Abstract
Human O6-alkylguanine-DNA alkyltransferase (AGT) repairs mutagenic O6-alkylguanine and O4-alkylthymine adducts in single-stranded and duplex DNAs. These activities protect normal cells and tumor cells against drugs that alkylate DNA; drugs that inactivate AGT are under test as chemotherapeutic enhancers. In studies using 6-carboxyfluorescein (FAM)-labeled DNAs, AGT reduced the fluorescence intensity by ~40% at binding saturation, whether the FAM was located at the 5′ or the 3′ end of the DNA. AGT protected residual fluorescence from quenching, indicating a solute-inaccessible binding site for FAM. Sedimentation equilibrium analyses showed that saturating AGT-stoichiometries were higher with FAM-labeled DNAs than with unlabeled DNAs, suggesting that the FAM provides a protein binding site that is not present in unlabeled DNAs. Additional fluorescence and sedimentation measurements showed that AGT forms a 1:1 complex with free FAM. Active site benzylation experiments and docking calculations support models in which the primary binding site is located in or near the active site of the enzyme. Electrophoretic analyses show that FAM inhibits DNA binding (IC50 ~ 76μM) and repair of DNA containing an O6-methylguanine residue (IC50 ~ 63μM). Similar results were obtained with other polycyclic aromatic compounds. These observations demonstrate the existence of a new class of non-covalent AGT-inhibitors. After optimization for binding-affinity, members of this class might be useful in cancer chemotherapy.
1. INTRODUCTION
O6-alkylguanine and O4-alkylthymine are mutagenic and cytotoxic residues that occur in DNA exposed to alkylating agents [1,2]. In many organisms, these modified bases are repaired by O6-alkylguanine-DNA alkyltransferase (AGT, also known as methylguanine methyltransferase, MGMT), a single-cycle (suicide) enzyme that mediates transfer of an alkyl group from a DNA base to an active site cysteine [3,4]. The alkyl-enzyme does not appear to be recycled; in eukaryotic cells it is ubiquitinated and rapidly degraded [5,6]. In addition to protecting normal cells, AGT’s repair activities also protect tumor cells against DNA-alkylating drugs. Clinical trials of AGT-inhibitors are underway, in attempts to increase the efficacy of these alkylating drugs in cancer chemotherapy [7-9], however we are only starting to learn how AGT interacts with and repairs DNA adducts.
Human AGT is a small, monomeric protein (Mr = 21,519) that is expressed constitutively in normal cells [10-12]. It is a member of a large family of DNA-modifying and –repair enzymes that act by flipping a target DNA base out of its stacked location in the duplex and binding it in an active site pocket [12-14]. Intriguingly, AGT binds single stranded and duplex DNAs with similar affinities (K·ω ~ 106 M−1) [15,16] and repairs single- and double-stranded DNAs with a modest preference for duplex [17,18]. Together these results suggest a mechanism that minimizes differences in the free energy changes associated with the base-flipping transition in single stranded and duplex substrates. Here we describe the interactions of AGT with short oligonucleotides in which the 5′-terminal or 3′-terminal residue is a 6-carboxyfluorescein (FAM) derivative. Our original aim was to use these DNAs in FRET-measurements of the separations between fluorophore-labeled AGT proteins and DNA ends. We were surprised to find that DNA-labeling with FAM perturbed the binding stoichiometries and affinities of target DNAs. The widespread use of fluorophores like FAM in studies of protein-DNA interaction, the potential of compounds like FAM to be useful probes of AGT structure and the possibility that molecules like FAM might become useful chemotherapeutic enhancers justify a more detailed characterization of these effects; the result of that characterization is offered below.
2. MATERIALS AND METHODS
2.1 Enzymes and Reagents
Restriction endonuclease NarI and T4 polynucleotide kinase were purchased from New England Biolabs. 6-Carboxyfluorescein (FAM; CAS Number 3301-79-9), 9-(2,2 dicyanovinyl)julolidine (DCVJ; CAS Number: 58293-56-4), guanosine 5′ monophosphate (CAS Number: 85-32-5) and acrylamide were from Sigma. 4,4′-bis(phenylamino)-[1,1′-Binaphthalene]-5,5′-disulfonic acid (bis-ANS, CAS Number 65664-81-5) was from Invitrogen. Thioflavin T (ThT; CAS Number 2390-54-7) was the kind gift of Dr. H. Levine (University of Kentucky). [γ-32P]ATP (6000 Ci/mmol) was from NEN Radiochemicals (Perkin Elmer). All other chemicals were reagent grade or better.
2.2 Protein purification
Recombinant human AGT protein in which the six C-terminal residues have been substituted with histidines was purified according to published protocols [4]. The purity of the protein was verified by SDS-gel electrophoresis [19,20]; by this criterion, sample purities of >95% were routine. Protein samples were ~96% active in a quantitative alkyltransferase assay described below. Samples were stored at −80°C until needed. AGT concentrations were measured spectrophotometrically using ε280 = 3.93 × 104 M−1cm−1 [16].
2.3 DNA samples
Oligonucleotides of 16, 24 and 26 residues (Table 1) were purchased from Invitrogen. The were purified by the supplier using reverse-phase HPLC, and after receipt, by phenol extraction (3X) followed by ether extraction and extensive dialysis against 10mM Tris (pH 8.0 at 20°C) buffer. Concentrations were measured spectrophotometrically using extinction coefficients provided by the manufacturers. The 24-mer oligonucleotide E (see Table 1) was labeled at its 5′ hydroxyl with 32P as described [21]. Unincorporated [γ-32P]ATP was removed by buffer exchange using Sephadex G-10 mini-spin columns equilibrated with 10 mM Tris (pH 8.0 at 21°C), 1 mM EDTA. DNA duplexes were prepared by mixing purified 5′-labeled oligonucleotide with 1.05-fold molar excesses of complementary unlabeled strands (oligonucleotide C or D), heating to 90°C for 1 min, then slowly cooling to room temperature. After annealing, the purities of duplex DNAs were tested by native polyacrylamide gel electrophoresis [22].
Table 1.
Oligodeoxyribonucleotides used in this study
| DNA | Sequence |
|---|---|
| 16-mers | |
| Oligo A | 5′- AGT CAG TCA GTC AGT C -3′ |
| 5′-FAM-Oligo A | 5′- (FAM)AGT CAG TCA GTC AGT C -3′ |
| 3′-FAM-Oligo A | 5′- AGT CAG TCA GTC AGT C(FAM) -3′ |
| 24-mers | |
| Oligo C | 5′- GGG TCA TTT GGC GCC TTT CGA TCC -3′ |
| Oligo D | 5′- GGG TCA TTT GGC GCC TTT CGA TCC -3′ |
| Residue positionsa | 1 2 3 4 5 6 |
| Oligo E (complements Oligos C and D) |
3′- CCC AGT AAA CCG CGG AAA GCT AGG -5′ |
| 26-mers | |
| Oligo F | 5′- AGT CAG TCA GTC AGT CAG TCA GTC AG -3′ |
| 5′-FAM-Oligo F | 5′- (FAM)AGT CAG TCA GTC AGT CAG TCA GTC AG -3′ |
| 3′-FAM-Oligo F | 5′- AGT CAG TCA GTC AGT CAG TCA GTC AG(FAM) -3′ |
NarI recognition sequence shown in large type. Residue shown in bold is O6-methylguanine.
