Abstract
Cell culture and the use of cell lines are routinely used in basic scientific research. It is therefore imperative for researchers to ensure the origin of the cell lines used and that they are routinely re-analysed for contamination and misidentification. Inter-species contamination is relatively frequent, and the most commonly used cell lines are of human, mouse and rat derivation. We have developed simple species specific primer assays based on genomic sequence differences in vomeronasal receptor gene family members to discriminate between human, mouse and rat DNA using standard agarose gel electrophoresis. Furthermore, these PCR assays are able to identify the species composition within an inter-species mixed population. This approach therefore provides a valuable tool to enable a rapid, simple and relatively inexpensive determination of the authentication and contamination of cell cultures.
Keywords: PCR, Vomeronasal receptors, Species specificity, Genomic DNA
Introduction
Cell lines which have acquired the ability to proliferate indefinitely are routinely cultured for use in laboratory studies. These continuous cell lines are repeatedly passaged, reliably cryopreserved and retain representative properties of the cells and tissues from which they were derived. These attributes have resulted in cell lines being widely used as model systems for the study of cellular processes in health and disease and are therefore commonly used within biomedical research and the pharmaceutical industry (Capes-Davis et al. 2010). Recently the robustness of cell cultures used in research has been under scrutiny as data have demonstrated that cross-contamination with other cells lines, misidentification of cell lines, and the use of cultures at high passage numbers has resulted in the generation of erroneous results (Hughes et al. 2007). Studies have highlighted the cross-contamination of various cell lines with well-established neoplastic cells. Indeed several human glioblastoma cell lines originally thought to be distinct have now either been shown to be identical or not of human origin (Chatterjee 2007; ECACC 2009). Moreover one of the first and most widely used human cell lines to be established, HeLa cells, has recently been demonstrated to have either contaminated many other cell lines or in many instances overgrown and replaced the original cells. A survey of 483 mammalian cell culturists indicated that 32% were unknowingly using HeLa cells whilst 9% were using HeLa contaminated cultures. As a result 220 research reports listed in the PubMed database (1969–2004) were identified which had used HeLa contaminated cultures. One of the underlying problems responsible for this misrepresentation was that over a third of the researchers surveyed indicated that their cells had been derived from another laboratory rather than from a repository (Buehring et al. 2004). Subsequently these research issues have received significant media attention bringing into question the credibility of in vitro scientific studies and in particular the waste of public and private resource as a consequence (Northam 2007). Clearly it is therefore essential that due diligence is paid to ensure the integrity of cell cultures used for research purposes (Stacey 2000).
To aid researchers in identifying contaminated cell lines a database of cross contaminated or misidentified cell lines has been established (Capes-Davis and Freshney 2009). However it is also imperative to routinely monitor cell lines for possible contamination using characteristics that authenticate their identity. Over recent decades several methods have been established for use in authenticating and characterising cell lines, including allozyme and isoenzyme analysis (Stacey et al. 1997; O’Brien et al. 1980), karyotyping analysis (Rush et al. 2002), human leukocyte antigen typing (O’Toole et al. 1983), immunophenotypic and immunocytochemical analysis (Quentmeier et al. 2001), DNA fingerprinting and short tandem repeat profiling (Stacey et al. 1992; Dirks et al. 2005; Yoshino et al. 2006; Azari et al. 2007). In general these methodologies can be relatively time consuming and expensive as well as requiring experienced personnel to undertake them. Such features can make these techniques impractical for routine application.
Currently the majority of cell lines used in research and development are derived from human and rodents. Yet despite the burgeoning information present in the genome databases for these species only a handful of PCR-based technologies for cell line authentification have been developed (Stacey et al. 1997; Parodi et al. 2002; Liu et al. 2003; López-Andreo et al. 2005; Steube et al. 2003, 2008). Whilst these approaches may be relatively inexpensive many of these assays utilise multiple primers or require interpretation of relatively complex gel banding patterns. These factors can hamper their application and the transfer of the technique between laboratories. The aim of this present study was to use comparative genome sequence information for human, mouse and rat species to develop specific PCR assays which provide relatively simple, robust and unequivocal data for cell line identification and authentification.
