CONSPECTUS
Nanoparticles conjugated with functional ligands are expected to have a major impact in medicine, photonics, sensing, and nanoarchitecture design. One major obstacle to realizing the promise of these materials, however, is the difficulty in controlling the ligand/nanoparticle ratio. This obstacle can be segmented into three key areas: First, many system designs have failed to account for the true heterogeneity of ligand/nanoparticle ratios that compose each material. Second, the field's accepted level of characterization of ligand-nanoparticle materials is deficient as it does not provide an accurate definition of material composition that is necessary to both understand the material-property relationships as well as to monitor the consistency of the material. In particular, many characterization assays only determine the mean ligand/nanoparticle ratio and do not provide the number and relative amount of the different ligand/nanoparticle ratios that compose a material. Third, some synthetic approaches that are presently in use may not produce consistent material as they are sensitive to reaction kinetics, and the synthetic history of the nanoparticle.
In this account we describe the recent advances that we have made in understanding the material composition of ligand-nanoparticle systems. Our work has been enabled by a model system using poly(amidoamine) dendrimers and two small molecule ligands. Using reverse phase high-pressure liquid chromatography (HPLC) we have successfully resolved and quantified the relative amount of each ligand/dendrimer ratio present. This type of information is rare for the entire field of ligand-nanoparticle materials because most analytical techniques have been unable to identify the components in the distribution.
Our experimental data indicates that the actual distribution of ligand-nanoparticle components is much more heterogeneous than commonly assumed. The mean ligand/nanoparticle ratio that is so often the only information known about a material is insufficient. This is because the mean does not provide information on the diversity of components in the material and often does not describe the most common component (the mode). Additionally, our experimental data has provided examples of material batches with the same mean ligand/nanoparticle ratio and very different distributions. This discrepancy indicates that the mean cannot be used as the sole metric to assess the reproducibility of a system. We further found that distribution profiles can be highly sensitive to the synthetic history of the starting material as well as slight changes in reaction conditions. We have incorporated the lessons from our experimental data into new systems designs to provide improved control over the ligand/nanoparticle ratios.
Keywords: nanoparticle, ligand conjugation, dendrimer, HPLC
Introduction
Nanoparticle-based platforms conjugated with functional ligands have been developed for a remarkably wide range of applications including nano-assemblies and structures,1 sensing,2,3 imaging and diagnostics,4,5 probes of biological structure,6 and targeted delivery.7–9 Conjugation of small molecule ligands to a nanoparticle with a large excess of attachment sites results in material that is composed of a distribution of particles with varying ligand/particle ratios. In our experience, the magnitude of this heterogeneity is frequently underestimated and explicit consideration of particle-ligand distributions is often not considered in platform design or fully appreciated when interpreting data derived from such systems. There remains a major need for improved characterization and definition of the material composition as well as a better understanding of the relationship between the components in the distribution and activity.
The arithmetic mean number of ligands is the most widely used parameter to characterize the distribution of ligand-nanoparticle components per particle. Although defining the ligand-nanoparticle composition by the arithmetic mean is an accepted standard of practice, this single value is insufficient to understand the material composition and ultimately predict the material activity. The problem is frequently compounded by the naive expectation that if a distribution of ligands exists, it will take the form of a normal Gaussian distribution centered at the arithmetic mean. This expectation results in a substantial misunderstanding of the actual material compositions, especially for low numbers of conjugated ligands (1–10). Furthermore, it is not reliable to assume material with the same mean number of ligands per particle necessarily has the same distribution. Finally, although values such as the median and the mode do provide additional information about the distribution, a comprehensive understanding requires characterization of the whole distribution.
This account provides our perspective on the problems that distribution in the ligand/particle ratio present to the field at large. This analysis is generally applicable to ligand-nanoparticle systems where in the ligand is conjugated with an excess of attachment sites on the nanoparticle and where the ligand is small relative to the nanoparticle such that site blocking is limited to the single attachment site. The generation 5 (G5) PAMAM dendrimer (Figure 1) plays a special role in these studies because of the very low polydispersity index (1.01) that can be obtained. The comparatively low degree of heterogeneity of this polymer platform facilitates characterization and isolation as a function of conjugated ligand. Other polymer platforms or nanoparticle systems showing a greater degree of heterogeneity in the base material would necessarily be more difficult to analyze in terms of the degree of ligand conjugation.
Figure 1.
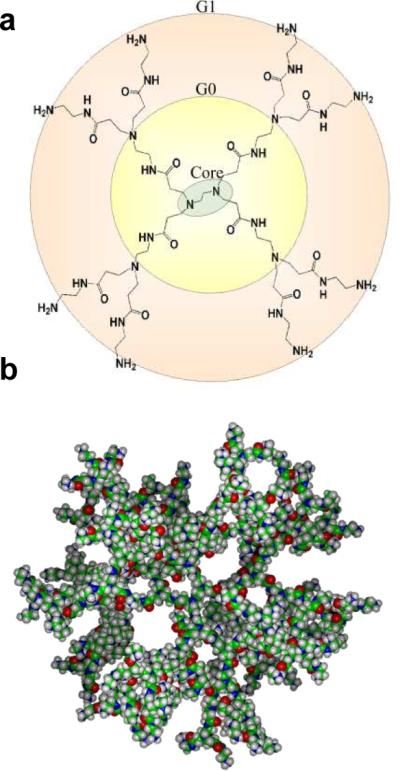
Poly(amidoamine) Dendrimer. a) Structure of a generation 1 (G1) PAMAM dendrimer. b) Space-filled model of an amine-terminated generation 5 (G5) PAMAM dendrimer. (Panel a reproduced in part from Kelly et al. J. Phys. Chem. B 2008, 112, 9337–9345.)