2.4 Mobility shift assays
Binding reactions were carried out at 20°C in 10 mM Tris (pH 7.6), 50 mM KCl and 5 mM 2-mercaptoethanol. Mixtures were equilibrated for 30 min. Duplicate samples incubated for longer periods gave identical results, indicating that equilibrium had been attained (result not shown). Electrophoresis was carried out in 20% polyacrylamide gels, as described [15]. Autoradiographic images were captured on storage phosphor screens (type GP, GE Healthcare) detected with a Typhoon phosphorimager and quantitated with Image-Quant software (GE Healthcare).
2.5 DNA alkyltransferase assays
The NarI endonuclease is inactive when substrate DNA contains an O6-methylguanine at position 2 of its cognate sequence [23] (numbering shown in Table 1). Cleavage is restored by DNA alkyltransferase activity provided by AGT. Oligonucleotides C and D containing the NarI sequence (Table 1) were annealed separately with complementary oligo E, as described above. Alkyltransferase reactions were carried out for 10 or 15 min (as indicated) with 1.1 μM AGT and 0.25μM duplex DNA dissolved in 20 mM Tris-acetate (pH 7.9 at 25°C), 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol. Reactions were stopped by addition of SDS (final concentration 0.2% w/v) followed by two extractions with water-saturated phenol and three extractions with water-saturated diethylether. Samples were incubated at room temperature under vacuum to evaporate ether before digestion with NarI. Samples were resolved by native gel electrophoresis [22]; electrophoretic distributions were recorded and quantified using a phosphorimager. In reactions carried out in AGT-excess, 98% of the DNA in our preparations was a substrate for both alkyltransferase and NarI activity. When this DNA was titrated with AGT, maximal repair was obtained when [AGT]/[competent DNA] > 1.04, corresponding to an alkyltransferase activity of 96%.
2.6 Analytical ultracentrifugation
Protein and DNA samples were dialyzed against 10 mM Tris (pH 7.6), 50 mM KCl and 5 mM 2-mercaptoethanol. Data sets were acquired at 4.0 ± 0.1°C in a Beckman XL-A centrifuge (Beckman, Fullerton, CA), using an AN60Ti rotor. Radial absorbance scans were taken at 260 nm (unlabeled DNA molecules) or 495 nm (FAM-labeled DNAs or free FAM). In sedimentation equilibrium experiments, rotor speeds were 15,000, 22,000 and 30,000 rpm and approach to equilibrium was considered to be complete when scans taken 6h apart were indistinguishable. Typically, equilibration times ≥24h met this criterion. Five scans were averaged for each sample at each wavelength and rotor speed.
Highly-cooperative binding to short DNAs with small numbers of binding sites can be described by the simple mechanism nP + D ⇆ PnD, in which free DNA (D) is in equilibrium with saturated protein-DNA complex (PnD) and intermediates with protein stoichiometries < n are not present at significant concentrations. At sedimentation equilibrium, the radial distribution of absorbance for such a system is given by equation 1.
| (1) |
Here A(r) is the absorbance at radial position r and αP, αD and αPnD are absorbances of protein, DNA and protein-DNA complex at the reference position (ro) and ε is a baseline offset that accounts for radial position-independent differences in the absorbances of different cell assemblies. The reduced molecular weights of AGT protein, DNA and protein-DNA complexes are given by , and . Here MP and MD are the molecular weights of protein and DNA, n is the protein:DNA ratio of the complex; ρ is the solvent density, z, the rotor angular velocity, R is the gas constant and T the temperature (Kelvin). The partial specific volume of AGT () was calculated by the method of Cohn and Edsall [24]; those of single stranded and duplex DNAs (; ) were obtained by interpolation of the data of Cohen and Eisenberg [25]. Partial specific volumes of protein-DNA complexes were estimated using Eq. 2.
| (2) |
Sedimentation velocity data was obtained at 4°C and 40,000 rpm with a 4 min scan interval and without signal averaging. Sedimentation coefficient distributions c(s) were obtained by direct boundary modeling using numerical solutions of the Lamm equation [26] implemented in the program SEDFIT [27], obtained from http://www.analyticalultracentrifugation.com/default.htm. Buffer density and viscosity values were calculated using the public domain program SEDNTERP, developed by D. Hayes, T. Laue and J. Philo [28], obtained from http://www.rasmb.bbri.org/.
2.7 Fluorescence analyses
Measurements were made with a Perkin Elmer LS55 fluorometer with sample temperature maintained at 4°C. Excitation and emission wavelengths were 496 nm and 520 nm, respectively. Anisotropy was calculated using equation 3.
| (3) |
Here IVV is the fluorescence intensity with excitation and emission polarizers oriented vertically, IVH is the intensity with excitation polarizer vertical and emission horizontal. The grating factor G = IHV/IHH corrects for instrumental polarization [29].
In fluorescence quenching studies, samples were titrated with 8 M acrylamide solution, directly in the cuvette. Fluorescence values were corrected for dilution effects. Data were analyzed using the Stern-Volmer equation (Eq. (4)):
| (4) |
where F0 and F are the fluorescence intensities in the absence and presence of quencher, respectively; KSV is the Stern Volmer constant and [Q] is concentration of quencher [30].
2.8. Docking calculations
Docking of 6-carboxyfluorescein to an AGT monomer (from PDB 1T38) was done with AutoDock Vina [31] and AutoDockTools [32]. The search region was defined interactively as a 50Å cube encompassing the entire AGT molecule, and rotational flexibility was allowed for appropriate ligand bonds. The exhaustiveness parameter in AutoDoc Vina was set to 80 to increase the probability of finding the scoring minimum. The default value of nine binding modes was used. Coordinates for 6-carboxyfluorescein were generated with the CORINA 3D web server (http://www.molecular-networks.com/online_demos/corina_demo). Figures were prepared using Pymol (W.L. DeLano, The PyMOL Molecular Graphics System, 2002, http://www.pymol.org).