Materials and methods
Cell lines and culture conditions
Mouse fibroblasts (3T3-Swiss) (Todaro and Green 1963) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 200 mM glutamine (Sigma, UK), 10% foetal calf serum (FCS) and 1% penicillin/streptomycin. Rat (WRC-256) (Hull 1953) and human (H400) (Prime et al. 1990) epithelial cells were cultured in DMEM/Ham’s Nutrient Mixture F12 supplemented with 10% FCS and 1% penicillin/streptomycin or 0.5 μg/mL hydrocortisone, respectively. Primary bone marrow cells were isolated from 100 to 120 g male Wistar rats which were freshly sacrificed by cervical dislocation. Rat femurs were dissected, cleaned of excess soft tissue and repeatedly flushed with DMEM supplemented with 10% FCS and 1% penicillin/streptomycin (Sigma, UK). All cells were cultured in 25 cm2 or 75 cm2 flasks in a humidified incubator with 5% carbon dioxide in air at 37 °C. Medium was changed every 3 days.
Isolation of high molecular weight genomic DNA
All genomic DNA isolations from cells and tissues were performed using the PureLink kit (Invitrogen, UK) using the protocol recommended by the manufacturer. For initial processing of cultures (including cell lines and primary rat bone marrow cells), ~80% confluence cells were detached using trypsin/EDTA (Gibco, UK), cells were pelletted by centrifugation and washed twice with phosphate-buffered saline (PBS). For murine genomic DNA, 20 mg of liver was removed from an adult male Swiss mouse and processed by finely mincing prior to DNA isolation. For human genomic DNA isolation, heparinized (17 IU/mL) venous blood was obtained and red blood cells removed using erythrocyte lysis (0.83% NH4Cl containing 1% KHCO3, 0.04% Na2EDTA.2H2O and 0.25% bovine serum albumin; 20 min). Isolated white blood cells were washed and re-suspended in PBS supplemented with glucose (1 mM) and cations (1 mM MgCl2, 1.5 mM CaCl2). The cell suspension was pelletted by centrifugation and all supernatant removed prior to DNA isolation. Subsequently all cell pellets and dissociated tissue were resuspended in 200 μL of Genomic Digestion Buffer (Invitrogen, UK) and incubated for 10 min at 55 °C prior to genomic DNA isolation using the PureLink kit (Invitrogen, UK) using the protocol supplied by the manufacturer. DNA was eluted in 50 μL of buffer and concentrations were determined from absorbance values at a wavelength of 260 nm using a BioPhotometer (Eppendorf, UK). DNA integrity was also confirmed by visualisation on 0.8% agarose gel electrophoresis containing 0.5 μg/mL ethidium bromide (Helena Biosciences, UK). Samples were stored at −20 °C prior to use.
Genomic PCR amplification
Details of human and rodent oligonucleotide primers and cycling conditions used in this study are provided within the Table 1. Vomeronasal pheromone receptor gene sequences were obtained from GenBank and their specificity was confirmed by searches of human and rodent genomic sequence databases using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Primers were designed using Primer-BLAST software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) and subsequently commercially obtained (Invitrogen, UK).
Table 1.