Our efforts in characterizing and defining ligand-nanoparticle distributions have been greatly facilitated by two ligand-nanoparticle systems for which the distribution of components can be resolved by high-performance liquid chromatography (HPLC). 3-(4-(prop-2-ynyloxy)phenyl)propanoic acid (Alkyne Ligand) and 3-(4-(2-azidoethoxy)phenyl)propanoic acid (Azide Ligand), shown in Figure 2, have characteristics similar to many commonly conjugated ligands including size and functional groups. The results of this work provide important information regarding questions of adequate characterization and also provide valuable insight into batch reproducibility challenges that several of these systems face.
Figure 2.
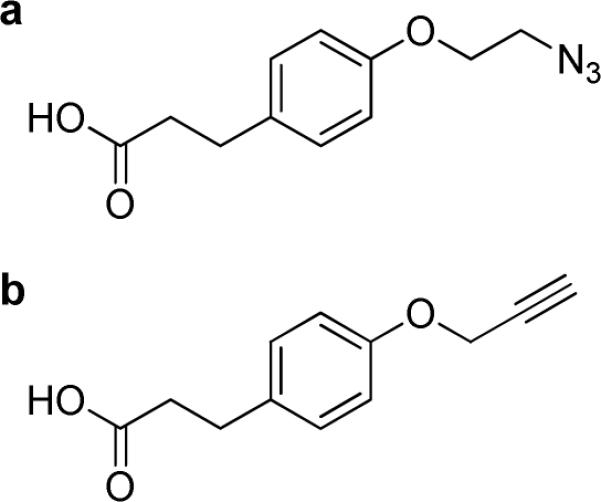
Structure of Azide (a) and Alkyne Ligands (b).
Distributions in ligand-nanoparticle systems: present but often overlooked
Distributions of ligand-nanoparticle components result because the synthetic methods used to conjugate ligand molecules to the particles are driven by random collision and attachment events. The distributions detailed in this account are those that are generated when an excess of attachment sites on the nanoparticle is present relative to the molar amount of ligand molecules being conjugated. Although the theory behind these reactions has long been described,10 very little experimental information exists that shows what these distributions look like in practice. Consequently, in practice, the heterogeneity, width, and functional impact of the distribution are grossly underestimated.
The lack of understanding of the actual distributions in ligand-nanoparticle systems is in large part due to an inability of analytical techniques to identify this form of heterogeneity. Several of the most common techniques are only capable of measuring the arithmetic mean number of ligands per particle. These methods include nuclear magnetic resonance (NMR) spectroscopy, ultraviolet-visible (UV-Vis) spectroscopy, Fourier Transform Infrared Spectroscopy (FTIR) and elemental analysis. While techniques such as gel permeation chromatography (GPC), HPLC, matrix assisted laser desorption ionization – time of flight mass spectrometry (MALDI-TOF MS), capillary electrophoresis (CE), and gel electrophoresis have the potential to resolve the distribution of ligand-nanoparticle components, the frequent outcome of these forms of characterization reveals no information about the distribution of components. This result is likely due to several factors including sub-optimized chromatographic conditions and/or the structural heterogeneity of the nanoparticle itself masking the distribution of ligand-nanoparticle components.
A common chromatographic result is the elution of the ligand-nanoparticle material in a single peak. In some cases, the chromatographic conditions have actually been tuned to produce idealized single peaks rather than multiple peaks corresponding to each of the different components in the distribution (Figure 3). In our experience, this single peak can lead to incorrect conclusions of homogeneous ligand-nanoparticle distributions. If the ligand-nanoparticle system is produced under the conditions described earlier, the single peak physically cannot be composed of a single ligand-nanoparticle component. The reality is that the single peak masks the actual distribution.
Figure 3.
For many ligand-nanoparticle systems, chromatographic-based characterization produces a single peak that fails to resolve the actual distribution of components. This single peak frequently causes the actual distribution of components to be significantly underestimated. If, in the ligand-nanoparticle conjugation reaction, there is an excess of attachment sites relative to the amount of ligand added the distribution follows a Poisson distribution.
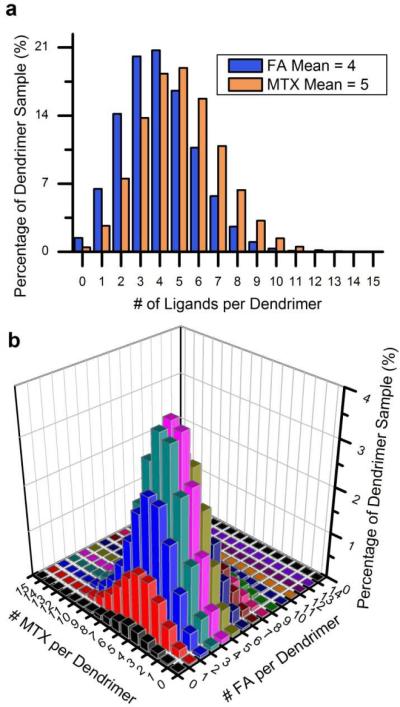
We have generated our share of chromatographic single peaks in our laboratory's research in dendrimer-based platforms for targeted drug delivery. In one such example,6 GPC chromatographs for PAMAM dendrimer conjugated with the dye AlexaFluor488 and with varying amounts of the targeting agent folic acid (means ranging from 0 to 15) contained only a single peak for each sample. In the HPLC elution traces for the same material, while two different peaks were observed, this level of peak resolution does not come close to identifying the actual components in the material. In reality, each of these materials is composed of hundreds of particles with different numbers of dye molecules and folic acid molecules.