3.0 RESULTS
3.1 AGT interacts with DNA-conjugated 6-carboxyfluorescein labels
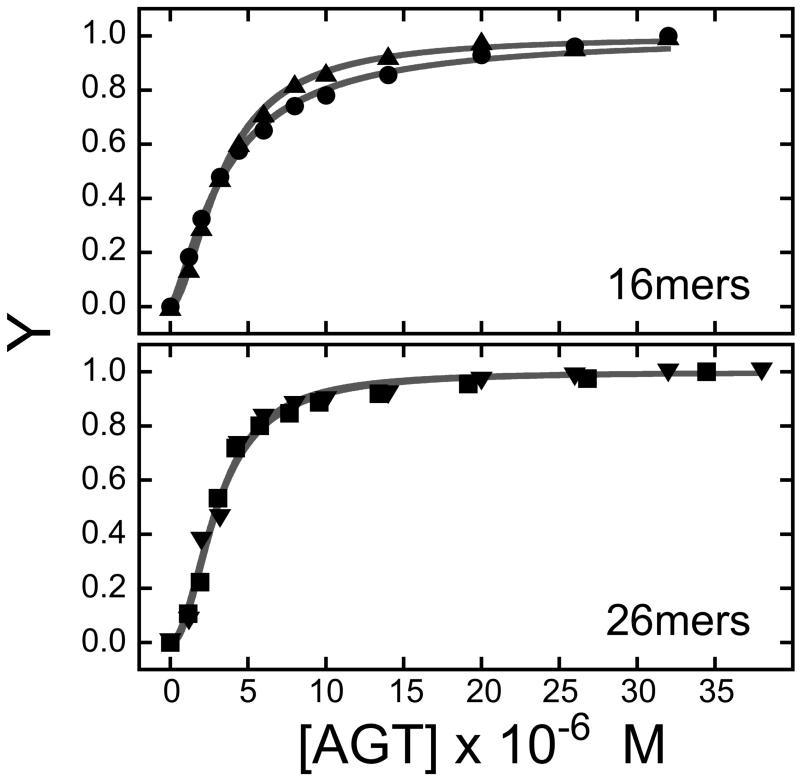
The fluorescence anisotropy assay is a standard method for detecting protein-DNA interactions [33]. If protein binding alters the rotational dynamics of the DNA-conjugated fluorophore and if the excited state lifetime of the fluorophore is similar to its rotational relaxation time, binding can be detected as a change in steady-state fluorescence anisotropy. Anisotropy assays for AGT binding to single stranded 16mer and 26mer DNAs that have been labeled at 5′ or 3′ ends with 6-carboxy fluorescein (FAM) are shown in Fig. 1A. The fractional saturation Y = (A − A0)/(Amax − A0) where A is the anisotropy at each titration step, A0 that of protein-free DNA and Amax that of the saturated complex. The smooth curves in Fig. 1A are fits to the data using Eq. 5.
| (5) |
Here, K is the apparent association constant, [A] is the concentration of free [AGT] and n is the Hill coefficient. These fits returned n = 1.36 ± 0.05 for 3′ labeled 16mer and n = 1.74 ± 0.04 for 5′-labeled 16mer DNAs (top panel) and n = 2.13 ± 0.14 for 3′ labeled 26mer and 2.02 ± 0.17 for 5′-labeled 26mer DNAs (bottom panel). AGT has previously been shown to form 4:1 complexes with the unlabeled 16mer sequence and 6:1 complexes with the unlabeled 26mer [15,34]. As expected for moderately-cooperative binding, the observed Hill coefficients are less than these stoichiometry values [35,36].
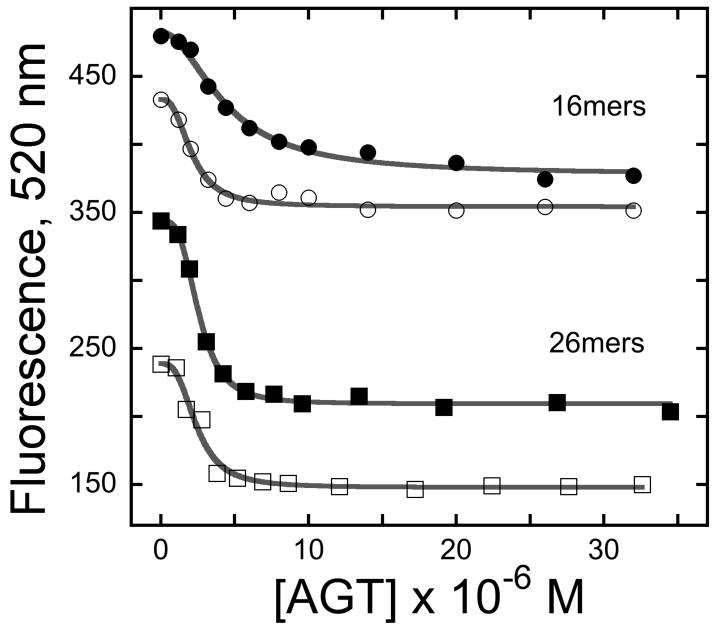
Figure 1. AGT binding to FAM-labeled DNAs detected by changes in fluorescence anisotropy and intensity.
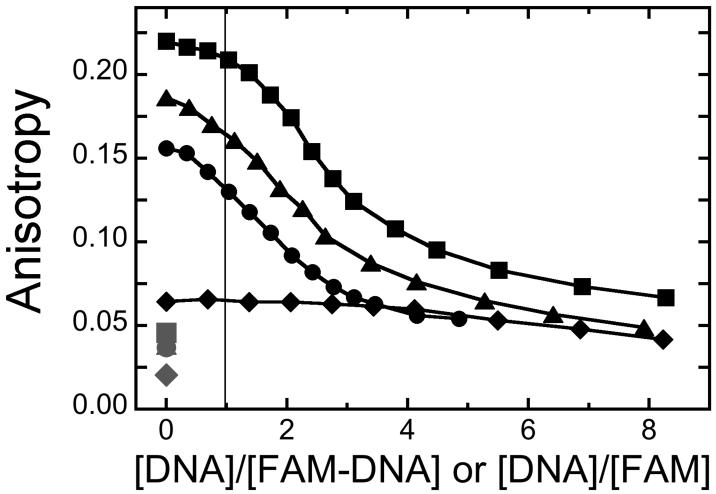
Panel A. Binding detected by changes in anisotropy. Graph of fractional binding saturation Y as a function of [AGT]. Samples contained 1.5 μM 5′-labeled 16-mer (▲) or 1.3 μM 3′-labeled 16-mer (●) or 1.2 μM 5′-labeled 26-mer (▼) or 1.1 μM 3′-labeled 26-mer (■) and the indicated concentrations of AGT in 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 5 mM 2-mercaptoethanol. Incubation was for 1h at 4°C. Fluorescence anisotropies were measured as described in Methods. The smooth curves are least-squares fits of Eq. 1 to each data set. Panel B. Binding detected by changes in emission intensity. Samples contained 1.5 μM 5′-labeled 16-mer (○) or 1.3 μM 3′-labeled 16-mer (●) or 1.2 μM 5′-labeled 26-mer (■) or 1.1 μM 3′-labeled 26-mer (□) and the indicated concentrations of AGT in 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 5 mM 2-mercaptoethanol. Incubation was for 1h at 4°C. Fluorescence intensities were measured with excitation and emission monochromators set at 496 nm and 520 nm, respectively. Panel C. Titration of AGT-saturated FAM-DNAs and AGT mixtures containing unconjugated FAM with unlabeled DNAs. Complexes contained 9μM AGT and 1.3 μM 3′-FAM-16-mer (●); 1.4 μM 5′-FAM-16-mer (▲); 1.1 μM 3′-FAM-26-mer (■); 0.9 μM 6-(FAM) (◆) in buffer containing 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 5 mM 2-mercaptoethanol. Samples were incubated 1h at 4°C, then titrated with unlabeled homologous DNAs. AGT solutions containing unconjugated FAM were titrated with unlabeled 16mer. Fluorescence values were corrected for dilution effects. The corresponding grey symbols show the anisotropies for FAM-DNAs and unconjugated FAM in the absence of AGT. The vertical reference line indicates a 1:1 molar ratio.