Species specific genes and associated human and rodent primer assays
| Gene name | Gene symbol | Species | Primer sequences (5′ → 3′) | Annealing temperature | Product size (bp) | Accession No. |
|---|---|---|---|---|---|---|
| Vomeronasal 1 receptor, 26 | V1rm1 | Rat | Forward: TGGCTTTCAGGCCACCAGGC Reverse: GCTCTGTCCTCAGGGGCAGGT |
66.5 °C | 387 | NM001009525 |
| Vomeronasal 1 receptor, 25 | V1rm2 | Rat | Forward: AGAAGAGTTACTGGCCCAAGGGACA Reverse: GGGGCTGAACGCTGGGAAGC |
66.5 °C | 408 | NM 001008921 |
| Vomeronasal 1 receptor, 200 | V1rh3 | Mouse | Forward: GGGAGGGGCCAGTGGCTACAT Reverse: TGCCACCAATCAACCAGAAGCCCA |
66 °C | 306 | NM_134212 |
| Vomeronasal 1 receptor, 210 | V1rh10 | Mouse | Forward: TTCAGGGTGCTATGGGAGGGGC Reverse: GCCCATCCCTGTGAATCAGCACA |
66 °C | 300 | NM134235 |
| Vomeronasal 1 receptor, 1 | VN1R1 | Human | Forward: TGGTCTGGGCCAGTGGCTCC Reverse: GAGTGTTTTCCTTGTCCTGCAGGCA |
67.5 °C | 332 | NM_020633 |
| Vomeronasal 1 receptor, 4 | VN1R4 | Human | Forward: TCGCACAGACACTGCGTGCA Reverse: ACACTGGGGTCACAGCTCATGAGA |
67.5 °C | 352 | NM_173857 |
PCR assays were performed using the BioMix PCR system (Bioline, UK). Each reaction mix contained 50 ng of DNA in 1 μL of RNase free water, 12.5 μL BioMix Red, 9.5 μL dH2O, and 1 μL each of 25 mM forward and reverse primers. PCRs were amplified in a Thermal Cycler (Mastercycler Gradient, Eppendorf) for 32 cycles. The programme consisted of an initial denaturation step for 5 min at 94 °C, followed by an amplification cycle consisting of 94 °C for 20 s, 20 s annealing at 66–67.5 °C (Table 1) and 68 °C extension for 20 s, ending with a final 10 min extension at 72 °C. Subsequently, 6 μL of each reaction was removed and the amplified product was separated and visualized under UV illumination on a 1.5% agarose gel containing 0.5 μg/ml ethidium bromide.
Results
Identification and validation of species specific PCR assays for cell line authentification
Recent publications have indicated that while pheromone receptors exhibit species conservation of gene sequence several family members are distinct to human, mouse and rat (Rat Genome Sequencing Project Consortium 2004; Shi et al. 2005; Grus et al. 2005). Subsequent BLAST analysis of the human, mouse and rat genomic sequences using selected vomeronasal receptor gene sequences retrieved from GenBank (Table 1) indicated their potential species specificity. Subsequently, for robustness, two primer assays per species were designed to amplify products using relatively high annealing temperatures to enable specific DNA sequence binding (Table 1).
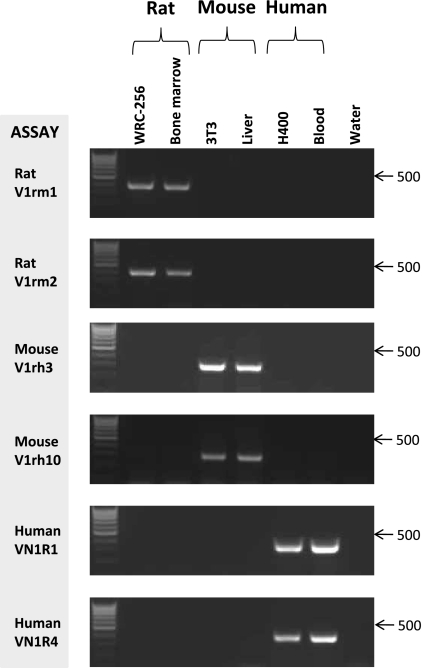
PCR analyses at a single cycle number (32) using genomic DNA derived from i) the cell lines, [WRC-256 (rat), Swiss 3T3 (mouse) and H400 (human)] and ii) control primary cells/tissues (rat bone marrow, mouse liver and human peripheral white blood cells) demonstrated that the assays designed amplified a specific product of the predicted size and that no cross species amplification was evident for any assay (Fig. 1). Data therefore indicated that in cell populations purely derived from a single species all assays supported culture clonality and authenticity.
Fig. 1.