Even for the two model ligand-dendrimer systems that can be successfully characterized by HPLC, all of the other characterization techniques (GPC, MALDI-TOF MS and NMR) have failed to identify the different components in the material. Figure 4a shows GPC light scattering signals for four of these ligand-dendrimer samples.11 The samples had mean ligand/dendrimer ratios between 0.4 and 1.5 (measured by NMR). Although all four samples elute as single peaks with similar peak shapes and retention times, the material was shown to have significantly different distributions. Sample A with a mean of 0.4 ligands per dendrimer was composed of 4 components, while Sample D had a mean of 1.5 ligands per dendrimer and was composed of 9 different ligand-dendrimer components. Similarly, characterization of the four ligand-dendrimer samples by MALDI-TOF MS (Figure 4b), provided no information about the distribution of components. In this case, the polydispersity in the dendrimer platform itself along with artifacts such as salt and matrix adducts, fragmentation and time-of-flight artifacts, prevents resolution of the different ligand-dendrimer ratios.
Figure 4.
Examples of ligand-nanoparticle distributions that fail to be resolved using standard analytical techniques (GPC and MALDI-TOF). Samples A–D are composed of a series of ligand-dendrimer conjugates. The mean numbers of ligands for each sample is 0.2, 0.6, 1.04, and 1.47, respectively. HPLC analysis determined the number of ligand-dendrimer components in each sample to be a) The GPC light scattering data of the four samples produces only single peaks. The different ligand-dendrimer distributions are indistinguishable by this technique. b) Similarly, MALDI-TOF characterization is unsuccessful at resolving the different ligand-dendrimer components. One of the challenges of this mass based technique is that the structural heterogeneity of the dendrimer alone masks the different ligand-dendrimer components. (From Mullen et al. Bioconjugate Chemistry, 2008)
Although the inadequate characterization exemplified in Figure 4 is common for most ligand-nanoparticle systems, several groups have successfully worked to characterize the distributions. A number of these studies have provided qualitative evidence of resolved ligandnanoparticle-nanoparticle distributions. The Alivisatos group has used both gel electrophoresis and anion exchange chromatography to resolve the distribution of Au DNA-nanoparticle components.12 Gel electrophoresis was also used by Sperling et al. to resolve nanoparticles with different numbers of PEG ligands.13 Because the resolution strategy was based on the incremental change in size from added PEG ligands, Sperling and co-workers were able to resolve components in both Au-PEG and CdSe/ZnS-PEG systems. A third technique, UPLC, was used by Cason and colleagues to resolve different components in generation 4 (G4) PAMAM dendrimer conjugated with biotin molecules.14 At least 7 different dendrimer-biotin components were resolved by this method. The same distribution was also resolved to a lesser extent using HPLC. HPLC has also been used by Lo and colleagues to resolve the ligand distribution in a PEG-conjugated triazine dendrimer system.15 Finally, in several detailed studies by the Murray group, mass spectrometry (MALDI and ESI-MS) was used to identify different gold nanoparticle components with different numbers of ligands.16
In addition to these qualitative studies, two studies have successfully quantified the distribution of nanoparticle components with different numbers of ligands. Working with quantum dots conjugated with fluorescently tagged proteins (ligand means of 8, 0.85, and 0.3 per particle), Casanova et al. used stepwise photobleaching to quantify the number of proteins bound to each nanoparticle and the ensemble distribution.17 Pons and co-workers used single particle FRET measurements to quantify the distribution of quantum dot-protein components.18 They reported experimental distributions consistent with Poisson distributions. Finally, Haken and colleagues used MALID-TOF to study ligand distributions in a PEG-conjugated enzyme system.19
These studies represent major progress in understanding the material composition of ligand-nanoparticle systems. At a minimum, they provide direct experimental evidence from a wide variety of ligand-nanoparticle systems that the conditions used to synthesize ligand-nanoparticle conjugates produce highly heterogeneous mixtures of components. These studies stand in marked contrast to analytical results that only provided the mean ligand-nanoparticle ratio and did not resolve the distribution. Motivated, in part, by the above studies, we sought to develop a comprehensive understanding of the actual distribution of components in ligand-nanoparticle materials.
Quantification of ligand-nanoparticle distributions using model ligand-dendrimer systems
As described in the introduction, our work in this area has been greatly facilitated by two small molecule ligands: the Alkyne and Azide Ligands. These molecules are similar in size to a wide number of functional ligands that have been conjugated to nanoparticles including siRNA, fluorescent tags, oligonucleotides, folic acid, peptides, therapeutic agents, and other small molecules. Additionally, the coupling chemistry employed to conjugate the Alkyne and Azide ligands to the dendrimer, amide coupling via activated esters formed by EDC or PyBOP, is a standard method for ligand-nanoparticle conjugation.20 Most importantly, reverse-phase HPLC conditions have been developed to resolve the distribution of components in samples of ligand-dendrimer conjugates. We have leveraged this resolution to provide a quantitative analysis of ligand-dendrimer conjugates with mean ligand/particle ratios ranging from 0.4 to 13.