When fluorescence anisotropy binding assays are performed, it is often assumed that the protein interacts with the DNA and not the fluorophore. As a test for interactions with the FAM residue, emission intensities were measured as functions of [AGT]. Large decreases in intensity were observed for both 16 mer and 26 mer DNAs, regardless of whether the FAM was located at the 5′ or the 3′ end of test oligonucleotides (Fig 1B). The emission intensity and of fluorescein are sensitive to environmental pH, polarity and the availability of hydrogen-bond partners [37,38]. In addition, at least one electronically-excited state of FAM is susceptible to quenching (see below). Our interpretation, supported by data given below, is that direct interaction with AGT reduces FAM emission through at least one of these mechanisms.
Protein interactions with DNA-conjugated dyes have the potential to change the affinities and/or stoichiometries observed for protein-DNA interactions. A competition assay provides a simple test of whether these quantities differ for FAM-labeled and unlabeled DNAs. Shown in Fig. 1C are anisotropy assays in which AGT complexes with labeled DNAs were titrated with homologous DNAs lacking labels. If stoichiometries and affinities are equal, then the mid-point of the DNA-exchange reactions should occur when concentrations of labeled and unlabeled DNAs are equal [39]. However, for all of the DNAs tested, a molar excess of unlabeled competitor was needed to reach the exchange reaction midpoint. This establishes that AGT affinities and/or stoichiometries for FAM-labeled DNAs are greater than those for the unlabeled homologues.
3.2 Binding stoichiometries are greater with FAM-labeled oligonucleotides than with unlabeled homologues
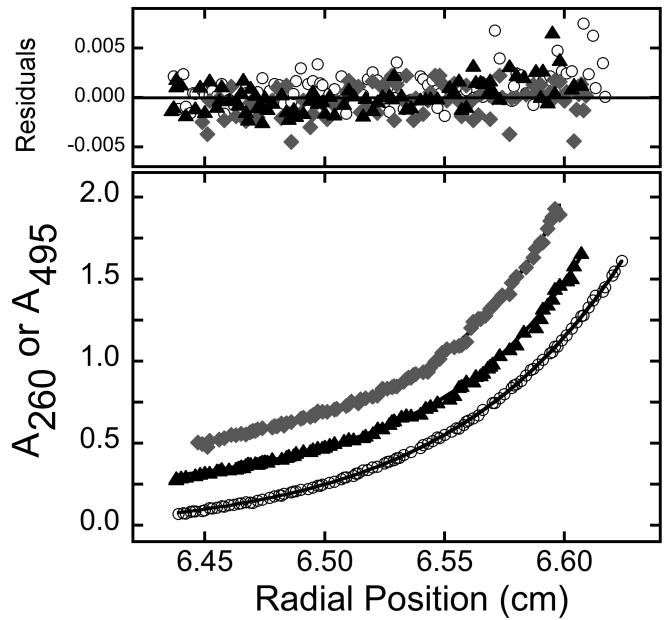
The binding competition results (above) raise the possibility that FAM labeling creates an extra binding site or sites for AGT. To test this notion, sedimentation equilibrium analyses were carried out on mixtures of labeled DNAs and AGT; parallel experiments were carried out using unlabeled DNAs. Data were collected at 15,000, 22,000 and 30,000 rpm (Fig. 2) and were fit using Eq. 1. Small residuals distributed symmetrically around zero indicate that the nP + D ⇆ PnD binding model embodied in Eq. 1 accounts well for the mass distributions in these samples. Unlabeled 16mer and 26mer DNAs gave binding stoichiometries (3.86 ± 0.08 and 5.89 ± 0.07 respectively) that were consistent with previously-reported values [15]. However, stoichiometries for FAM-labeled molecules were significantly larger than those for unlabeled DNAs (for example, 5.06 ± 0.09 for 3′-FAM-16mer and 6.59 ± 0.20 for 5′-FAM 26mer; all results are summarized in Table 2). These stoichiometries are also significantly larger than ones predicted on the basis of the 4nt/protein binding site size observed with a wide range of DNA lengths and sequences [15,40]. Together, these results indicate that FAM-labeled DNAs allow novel AGT-interactions that are not available with unlabeled substrates.
Figure 2. Sedimentation equilibrium analyses of samples containing AGT, DNA and AGT-DNA complexes.
Samples contained 19 μM wt-AGT and 1.3 μM of unlabeled single-stranded 16mer (○), 3′-FAM-labeled single-stranded 16mer (◆) or 5′-FAM-labeled single-stranded 16mer (▲), in buffer consisting of 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 5 mM 2-mercaptoethanol. Samples were brought to sedimentation equilibrium at 15,000 rpm. Radial scans (bottom panel) were taken at 260nm (unlabeled DNA) or 495 nm (FAM-labeled DNAs). The smooth curves correspond to fits of Eq. 1 to the data. The upper panel shows fitting residuals to the three data sets.
Table 2.
Stoichiometries of saturated AGT-DNA complexes measured by analytical ultracentrifugation
| DNA | Stoichiometrya |
|---|---|
| Unmodified 16-mer Oligo A | 3.86 ± 0.08 |
| 3′-(FAM)-16-mer Oligo A | 5.06 ± 0.09 |
| 5′-(FAM)-16-mer Oligo A | 4.53 ± 0.11 |
| Unmodified 26-mer | 5.89 ± 0.07 |
| 3′-(FAM)-26-mer Oligo F | 6.38 ± 0.21 |
| 5′-(FAM)-26-mer Oligo F | 6.59 ± 0.20 |
Mean ± SEM for ≥3 independent determinations.