PCR amplification with species specific primer pairs. PCR analysis of genomic DNA from rat, mouse and human cell lines and primary tissue using species specific primer pairs. PCRs assays used and source of input DNA (50 ng each) are shown. Amplified products generated at 32 cycles were detected by ethidium bromide staining of 1.5% agarose gels and UV transillumination. Blood refers to human peripheral white blood cells. RNase free water was used as negative control. Arrows indicate the DNA molecular weight marker of 500 bp (Hyperladder IV; Bioline)
Sensitivity of PCR assays for detection of mixed species populations
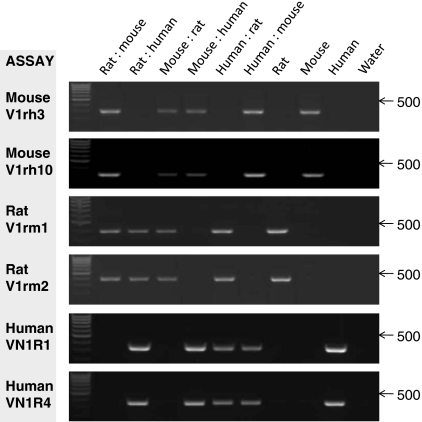
To determine whether the assays developed were able to detect mixed populations all combinations of species DNA were mixed at ratios of 5 ng : 45 ng and used for input in all PCRs. Data clearly demonstrated that with only a relatively minimal 10%/5 ng DNA inclusion all assays were robust and sensitive and able to detect minor contamination (Fig. 2).
Fig. 2.
Verification of the ability of the assay to detect interspecies cross contamination. DNA from each species was mixed at a ratio of 5 ng:45 ng (order as labelled) or used as 50 ng input from a single species. DNA used was: WRC-256 (rat); 3T3 (mouse); H400 (human). PCRs were performed with the primer assays indicated. Amplified products generated at 32 cycles were detected by ethidium bromide staining of 1.5% agarose gels and UV transillumination. RNase free water was used as negative control. Arrows indicate the DNA molecular weight marker of 500 bp (Hyperladder IV; Bioline)
Discussion
Several studies over the past decade have demonstrated the problem of cell line contamination which generally arises due to poor laboratory practice (Capes-Davis et al. 2010). Reported reasons include cell spreading via aerosols, use of unplugged pipettes, sharing media/reagents between cell lines and the use of mitotically inactivated feeder cells or conditioned medium, which have not had originating cells removed (van Pelt et al. 2003). Mislabelling, during either active culture handling or due to poor inventory records, is another source of misidentification (Capes-Davis et al. 2010). For future authentification and identification clear documentation of the cell line developed and robust assays are necessary for use in originating and recipient laboratories (Stacey 2000; Schmitt and Pawlita 2009).
Inter-species culture contamination is a common occurrence with the most frequently used cell lines being of human, mouse and rat origin (Steube et al. 2008). While assays based on karyotyping, fluorescent in situ hybridization and isozyme analysis (Nardone 2007) have been developed to distinguish between cell lines from different species their application and interpretation can require significant expertise and costs which can inhibit their uptake. In-house approaches using relatively simple inexpensive PCR technologies are desirable however limitations remain with published approaches which can require complex gel banding pattern interpretation, multiple-primer/nested assays and/or application of complementary approaches for data confirmation. Many of the previous reports have also not demonstrated technique sensitivity or ability to detect contamination (Liu et al. 2003; Ono et al. 2007; Ramya et al. 2009; Higgins et al. 2010; Parodi et al. 2002). Inexpensive assays for the identification and authentication of cell lines are therefore clearly necessary and should be relatively easily developed using the vast DNA databases as is demonstrated here. The simple assays reported here require minimal expertise for application and data interpretation and use standard molecular biology equipment. By combining exact amounts of DNA it was also able to sensitively detect minimal species contaminations.
Challenges still however remain for intra-species authentification and contamination detection. Techniques, such as genetic fingerprinting and immunocytological approaches can identify intraspecies contaminations as long as a known pure original stock is available as a comparator. It is however envisaged that high-throughput transcriptomic approaches may provide more robust future solutions which enable confirmation of the molecular phenotype of the cell lines providing assurances that they are representative of the tissue from which they were derived and are not contaminated.
The vomeronasal receptor PCR assays reported here provide a sensitive, accurate, rapid and cost effective approach for determining cell line authentification and inter-species contamination. In addition this test requires a relatively small number of cells, can utilise genomic DNA isolation kits from a range of suppliers and can easily be performed on a routine basis requiring only standard laboratory expertise.