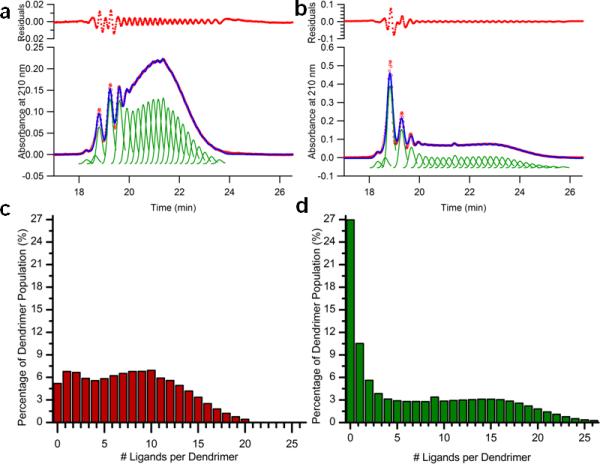
A representative HPLC trace of one such ligand-dendrimer conjugate can be found in Figure 5a. The dendrimer sample in this figure had a mean of 2.7 ± 0.3 Azide Ligands per dendrimer as calculated by combining information from 1H NMR, GPC and potentiometric titration. Five different peaks (labeled 0–4) can be clearly seen in the trace with an additional tailing region between 20 and 22 minutes. Peak 0, at approximately 18.8 minutes, had the same retention time as the parent unmodified dendrimer. The peaks with longer retention times as well as the “tailing” region, beginning at 20 minutes, were found to contain dendrimer particles with increasing numbers of conjugated ligands. A peak fitting method, using multiple copies of a fitting peak, shown in Figure 5b in green, made it possible to deconstruct the entire HPLC trace, including the tailing region, and quantify the relative amount of each ligand-dendrimer component. The shape of the fitting peak was developed based on the peak shape for the unmodified dendrimer. The distribution of ligand-dendrimer components, quantified by the peak fitting analysis, is plotted in Figure 5c. With this analysis, the arithmetic mean ligand/dendrimer ratio was determined to be 2.8 ± 0.1, which is identical (within experimental error) to the arithmetic mean determined by the combined NMR, GPC and potentiometric titration analysis (2.7 ± 0.3). With the entire distribution quantified, additional parameters such as the median (2), the mode (1) and the number of components in the distribution (12) were obtained.
Figure 5.
Peak fitting analysis deconstructs the HPLC trace of a ligand-dendrimer sample to provide the quantified distribution of ligand-dendrimer components. a) The HPLC trace at 210 nm for a ligand-dendrimer sample with a mean of 2.7 Alkyne Ligands per dendrimer is shown in red. Peak 0 had the same retention time as the un-modified parent dendrimer. b) Peak fitting analysis deconstructs the HPLC trace to provide the relative concentration of each ligand-dendrimer component. HPLC data is shown in red dots and the multiple copies of the fitting peak are shown in green. The shape of the fitting peak is based on the HPLC peak shape for the un-modified parent dendrimer. The summation of the fitting peaks is in blue. c) The relative amount of each ligand-dendrimer component in the distribution.
In two initial studies,11,21 distributions were quantified for samples produced using 100% amine terminated PAMAM dendrimer (G5-(NH2)112) and samples produced with partially acetylated dendrimer (G5-Ac80-(NH2)34). The partial acetylation of the dendrimer prior to the conjugation of biologically active ligands has been a key design approach to prevent non-specific interactions between biological systems and the final device.
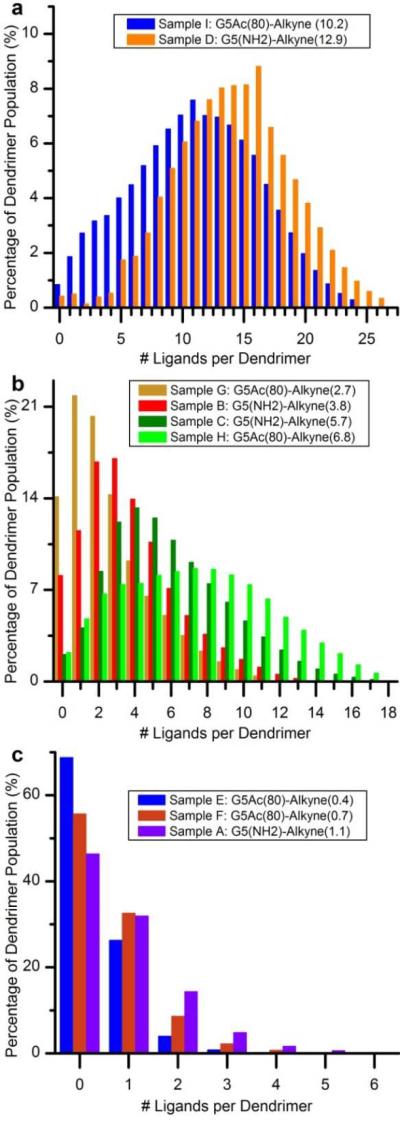
Quantified distributions for some of these ligand-dendrimer conjugates are shown in Figure 6, grouped in 3 different ranges of ligand means: 0.4 to 1.1, 2.7 to 6.8 and 10.2 to 12.9. The combined results from these two studies lead to two important observations. First, the distributions of components in these samples are very heterogeneous. The sample with a mean of 1.1 ligands (Figure 6c), for example, is composed of 6 different components with the mode, a dendrimer with 0 ligands, comprising over 45% of the entire material. Note that if this ligand were biologically active and being used in a drug delivery system, almost half of the material would not have the desired conjugate. This is a significant amount of inactive material, which will have no positive benefit, to inject into a patient. In the sample with a ligand mean of 12.9 (Figure 6a), the distribution is made up of 27 different components. No individual component comprises more than 9% of the total material. Comparisons of each sample against the Poissonian distribution with the same ligand mean revealed the distribution to be skewed and slightly more heterogeneous than the theoretically expected result. Specifically, in all samples, the components at both extremes of the distribution were present in larger quantities than in the Poisson distribution and the components close to the mean were present in slightly smaller quantities. These experimentally quantified distributions are also significantly more heterogeneous than a narrow Gaussian distribution with a standard deviation of 1 or 2.