3.3 AGT protects DNA-linked FAM from acrylamide quenching
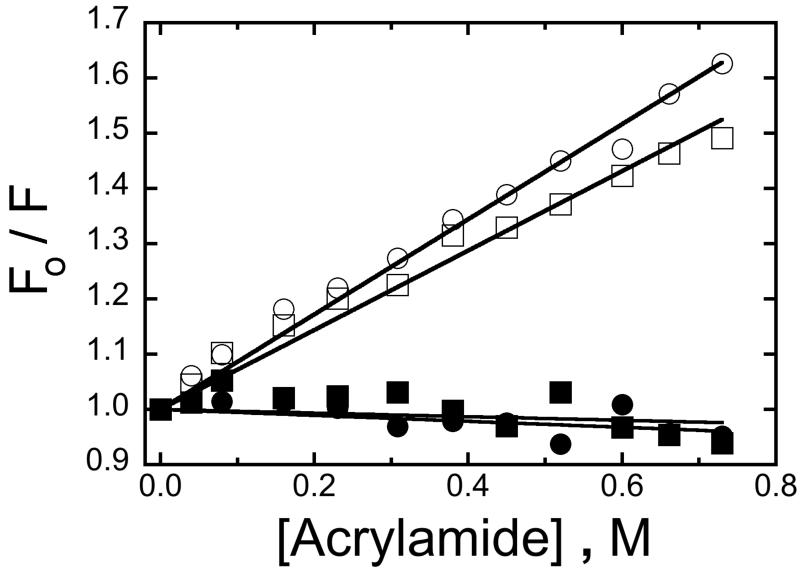
FAM might interact with AGT on the protein’s outer surface or it might be bound in an internal site such as the alkyltransferase cleft. Surface binding sites are expected to be accessible to small solutes while internal sites should be much less so. Fluorescence quenching by acrylamide monomer, a small, neutral solute, tests this prediction. Guided by titration data (Figs 1A and B), saturated AGT-DNA complexes were prepared with 5′- and 3′-FAM-labeled 16mer DNAs. The dependence of fluorescence intensity on [acrylamide] for these samples is given by the Stern-Volmer plot shown in Figure 3. In the absence of AGT, FAM fluorescence is readily quenched and the nearly-linear dependences of F0/F on [acrylamide] are consistent with a homogeneous quenching mechanism [30,41]. In contrast, the fluorescence from saturated AGT complexes (filled symbols) is effectively unquenched over the entire [acrylamide] range. We interpret this as evidence that AGT strongly limits acrylamide access to FAM; the simplest model consistent with this outcome is one in which FAM is bound in an internal site on AGT. Further evidence that FAM is bound near the alkyltransferase site is given below.
Figure 3. Stern-Volmer plots for acrylamide quenching of fluorescence from FAM-labeled 16-mer-DNAs.
Symbols: DNAs carrying 3′-labels (○, ●); DNAs carrying 5′-labels (□, ■). Data from samples containing AGT are represented by filled symbols; data from samples containing no AGT are represented by open symbols. Samples contained 0.6 μM FAM-labeled DNA and where indicated, 14μM AGT, in buffer containing 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 5 mM 2-mercaptoethanol. Samples were incubated for 1h at 4°C prior to measurement at the same temperature. The smooth lines are fits to the Stern-Volmer equation (Eq. 3).
3.4 AGT binds free FAM
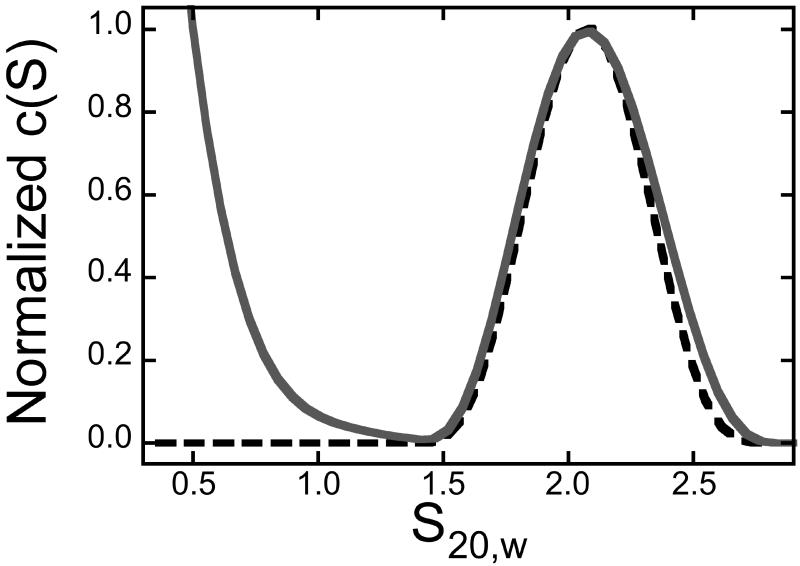
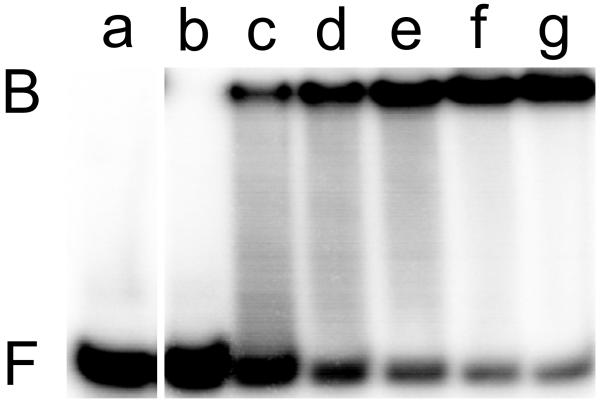
The results given above do not indicate whether DNA contacts are necessary for the AGT-FAM interaction. To test for DNA-independent FAM binding, sedimentation velocity measurements were made on solutions containing FAM and AGT protein (Fig 4A). The c(s) distribution detected at 495 nm (where FAM absorbs but protein does not) has high values at S20,w < 0.5, corresponding to free dye and at S20,w = 2.07, coincident with the S20,w of AGT alone, detected at 280 nm. The superposition of s-value distributions for free protein and dye-protein complex indicates that dye is bound by the protein and also shows that dye binding is not accompanied by significant changes in protein conformation or aggregation state.
Figure 4. Binding of FAM to AGT.