References
- Azari S, Ahmadi N, Tehrani MJ, Shokri F. Profiling and authentication of human cell lines using short tandem repeat (STR) loci: report from the National Cell Bank of Iran. Biologicals. 2007;35:195–202. doi: 10.1016/j.biologicals.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Buehring GC, Eby EA, Eby MJ. Cell line cross-contamination: how aware are mammalian cell culturists of the problem and how to monitor it? In Vitro Cell Dev Biol Anim. 2004;40:211–215. doi: 10.1290/1543-706X(2004)40<211:CLCHAA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Capes-Davis A, Freshney RI (2009) Database of cross-contaminated or misidentified cell lines. European collection of cell cultures. http://www.hpacultures.org.uk/media/E50/3B/CellLineCrossContaminationsv60.pdf
- Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- Chatterjee R. Cell biology. Cases of mistaken identity. Science. 2007;315:928–931. doi: 10.1126/science.315.5814.928. [DOI] [PubMed] [Google Scholar]
- Dirks WG, Faehnrich S, Estella IA, Drexler HG. Short tandem repeat DNA typing provides an international reference standard for authentication of human cell lines. ALTEX. 2005;22:103–109. [PubMed] [Google Scholar]
- ECACC (2009) Cross contaminated and misidentified cell lines. Eur Collect Cell Cult. http://www.hpacultures.org.uk/services/celllineidentityverification/misidentifiedcelllines.jsp
- Grus WE, Shi P, Zhang YP, Zhang J. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc Natl Acad Sci USA. 2005;102:5767–5772. doi: 10.1073/pnas.0501589102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins SC, Steingrimsdottir H, Pilkington GJ. Human, mouse or rat? Species authentication of glioma-derived cell cultures. J Neurosci Methods. 2010;194:139–143. doi: 10.1016/j.jneumeth.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Hughes P, Marshall D, Reid Y, Parkes H, Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques. 2007;43:575–578. doi: 10.2144/000112598. [DOI] [PubMed] [Google Scholar]
- Hull RN. Establishing long-term cultures of mammalian normal, solid tumor, and ascites tumor cells on glass. Science. 1953;117:223–225. doi: 10.1126/science.117.3035.223. [DOI] [PubMed] [Google Scholar]
- Liu MY, Lin SC, Liu H, Candal F, Vafai A. Identification and authentication of animal cell culture by polymerase chain reaction amplification and DNA sequencing. In Vitro Cell Dev Biol Anim. 2003;39:424–427. doi: 10.1290/1543-706X(2003)039<0424:IAAOAC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- López-Andreo M, Lugo L, Garrido-Pertierra A, Prieto MI, Puyet A. Identification and quantitation of species in complex DNA mixtures by real-time polymerase chain reaction. Anal Biochem. 2005;339:73–82. doi: 10.1016/j.ab.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Nardone RM. Eradication of crosscontaminated cell lines: a call for action. Cell Biol Toxicol. 2007;23:367–372. doi: 10.1007/s10565-007-9019-9. [DOI] [PubMed] [Google Scholar]
- Northam G (2007) How cancer studies wasted cash. BBC News Channel. http://news.bbc.co.uk/1/hi/programmes/file on 4/7104070.stm
- O’Brien SJ, Shannon JE, Gail MH. A molecular approach to the identification and individualization of human and animal cells in culture: isozyme and allozyme genetic signatures. In Vitro. 1980;16:119–135. doi: 10.1007/BF02831503. [DOI] [PubMed] [Google Scholar]
- Ono K, Satoh M, Yoshida T, Ozawa Y, Kohara A, Takeuchi M, Mizusawa H, Sawada H. 0 Species identification of animal cells by nested PCR targeted to mitochondrial DNA. In Vitro Cell Dev Biol Anim. 2007;43:168–175. doi: 10.1007/s11626-007-9033-5. [DOI] [PubMed] [Google Scholar]
