belong to different IPI database proteins) and not used for the unambiguous identification in this work. Electron-transfer dissociation may be an alternative method for improving the fragmentation of disulfide-containing peptides (14,24), and we are now evaluating different dissociation methods for analysis of blood plasma peptidome peptides including those containing disulfide bonds.
High temperature, alkaline pH, and air-catalyzed oxidation can lead to changes of disulfides to form the lysinoalanyl crosslink, undergo hydrolysis to yield sulfhydryl groups and other products, and initiate thiol-disulfide interchanges (25). In this work, we analyzed disulfides-containing peptides in the blood plasma peptidome without implement of special protections of the disulfide bonds. We examined the disulfide bridges identified with reference to the UniProt protein structural information (http://www.uniprot.org/). All disulfide bridges identified are in agreement with the UniProt information except for the VN dimer, for which no information for VN Cys473 is available. Blood VN dimer is well known and it is reasonable for observation that the disulfide bridge is responsible for the dimer as free cysteine is difficult to be retained in the protein crosslink polymers. The process of the disulfide formation, i.e., the disulfide bond leading to formation of dimer or first formation of dimer and then the disulfide bond through interchange, keeps unknown.
Acknowledgments
Portions of this research were supported by the U.S. Department of Energy Office of Biological and Environmental Research (DOE/BER) and the NIH National Center for Research Resources (RR18522). Work was performed in the Environmental Molecular Science Laboratory, a DOE/BER national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO-1830.
References
- 1.Christis C, Lubsen NH, Braakman I. Protein folding includes oligomerization – examples from the endoplasmic reticulum and cytosol. FEBS J. 2008;275:4700–4727. doi: 10.1111/j.1742-4658.2008.06590.x. [DOI] [PubMed] [Google Scholar]
- 2.Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- 3.Dranoff G. Targets of protective tumor immunity. Ann N Y Acad Sci. 2009;1174:74–80. doi: 10.1111/j.1749-6632.2009.04938.x. [DOI] [PubMed] [Google Scholar]
- 4.Hogg PJ. Contribution of allosteric disulfide bonds to regulation of hemostasis. J Thromb Haemost. 2009;7:12–16. doi: 10.1111/j.1538-7836.2009.03364.x. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura T, Lipton SA. Cell death: protein misfolding and neurodegenerative diseases. Apoptosis. 2009;14:455–468. doi: 10.1007/s10495-008-0301-y. [DOI] [PubMed] [Google Scholar]
- 6.Guo Q, Manolopoulou M, Bian Y, Schilling AB, Tang W-J. Molecular basis for the recognition and cleavages of IGF-II, TGF-α, and amylin by human insulin-degrading enzyme. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wess J, Han S-J, Kim S-K, Jacobson KA, Li JH. Conformational changes involved in G-protein-coupled-receptor activation. Trends Pharmacol Sci. 2008;29:616–625. doi: 10.1016/j.tips.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones TA, Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 9.Klaus W, Broger C, Gerber P, Senn H. Determination of the disulphide bonding pattern in proteins by local and global analysis of nuclear magnetic resonance data. Application to flavoridin. J Mol Biol. 1993;232:897–906. doi: 10.1006/jmbi.1993.1438. [DOI] [PubMed] [Google Scholar]
- 10.Gorman JJ, Wallis TP, Pitt JJ. Protein disulfide bond determination by mass spectrometry. Mass Spectrom Rev. 2002;21:183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 11.Nair SS, Nilsson CL, Emmett MR, Schaub TM, Gowd KH, Thakur SS, Krishnan KS, Balaram P, Marshall AG. De Novo sequencing and disulfide mapping of a bromotryptophan-containing conotoxin by Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2006;78:8082–8088. doi: 10.1021/ac0607764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res. 2008;7:138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tipton JD, Carter JD, Mathias JD, Emmett MR, Franucci GE, Marshall AG. Sequential proteolysis and high-field FTICR MS to determine disulfide connectivity and 4-maleimide TEMPO spin-label location in L126C GM2 activator protein. Anal Chem. 2009;81:7611–7617. doi: 10.1021/ac9009935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu SL, Jiang H, Lu Q, Dai S, Hancock WS, Karger BL. Anal Chem. 2009;81:112–122. doi: 10.1021/ac801560k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi S, Jeong J, Na S, Lee HS, Kim H-Y, Lee K-J, Paek E. New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra. J Proteome Res. 2010;9:626–635. doi: 10.1021/pr900771r. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Liu T, Tolić N, Petritis BO, Zhao R, Moore RJ, Purvine SO, Camp DG, II, Smith RD. Strategy for degradomic-peptidomic analysis of human blood plasma. J Proteome Res. doi: 10.1021/pr901083m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Tolić N, Hixson KK, Purvine SO, Anderson GA, Smith RD. De novo sequencing of unique sequence tags for discovery of post-translational modifications of proteins. Anal Chem. 2008;80:7742–7754. doi: 10.1021/ac801123p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen TE, Dudek-Wojciechowska G, Sottrup-Jensen L, Magnusson S. Primary structure of antithrombin-III (heparin cofactor). Partial homology between alpha-1-antitrypsin and antithrombin-III. In: Collen D, Wiman B, Verstraete M, editors. The physiological inhibitors of blood coagulation and fibrinolysis. Elsevier; Amsterdam: 1979. pp. 43–54. [Google Scholar]
- 19.Rabiet MJ, Blashill A, Furie B, Furie BC. Prothrombin fragment 1•2•3, a major product of prothrombin activation in human plasma. J Biol Chem. 1986;261:13210–13215. [PubMed] [Google Scholar]
- 20.Dahlbäck B, Podack ER. Characterization of human S protein, an inhibitor of the membrane attack complex of complement. Demonstration of a free reactive thiol group. Biochemistry. 1985;24:2368–2374. doi: 10.1021/bi00330a036. [DOI] [PubMed] [Google Scholar]
- 21.Jörnvall H, Fish WW, Björk I. The thrombin cleavage site in bovine antithrombin. FEBS Lett. 1979;106:358–362. doi: 10.1016/0014-5793(79)80532-3. [DOI] [PubMed] [Google Scholar]
- 22.Wiman B. Primary structure of peptides released during activation of human plasminogen by urokinase. Eur J Biochem. 1973;39:109. doi: 10.1111/j.1432-1033.1973.tb03096.x. [DOI] [PubMed] [Google Scholar]
- 23.Janatova J. Detection of disulfide bonds and localization of interchain linkages in the their (C3) and the forth (C4) components of human complement. Biochem J. 1986;233:819–825. doi: 10.1042/bj2330819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueberheide BM, Fenyö D, Alewood PF, Chait BT. Rapid sensitive analysis of cysteine rich peptide venom components. Proc Natl Acad Sci. 2009;106:6910–6915. doi: 10.1073/pnas.0900745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaisgood HE. The importance of disulfide bridging. Biotechnol Adv. 2005;23:71–73. doi: 10.1016/j.biotechadv.2004.09.004. [DOI] [PubMed] [Google Scholar]