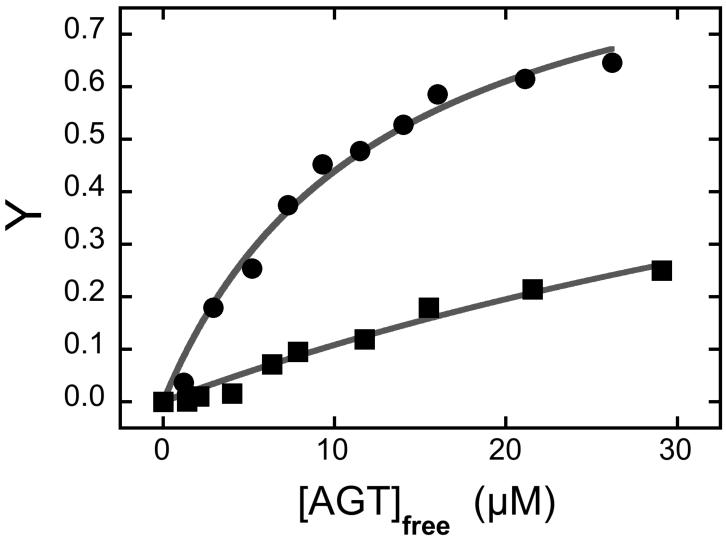
Panel A. Binding detected by sedimentation velocity analysis. Samples containing FAM (1.6 μM) and AGT (14μM) were equilibrated and analyzed at 4°C in 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 5 mM 2-mercaptoethanol. Sedimentation velocity data were acquired at 40,000 rpm. Distributions of c(s) were normalized against the amplitude of the S20,w = 2.07 peak, to facilitate comparison. Symbols: solid curve, data for FAM, obtained at 495nm; dashed curve, data for AGT sedimented in the absence of FAM, obtained at 280 nm. Panel B. Binding of FAM with AGT detected by fluorescence anisotropy. Samples containing FAM (0.9 μM) and AGT (0-27μM) in 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 5 mM 2-mercaptoethanol buffer were equilibrated for 1h at 4°C before measurement. Fluorometry was performed as described in Methods. Fractional saturation Y = (A − A0)/(Amax − A0) where A is the anisotropy at each titration step, A0 that of protein-free DNA and Amax that of the saturated complex. Inset: the corresponding fluorescence intensities.
To determine the affinity with which FAM binds AGT, a fixed concentration of free dye was incubated with increasing concentrations of AGT. Significant increases in fluorescence anisotropy and decreases in intensity accompanied AGT addition (Fig. 4B). These changes parallel those seen when AGT binds FAM-DNA adducts. Using the fluorescence anisotropy data, the dependence of the fractional binding saturation Y on [AGT] could be analyzed using Eq. 5, with n = 1 (corresponding to a 1:1 complex). This gave an apparent binding affinity K = 7.84 ± 0.24 × 104 M−1 (corresponding Kd = 1.27 ± 0.04 × 10−5 M) that is only 10-fold less than the monomer-equivalent association constant found for AGT binding to single-stranded DNAs [15,42]. Thus, interactions with FAM alone appear to be strong enough to perturb binding affinity measurements (such as the partition measurement shown in Fig. 1C) and stoichiometry measurements made in the presence of >10μM AGT, like those shown in Fig. 2.
3.5 FAM and DNA compete for binding to AGT
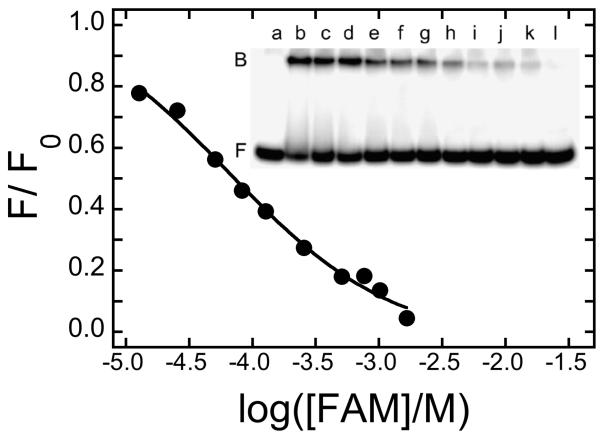
The quenching results (Fig. 3) show that FAM is bound in a quencher-inaccessible site in the AGT structure. One obvious candidate is the cleft that contains the site of alkyltransferase activity. Bound at that site, a molecule of FAM might affect DNA binding and/or alkyltransfer activity. To test the first possibility, mobility shift assays were performed in which AGT-DNA mixtures were titrated with FAM. Shown in Fig. 5, DNA binding by AGT decreased with increasing [FAM]. Analysis of the dependence of DNA binding on [FAM] shows that the IC50 for FAM-inhibition of DNA binding (7.63 ± 0.61 × 10−5 M) was slightly greater than the value of Kd found for the AGT-FAM interaction in the absence of DNA (1.27 ± 0.04 × 10−5 M). This is the expected outcome if DNA binding and FAM binding are competitive.
Figure 5. Inhibition of DNA binding by FAM.
Electrophoretic mobility shift assay (inset) carried out as described in Methods. Samples contained 32P-labeled duplex 24mer (0.17μM), AGT, either 0μM (lane a) or 3.9μM (lanes b-l) and FAM (lanes c-l) at concentrations ranging from 12.7μM to 1.65mM in buffer containing 10 mM Tris (pH 7.6), 50 mM KCl and 5 mM 2-mercaptoethanol. Samples were equilibrated at 20°C for 30 min before electrophoresis on 20% native gels. Main Panel: Fraction of binding activity remaining as a function of [FAM]. The smooth curve is a fit to F/F0 = 1 − (B[FAM]/(1 + B[FAM])), where F0 is the fraction bound in the absence of FAM, F is the fraction bound at a given [FAM] and 1/B is the apparent IC50 for FAM-inhibition of DNA binding.
Competitive-binding models also predict that FAM bound by AGT in the absence of DNA should be released as DNA concentration is increased. As shown in Fig. 1C, the fluorescence anisotropy of an AGT-FAM mixture decreases with increasing [DNA]. A parallel increase in fluorescence intensity was also observed (not shown). These are the changes expected for a net decrease in the mole fraction of FAM that is bound to AGT. Together the [DNA]-dependent release of FAM by AGT and the [FAM]-dependent inhibition of DNA binding are most simply accounted for by models in which FAM and DNA compete for AGT binding. Such competition would be expected if FAM were bound at or near the protein’s DNA binding surface.