- O’Toole CM, Povey S, Hepburn P, Franks LM. Identity of some human bladder cancer cell lines. Nature. 1983;301:429–430. doi: 10.1038/301429a0. [DOI] [PubMed] [Google Scholar]
- Parodi B, Aresu O, Bini D, Lorenzini R, Schena F, Visconti P, Cesaro M, Ferrera D, Andreotti V, Ruzzon T (2002) Species identification and confirmation of human and animal cell lines: a PCR-based method. Biotechniques 32: 432–434, 436, 438–440 [DOI] [PubMed]
- Prime SS, Nixon SV, Crane IJ, Stone A, Matthews JB, Maitland NJ, Remnant L, Powell SK, Game SM, Scully C. The behaviour of human oral squamous cell carcinoma in cell culture. J Pathol. 1990;160:259–269. doi: 10.1002/path.1711600313. [DOI] [PubMed] [Google Scholar]
- Quentmeier H, Osborn M, Reinhardt J, Zaborski M, Drexler HG. Immunocytochemical analysis of cell lines derived from solid tumors. J Histochem Cytochem. 2001;49:1369–1378. doi: 10.1177/002215540104901105. [DOI] [PubMed] [Google Scholar]
- Ramya R, Nagarajan T, Sivakumar V, Senthilkumar RL, Bala Obulapathi B, Thiagarajan D, Srinivasan VA. Identification of cross-contaminated animal cells by PCR and isoenzyme analysis. Cytotechnology. 2009;61:81–92. doi: 10.1007/s10616-009-9245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Rush LJ, Heinonen K, Mrozek K, Wolf BJ (2002) Comprehensive cytogenetic and molecular genetic characterization of the TI-1 acute myeloid leukemia cell line reveals cross-contamination with K-562 cell line. Blood 99:1874–1876 [DOI] [PubMed]
- Schmitt M, Pawlita M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res. 2009;37:e119. doi: 10.1093/nar/gkp581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Bielawski JP, Yang H, Zhang YP. Adaptive diversification of vomeronasal receptor 1 genes in rodents. J Mol Evol. 2005;60:566–576. doi: 10.1007/s00239-004-0172-y. [DOI] [PubMed] [Google Scholar]
- Stacey GN. Cell contamination leads to inaccurate data: we must take action now. Nature. 2000;403:356. doi: 10.1038/35000394. [DOI] [PubMed] [Google Scholar]
- Stacey GN, Bolton BJ, Morgan D, Clark SA, Doyle A. Multilocus DNA fingerprint analysis of cell banks: stability studies and culture identification in human B-lymphoblastoid and mammalian cell lines. Cytotechnology. 1992;8:13–20. doi: 10.1007/BF02540025. [DOI] [PubMed] [Google Scholar]
- Stacey GN, Hoelzl H, Stephenson JR, Doyle A. Authentication of animal cell cultures by direct visualization of repetitive DNA, aldolase gene PCR and isoenzyme analysis. Biologicals. 1997;25:75–85. doi: 10.1006/biol.1996.0062. [DOI] [PubMed] [Google Scholar]
- Steube KG, Meyer C, Uphoff CC, Drexler HG. A simple method using beta-globin polymerase chain reaction for the species identification of animal cell lines: a progress report. In Vitro Cell Dev Biol Anim. 2003;39:468–475. doi: 10.1290/1543-706X(2003)039<0468:ASMUGP>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Steube KG, Koelz AL, Drexler HG. Identification and verification of rodent cell lines by polymerase chain reaction. Cytotechnology. 2008;56:49–56. doi: 10.1007/s10616-007-9106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelt JF, Decorte R, Yap PS, Fevery J. Identification of HepG2 variant cell lines by short tandem repeat (STR) analysis. Mol Cell Biochem. 2003;243:49–54. doi: 10.1023/A:1021653506123. [DOI] [PubMed] [Google Scholar]
- Yoshino K, Iimura E, Saijo K, Iwase S, Fukami K, Ohno T, Obata Y, Nakamura Y. Essential role for gene profiling analysis in the authentication of human cell lines. Hum Cell. 2006;19:43–48. doi: 10.1111/j.1749-0774.2005.00007.x. [DOI] [PubMed] [Google Scholar]