Figure 6.
Quantified ligand-dendrimer distributions for samples with ligand means ranging from 0.4 to 12.9. Samples were prepared using either a partially acetylated dendrimer (G5-Ac80-(NH2)34) or a 100% amine terminated dendrimer (G5-(NH2)112) a) Ligand-dendrimer distributions for samples with mean ligand/dendrimer ratios of 10.2 and 12.9. b) Distributions for samples with mean ligand/dendrimer ratios between 2.7 and 6.8. c) Distributions for ligand-dendrimer samples with mean ligand/dendrimer ratios ranging from 0.4 to 1.1. (From Mullen et al. ACS Nano 2010)
The second key observation is that pre-existing distributions increase the heterogeneity of subsequent ligand distributions. This observation was made by comparing distributions in ligand-dendrimer material made with the G5-(NH2)112 dendrimer with the distributions in the material made with the partially acetylated dendrimer. Increased skewing from the Poisson distribution was observed for the partially acetylated material. Because the acetic anhydride is itself a very small ligand in the partial acetylation reaction and because an excess of modification sites on the dendrimer (primary amines) are present relative to the amount of acetic anhydride added, the reaction results in a distribution of dendrimer with different numbers of acetyl groups. In the subsequent Alkyne Ligand conjugation, the pre-existing distribution of acetyl groups per dendrimer increases the heterogeneity of the Alkyne Ligand distribution. In addition, dendrimers with larger numbers of acetyl groups, and few amines available for conjugation, are more likely to suffer from site blocking effects which also inducing variation from the simple Poisson distribution. This observation is very important because one of the advantages of using nanoparticles as platforms is that multiple copies of several different functional ligands can be conjugated to the same particle to obtain synergistic properties. In our own work, we frequently combine multiple copies of a targeting agent, a therapeutic agent, and a dye or imaging agent to the same dendrimer. With each conjugation step, however, the pre-existing distribution becomes increasingly heterogeneous. Additionally, simply changing the sequence in which different ligands are added to the nanoparticle could change the final material.
In a third study, the effect of the mass transport quality during the partial acetylation reaction on subsequent ligand-dendrimer distributions was investigated.22 This study was motivated by an observation in our laboratory that ligand-dendrimer distribution profiles had a dependence on the batch of partially acetylated dendrimer. These different batches were produced under supposedly the same experimental conditions and yet they introduced a high level of variability into the system. Figure 7 displays the distribution analysis of two different ligand-dendrimer samples. Both samples had identical mean ligand/dendrimer ratios (6.6 ± 0.7 and 6.8 ± 0.7). The parent partially acetylated dendrimer for each sample was produced under different reaction conditions. For the ligand-dendrimer sample in panel a and c, the parent batch of partially acetylated dendrimer was produced with optimal mass transport conditions (dilution of acetic anhydride, rate of addition, and mixing efficiency). The parent dendrimer for the ligand-dendrimer sample in panel b and d was produced with sub-optimal mass transport conditions. Two dramatically different distribution profiles result as a direct consequence of the changes in mass transport. These differences were extreme versions of the variability observed across batches of materials that were intended to have the same reaction conditions. Alarmingly, the characterization methods used to assess both the batches of partially acetylated dendrimer, HPLC and NMR, as well as the NMR characterization of the ligand-dendrimer samples found no differences between the materials. These difficulties in synthesis associated with poor mass transport would be expected to become exacerbated when attempting to scale up such a process and may have contributed to the failure of this material to be successfully produced for phase I clinical trials.23
Figure 7.
Quantified distributions of two ligand-dendrimer conjugates with mean ligand/dendrimer ratios of 6.6 ± 0.7 (a and c) and 6.8 ± 0.7 (b and d). Although the samples have the same arithmetic means, they have dramatically different distributions of ligand-dendrimer components. The dendrimer samples were produced using two different batches of partially acetylated dendrimer and the differences in ligand-dendrimer distributions were found to be caused by differences in dendrimer-acetyl group distributions in the parent partially acetylated dendrimer batches. For the sample in panel a and c, the parent partially acetylated dendrimer was produced under effective mass transport conditions. The parent dendrimer for the sample in b and d was produced with ineffective mass transport during the partial acetylation reaction. Residuals are ×10−6. (From Mullen et al. Macromolecules 2010)
The inadequacies of the arithmetic mean
The combined knowledge gained from these three studies11,21,22 has provided critical insight into the heterogeneous distributions that are present in ligand-nanoparticle systems. Perhaps one of the most important lessons from these studies is that efforts to characterize these nanomaterials that only determine the mean nanoparticle/ligand ratio are wholly insufficient. The mean value alone provides no information about the number of different components in the material or the relative amount of each component in the distribution. For a simple approximation, a Poisson distribution could be considered in the case where only the mean number of ligands is known. This form of distribution, however, is the “best case” distribution in terms of material homogeneity and is only accurate for particles containing many potential reaction sites as compared to the total number ligands conjugated. All of our experimental data has found the actual distributions in these materials to be more heterogeneous than Poisson. This arises from at least two factors including site-blocking effects and autocatalysis. The mean is also not capable of telling if the material is being produced consistently. The possibility that two samples can have the same mean number of ligands and yet have completely different distributions should no longer be considered unlikely. Finally, attempting to investigate composition-property relationships using the mean number of ligands as the representative material parameter is inadequate. In systems that conjugate multiple different ligand molecules to the nanoparticle, the component that has the same number of ligands as the mean number is likely to make up no more than 4% of the material.21 Figure 8, showing the multiplicative effect of two Poisson distributions, illustrates the wide variety of species present when two different ligands are attached to the same particle. Note that the implications of this are quite severe if one of the ligands is a targeting molecule and the other is a drug or a imaging agent. Many particles with a lot of targeting molecules will have little to no drug or imaging agent and particles with a lot of drug or imaging agent will have little to no targeting agent. Both of these cases make the material ineffective at best, and added toxicity with no benefit at worst, and misleading when interpreted as an imaging agent.