3.6 FAM inhibits alkyltransferase activity
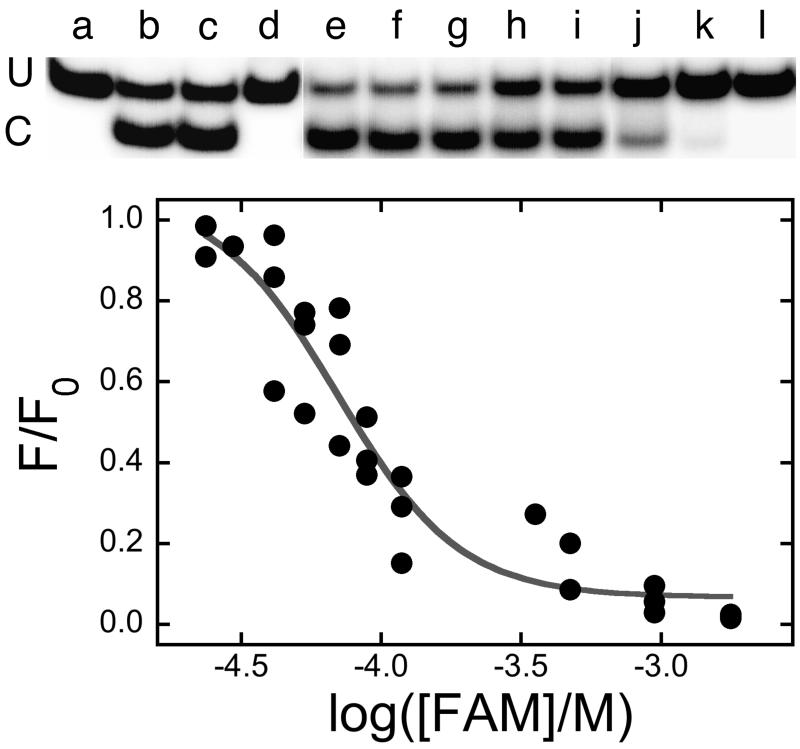
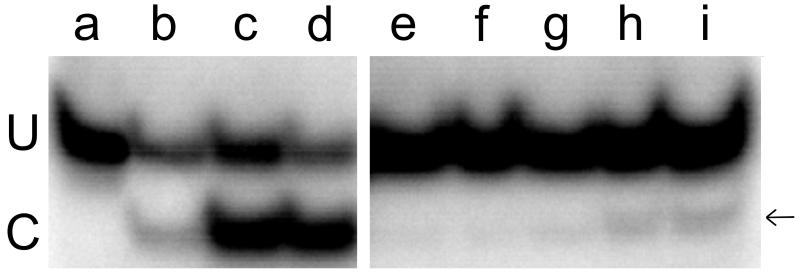
If FAM is bound in the active site cleft, it might inhibit DNA repair by blocking the entry of a DNA base or by interfering with the interaction of the active site nucleophile (C145 in the human enzyme) with the target base. A DNA alkyltransferase assay (Fig. 6) can test this possibility. This assay takes advantage of the observation that NarI endonuclease is inactive against substrates in which the guanine residue at position 2 in its recognition sequence carries an O6-methyl group [23]. Quantitative cleavage was restored if the DNA was first treated for 10 min with a molar excess of AGT (top panel). Reactions carried out for 30 min gave equivalent results indicating that the reactions had reached completion after 10 min (not shown). Inclusion of FAM in parallel reactions caused a concentration-dependent inhibition of methyltransferase activity, with an IC50 = 6.3 ± 1.6 × 10−5 M (bottom panel). This value is only 5-fold larger than the Kd estimated for FAM-binding to AGT (Ka = 7.8 × 104 M−1, Kd = 1.3 × 10−5 M, see above) and is in excellent agreement with the IC50 for FAM-inhibition of DNA binding (7.63 ± 0.61 × 10−5 M). This outcome is consistent with a functional overlap of the sites of DNA repair and FAM binding as predicted by models in which FAM occupies the nucleotide binding pocket.
Figure 6. Inhibition of DNA-alkyltransferase activity by FAM.
The assay is based on the fact that duplex DNAs are not susceptible to NarI cleavage if the residue at position 2 in the NarI sequence is O6-methylguanine, but transfer of the methyl group to AGT restores DNA cleavage susceptibility. Top: electrophoretic resolution of uncut (U) and cut (C) DNAs in a 20% polyacrylamide gel. All samples initially contained duplex DNA at a final concentration of 0.25μM (duplex). Where indicated, samples were treated with AGT (1.1μM) for 10 min; reactions were stopped by addition of SDS (final concentration 0.2% w/v). Following the work-up described in Methods, samples were treated with NarI. Samples contained (a) unmethylated 24mer without further treatment; (b) unmethylated 24mer digested with 5U Nar I for 1h; (c) unmethylated 24mer digested with 5U Nar I for 1h in the presence of a FAM concentration sufficient to completely inhibit AGT activity (1.8 mM); (d) methylated 24mer without treatment; (e) methylated 24mer treated with AGT, then 10U Nar I for 1h. Samples f-l contained methylated 24mer treated with AGT in the presence of 23μM, 41μM, 53μM, 71μM, 88μM, 118μM and 470μM FAM, respectively; after work-up they were incubated with 10U Nar I for 1h. This figure is a composite: lanes a-d and e-l are from separate gels. Bottom: the fraction of DNA repaired as a function of FAM concentration. Repair was measured by Nar I cleavage susceptibility. The data shown is from the experiment shown in the top panel and two similar experiments. The smooth curve is a fit to the equation F/F0 = 1 − (B[FAM]/(1 + B[FAM])), where F0 is the fraction of DNA repaired in the absence of FAM, F is the fraction repaired at a given [FAM] and 1/B is the apparent IC50 for FAM-inhibition of DNA repair.
3.7 Benzylation of active site residue C145 inhibits FAM binding
O6-benzylguanine (Fig 7, structure 1) is a well-characterized inhibitor of AGT that is undergoing clinical trial as an adjuvant to alkylating chemotherapy agents [7-9]. Nucleophilic attack by C145 in the active site results in transfer of the benzyl moiety to the C145 sulfur, permanently inactivating the enzyme [43,44]. We predicted that the bulky C145-benzyl adduct would inhibit FAM binding if the dominant binding site is in the active site cleft. To test this prediction, AGT was modified with O6-benzylguanine under conditions (37°C, 50mM Tris, pH 7.6, 1mM EDTA, 5 mM DTT) that give quantitative modification of C145 and no detectable modification at other residues [45,46]). Following this treatment, the AGT was <1% active in DNA repair but remained fully-competent in DNA binding (Fig. 7). Analysis of the binding returned an apparent stoichiometry of n = 5.5 ± 0.2, in agreement with the limiting value of n = 6 expected for a 24mer DNA [16] and a monomer-equivalent Kd of 8.2 ± 1.1 μM, in good agreement with a value obtained with a duplex 16mer DNA [42].
Figure 7. Benzylation of active site residue C145 inhibits FAM binding.