Figure 8.
Illustration of a) Poisson distributions for a particle containing 128 functional surface sites and averages of 4 and 5 conjugated ligands b) the distribution on a single dendrimer containing 128 surface functional sites resulting from the product of the averages of 4 and 5 ligands per dendrimer. (From Mullen et al. ACS Nano 2010)
Challenges for reproducible synthesis
In providing this account, it is not our intention to contend that distributions in ligandnanoparticle materials are a fatal flaw. Certainly, there are many examples of materials that have been composed of heterogeneous mixtures of components and yet have made important societal and industrial impacts. The entire field of polymers, for example, has confronted this challenge for more than a century. From a commercial translation perspective, however, ligandnanoparticle distributions are major challenge to reproducibility. This is mainly because controls have not been incorporated to ensure that the distribution is reproduced in each manufactured batch. As described above, batches of material with consistent ligand means can have very different distributions. These can be caused by small changes in the starting material, mass transport conditions and reaction kinetics. These changes, however, will almost certainly not be detected by the standard levels of characterization being used by the field. There remains a great deal of progress to be made in order to identify the tolerable levels of variability in these materials as well as ways to implement control. It is likely that these distribution tolerances will be very application and material specific.
To address these challenges, major improvements will need to be made in the ability to generate and characterize ligand-nanoparticle distributions that are consistent across multiple batches. For dendrimer-based systems, the Azide and Alkyne Ligand, can be used during process development and scale-up to ensure distribution reproducibility. The Azide and Alkyne ligands can be temporarily substituted for functional ligands when the actual ligand-nanoparticle distribution cannot be directly resolved. These two model ligand systems can be used to test the effectiveness of the reaction conditions (mixing, temperature, reagent addition rate etc.) as well as to monitor the distribution dependence on variations in the starting dendrimer material (number of end groups, amount of trailing generations and dimer).
If future synthetic strategies to produce functional dendrimer-based platforms continue to employ stochastic conditions, our results indicate that distributions can be more reproducible and homogeneous if the partial acetylation step is eliminated and functional ligands are instead conjugated directly to the 100% amine terminated dendrimer. Not only does this approach bypass the poor control present in the partial acetylation reaction, it also may make functional ligand distributions less sensitive to varying amounts of dendrimer defect structures in different commercial lots.
New platform designs
In order to address the challenges inherent in systems with heterogeneous distributions of functional components, several groups have worked to design ligand-nanoparticle systems with improved distribution control. These systems include PAMAM dendrons,24 dendrimer with single orthogonal modification sites,25,26 and gold nanoparticles with precise numbers of ligands.12,13,27,28
Most recently our laboratory has focused on developing PAMAM dendrimer with precise numbers of `click' modifiable ligands.29 Using semi-preparative HPLC, we have been successful in isolating nine different ligand-dendrimer components with 0–8 Ligands from a liganddendrimer distribution. Analytical HPLC characterization of these “Precision Dendrimer” can be found in Figure 9. Using peak fitting, the degree of purity for each of these components was found to be 80% or greater, representing an order of magnitude increase in purity.
Figure 9.
Dendrimer with precise numbers of ligands isolated from a distribution of ligand-dendrimer components. A total of nine different Precision Dendrimer were successfully isolated a) Analytical HPLC characterization of the Precision Dendrimer taken at the isolated concentration. b) Normalized analytical HPLC traces for the nine different Precision Dendrimer. Blue lines show the relationship between the isolated material and the distribution of ligand-dendrimer components. c) Peak fitting found the degree of purity for each isolated dendrimer component to be 80% or greater. Residuals are ×10−6. (From Mullen et al. Chemistry A European Journal 2010)
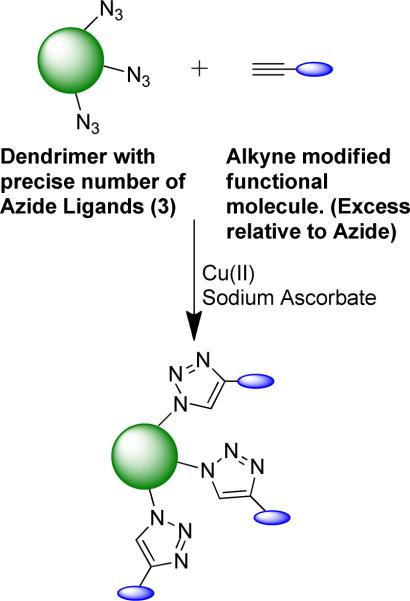
The dendrimer with precise numbers of Azide Ligands, described in Figure 9, offers a truly exciting number of new opportunities. This material can be used as a tool kit to investigate the activity of different functional ligand-dendrimer components. To accomplish this, the functional ligands of interest will need to be modified with a terminal alkyne group and then coupled, using `click' chemistry, to the dendrimer with precise numbers of Azide Ligands (Figure 10). In this coupling reaction, an excess of alkyne-modified functional ligands can be added relative to the number of Azide Ligands in order to produce non-stochastic conditions and ensure that all Azide Ligands are modified. From an engineering design perspective, this information will be valuable for identifying the most active ligand-dendrimer components as well as establishing the tolerable variations in distributions between batches of material that still maintain the same properties. With the most active components identified, this material should also be considered for use as an actual product (a therapeutic in the case of a drug delivery application).
Figure 10.
General strategy to produce dendrimer with precise numbers of functional molecules. The dendrimer with a precise number of Azide Ligands, isolated from a stochastic distribution using HPLC can be further modified with functional molecules using `click' chemistry.
The major feature of this material is reproducibility of properties and activity (and quite possibly, dramatically increased efficacy). Due to the highly reproducible separation of the ligand-dendrimer components on the HPLC column, dendrimer with precise numbers of Alkyne or Azide ligands can be obtained with great consistency. This is a major advantage of this approach because column-loading limitations can be overcome by isolating material over multiple runs. Second, the precision dendrimer have great potential to be a versatile platform that can be modified by a wide range of functional molecules via the click reaction to obtain dendrimer with precise numbers of functional molecules. As such this approach has great potential to solve at least one major existing need in the field of targeted drug delivery. Although the current isolation method that produces the material is inefficient (specific components are isolated from a stochastic distribution), this drawback may be outweighed by its benefits and furthermore, a number of strategies currently exist in the literature that could be leveraged to improve the conservation of material.
Conclusion
In conclusion, ligand-nanoparticle materials are composed of heterogeneous distributions that are widely unappreciated. Where possible, the present reliance on the mean number of ligands per dendrimer should be replaced with a full characterization of the entire distribution of ligand/particle ratios. Furthermore, without the introduction of sufficient controls in platform designs, distributions are a major obstacle to successful translation of ligand-nanoparticle systems.
ACKNOWLEDGEMENT
The authors thank the members of the Michigan Nanotechnology Institute for Medicine and Biological Sciences (MNIMBS) for their contributions to the projects described. We would especially like to acknowledge our colleagues J. R. Baker and B. G. Orr for many stimulating discussions. This work was supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under Award 1 R01 CA119409 and Department of Defense DARPA award W911NF-07-1-0437.
Biography
Douglas G. Mullen was born in Hartford, CT, in 1982. He obtained his B.S.E. in Mechanical Engineering from Duke University in 2005. He then began his doctoral work at the University of Michigan, joining the Banaszak Holl group and the Michigan Nanotechnology Institute for Medicine and Biological Sciences. In 2007, he obtained a M.S.E in Macromolecular Science and Engineering and completed his Ph.D. in Macromolecular Science and Engineering in 2010. He is presently the Dick Sarns Medical Innovation Fellow at the University of Michigan Medical Innovation Center.
Mark M. Banaszak Holl was born in Ithaca, NY, in 1964. He graduated from the University of Chicago in 1986 and obtained his Ph.D. degree at Cornell University in 1991. He spent 1 year as postdoctoral fellow at the IBM T. J. Watson research laboratory in Yorktown Heights, NY, prior to accepting a position as an assistant professor of chemistry at Brown University (1992–1995). He accepted a position at the University of Michigan in 1995 where he is currently Professor of Chemistry, Biomedical Engineering, and Macromolecular Science and Engineering. He is also Associate Vice-President for Research.
References
- (1).Maye MM, Nykypanchuk D, Cuisinier M, van der Lelie D, Gang O. Stepwise surface encoding for high-throughput assembly of nanoclusters. Nature Mat. 2009;8:388–391. doi: 10.1038/nmat2421. [DOI] [PubMed] [Google Scholar]
- (2).Jain KK. Nanotechnology in clinical laboratory diagnostics. Clin. Chim. Acta. 2005;358:37–54. doi: 10.1016/j.cccn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- (3).Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- (4).Landmark KJ, DiMaggio S, Ward J, Kelly C, Vogt S, Hong S, Kotlyar A, Myc A, Thomas TP, Penner-Hahn JE, Baker JR, Holl MMB, Orr BG. Synthesis, characterization, and in vitro testing of superparamagnetic iron oxide nanoparticles targeted using folic acid-conjugated dendrimers. ACS Nano. 2008;2:773–783. doi: 10.1021/nn800034w. [DOI] [PubMed] [Google Scholar]
- (5).Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, Mirkin CA. Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy. Proc. Nat. Acad. Sci. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Holl MMB. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chemistry and Biology. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- (7).Peer D, Karp JM, Hong S, FaroKhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotech. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- (8).Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Design considerations for tumour-targeted nanoparticles. Nat. Nanotech. 2010;5:42–7. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Patri AK, Majoros IJ, Baker JR. Dendritic polymer macromolecular carriers for drug delivery. Curr. Opin. Chem. Biology. 2002;6:466–471. doi: 10.1016/s1367-5931(02)00347-2. [DOI] [PubMed] [Google Scholar]
- (10).Beichelt FE, Fatti LP. Stochastic processes and their applications. Taylor & Francis; London: 2002. [Google Scholar]
- (11).Mullen DG, Desai AM, Waddell JN, Cheng X-M, Kelly CV, McNerny DQ, Majoros IJ, Baker JR, Sander LM, Orr BG, Banaszak Holl MM. The implications of stochastic synthesis for the conjugation of functional groups to nanoparticles. Bioconjugate Chem. 2008;19:1748–52. doi: 10.1021/bc8002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Claridge SA, Liang HYW, Basu SR, Frechet JMJ, Alivisatos AP. Isolation of discrete nanoparticle - DNA conjugates for plasmonic applications. Nano Letters. 2008;8:1202–1206. doi: 10.1021/nl0802032. [DOI] [PubMed] [Google Scholar]
- (13).Sperling RA, Pellegrino T, Li JK, Chang WH, Parak WJ. Electrophoretic separation of nanoparticles with a discrete number of functional groups. Adv. Funct. Mat. 2006;16:943–948. [Google Scholar]
- (14).Cason CA, Oehrle SA, Fabre TA, Girten CD, Walters KA, Tomalia DA, Haik KL, Bullen HA. Improved methodology for monitoring poly(amidoamine) dendrimers surface transformations and product quality by ultra performance liquid chromatography. J. Nanomat. 2008 doi:10.1155/2008/456082. [Google Scholar]
- (15).Lo ST, Stern S, Clogston JD, Zheng JW, Adiseshaiah PP, Dobrovolskaia M, Lim JD, Patri AK, Sun XK, Simanek EE. Biological Assessment of Triazine Dendrimer: Toxicological Profiles, Solution Behavior, Biodistribution, Drug Release and Efficacy in a PEGylated, Paclitaxel Construct. Molec. Pharm. 2010;7:993–1006. doi: 10.1021/mp100104x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tracy JB, Kalyuzhny G, Crowe MC, Balasubramanian R, Choi JP, Murray RW. Poly(ethylene glycol) ligands for high-resolution nanoparticle mass spectrometry. J. Am. Chem. Soc. 2007;129:6706–6707. doi: 10.1021/ja071042r. [DOI] [PubMed] [Google Scholar]
- (17).Casanova D, Giaume D, Moreau M, Martin JL, Gacoin T, Boilot JP, Alexandrou A. Counting the number of proteins coupled to single nanoparticles. J. Am. Chem. Soc. 2007;129:12592–12593. doi: 10.1021/ja0731975. [DOI] [PubMed] [Google Scholar]
- (18).Pons T, Medintz IL, Wang X, English DS, Mattoussi H. Solution-phase single quantum dot fluorescence resonance energy transfer. J. Am. Chem. Soc. 2006;128:15324–15331. doi: 10.1021/ja0657253. [DOI] [PubMed] [Google Scholar]
- (19).Hakem IF, Leech AM, Johnson JD, Donahue SJ, Walker JP, Bockstaller MR. Understanding Ligand Distributions in Modified Particle and Particlelike Systems. J. Am. Chem. Soc. 2010;132:16593–16598. doi: 10.1021/ja107139c. [DOI] [PubMed] [Google Scholar]
- (20).Hermanson GT. Bioconjugate Techniques. 2nd ed. Elsevier Academic Press; Amsterdam: 2008. [Google Scholar]
- (21).Mullen DG, Fang M, Desai AM, Baker JR, Orr BG, Banaszak Holl MM. A quantitative assessment of nanoparticle-ligand distributions: implications for targeted drug and imaging delivery in dendrimer conjugates. ACS Nano. 2010;4:657–670. doi: 10.1021/nn900999c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mullen DG, Borgmeier EL, Fang M, McNerny DQ, Desai A, Baker JR, Orr BG, Holl MMB. Effect of Mass Transport in the Synthesis of Partially Acetylated Dendrimer: Implications for Functional Ligand-Nanoparticle Distributions. Macromolecules. 2010;43:6577–6587. doi: 10.1021/ma100663c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kukowska-Latallo JF, Candido KA, Cao ZY, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR. Nanoparticle targeting of anticancer drug improves therapeutic response in animal model of human epithelial cancer. Cancer Res. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- (24).McNerny DQ, Kukowska-Latallo JF, Mullen DG, Wallace JM, Desai AM, Shukla R, Huang BH, Holl MMB, Baker JR. RGD Dendron Bodies; Synthetic Avidity Agents with Defined and Potentially Interchangeable Effector Sites That Can Substitute for Antibodies. Bioconjugate Chem. 2009;20:1853–1859. doi: 10.1021/bc900217h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Goyal P, Yoon K, Weck M. Multifunctionalization of dendrimers through orthogonal transformations. Chem. Eur. J. 2007;13:8801–8810. doi: 10.1002/chem.200700129. [DOI] [PubMed] [Google Scholar]
- (26).Yoon K, Goyal P, Weck M. Monofunctionalization of dendrimers with use of microwave-assisted 1,3-dipolar cycloadditions. Org. Lett. 2007;9:2051–2054. doi: 10.1021/ol062949h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chak CP, Xuan SH, Mendes PM, Yu JC, Cheng CHK, Leung KCF. Discrete Functional Gold Nanoparticles: Hydrogen Bond-Assisted Synthesis, Magnetic Purification, Supramolecular Dimer and Trimer Formation. ACS Nano. 2009;3:2129–2138. doi: 10.1021/nn9005895. [DOI] [PubMed] [Google Scholar]
- (28).Zanchet D, Micheel CM, Parak WJ, Gerion D, Alivisatos AP. Electrophoretic isolation of discrete Au nanocrystal/DNA conjugates. Nano Letters. 2001;1:32–35. [Google Scholar]
- (29).Mullen DG, Byrne EL, Desai A, van Dongen MA, Barash M, Cheng XM, Baker JR, Holl MMB. Isolation and characterization of dendrimer with precise numbers of functional groups. Chem. Eur. J. 2010;16:10675–10678. doi: 10.1002/chem.201001175. [DOI] [PMC free article] [PubMed] [Google Scholar]