AGT samples were treated with O6-benzylguanine as described [45,46]. Panel A: AGT repair of a duplex 24-mer containing O6-methylguanine-modified NarI sequence, assayed by NarI cleavage as described for Fig. 6. Samples contained 36nM DNA. Sample a, negative control (the DNA was not treated with AGT); samples b-d, positive controls containing 0.12μM, 0.22μM and 0.73μM unmodified AGT, respectively. Samples e-i contained benzyl-AGT at concentrations of 0.23μM, 0.77μM, 2.2μM, 5.5μM and 12.8μM, respectively. Band designations: U, uncut; C, cut. The arrow denotes the position of the faint band of cut DNA present in the benzyl-AGT-treated samples, evidence of a small residual (<1%) of AGT activity. Panel B: Titration of 32P-labeled 24-mer DNA with benzyl-AGT. Samples a-g contained 8.25 nM duplex DNA and 0μM, 2.95μM, 6.6μM, 9.9μM, 13.2μM, 16.5μM and 21.5μM benzyl-AGT, respectively. Samples were equilibrated at 20 ± 1°C for 30 min in 20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol buffer prior to resolution on a 20% polyacrylamide gel. Panel C: Binding of FAM to unmodified and benzyl-AGT, detected by fluorescence anisotropy. Fractional saturation Y = (A − A0)/(Amax − A0) is graphed as a function of AGT concentration. Symbols: (●) binding to unmodified AGT; (■) binding to benzyl-AGT. The smooth curves are fits to single-site binding isotherms (Eq. 5), returning Kd(apparent) = 1.27 ± 0.17 × 10−5M for unmodified AGT and Kd(apparent) = 8.22 ± 3.18 × 10−5M for benzyl-AGT.
Fluorescence anisotropy measurements were used to detect FAM binding to unmodified and benzyl-AGTs (Fig. 7C). Although full saturation was not reached in either titration, it is clear that binding densities are less for benzyl-AGT than for the unmodified protein at equivalent AGT concentrations. Satisfactory fits of the fractional saturation Y = (A − A0)/(Amax − A0) with a single-site isotherm (Eq. 5) show that the data are consistent with 1:1 stiochiometries and return values of Kd(apparent) = 1.27 ± 0.17 × 10−5M for unmodified AGT and Kd(apparent) = 8.22 ± 3.18 × 10−5M for benzyl-AGT. This difference in binding affinities is consistent with the prediction that the C145-benzyl adduct would inhibit FAM binding and supports the notion that the dominant binding site for FAM is located in or near the active site cleft.
3.8 Docking calculations predict that the active site pocket is the dominant FAM binding site
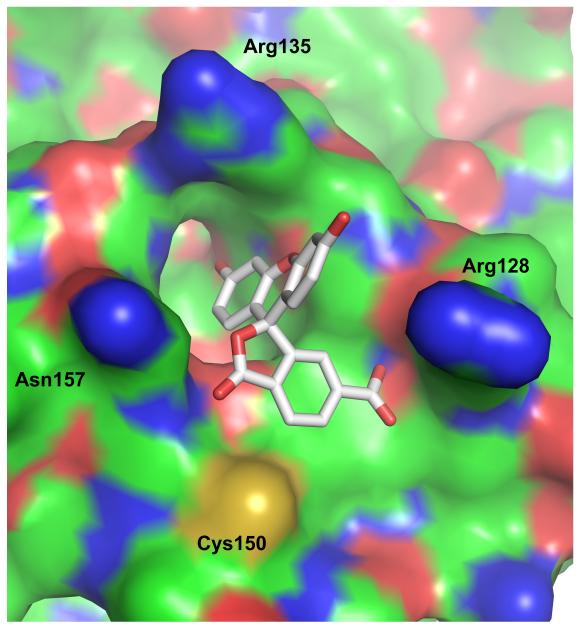
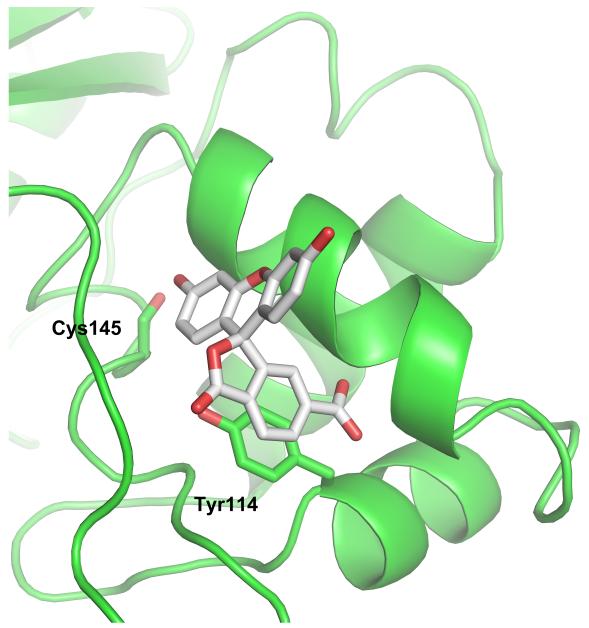
Simulations were carried out using a coordinate set based on the crystal structure of human AGT (PDB 1T38) [4]. In 9 simulation cycles, the 7 top-scoring alignments positioned the FAM molecule in the active site cleft. The two lowest scoring alignments placed the FAM at other sites on the protein surface. In the top-scoring solution obtained for FAM, the 3-ring dihydroxyxanthene moiety penetrates the cleft and the carboxyl-benzofuranone moiety is stacked on Tyr 114 (Fig. 8A, B). This positions the carboxylic acid to make charge- and hydrogen bond-interactions with Arg 128. This model accounts for several experimental observations. The burial of the dihydroxyxanthene group in the largely non-polar cleft should protect it from collisional encounter with fluorescent quenchers such as acrylamide (Fig. 3), while quenching interactions with Tyr 114 and/or Tyr 158 residues in the active site may lower the fluorescence intensity of the bound dye compared to the free species (seen in Figs. 1B and 4B). The location of the dye at the mouth of the active site cleft is consistent with FAM inhibition of DNA binding and repair (Figs 5 and 6) while its penetration into the cleft will bring it into steric clash with a benzylated C145 residue, accounting for the reduction in binding affinity observed with this protein-derivative (Fig. 7C). The ability to account for such a range of disparate evidence argues strongly in favor of this cleft-binding model of FAM interaction with AGT.
Figure 8.
Docking simulations predict that FAM binds the active site cleft. Both views are representation of the highest scoring alignment of FAM with AGT. Residue numbers correspond to the sequence of the human enzyme. Panel A: space-filling view of the DNA-binding surface of AGT showing the entrance to the active site cleft. The FAM structure is shown in stick-format. Most protein surface atoms are colored according to formal charge (positive, blue; uncharged, green; negative, red). The sulfur atom of residue C150 is colored yellow. Panel B: Cartoon representation of the active site cleft showing secondary structure elements, emphasizing proximity of FAM to the C145 and Y114 residues. Simulations were performed with AutoDock Vina [31] and AutoDockTools [32], using AGT coordinates from PDB 1T38 and FAM coordinates generated with the CORINA 3D web server (http://www.molecularnetworks.com/online_demos/corina_demo).
3.9 Other polycyclic aromatic compounds inhibit DNA binding and repair activities
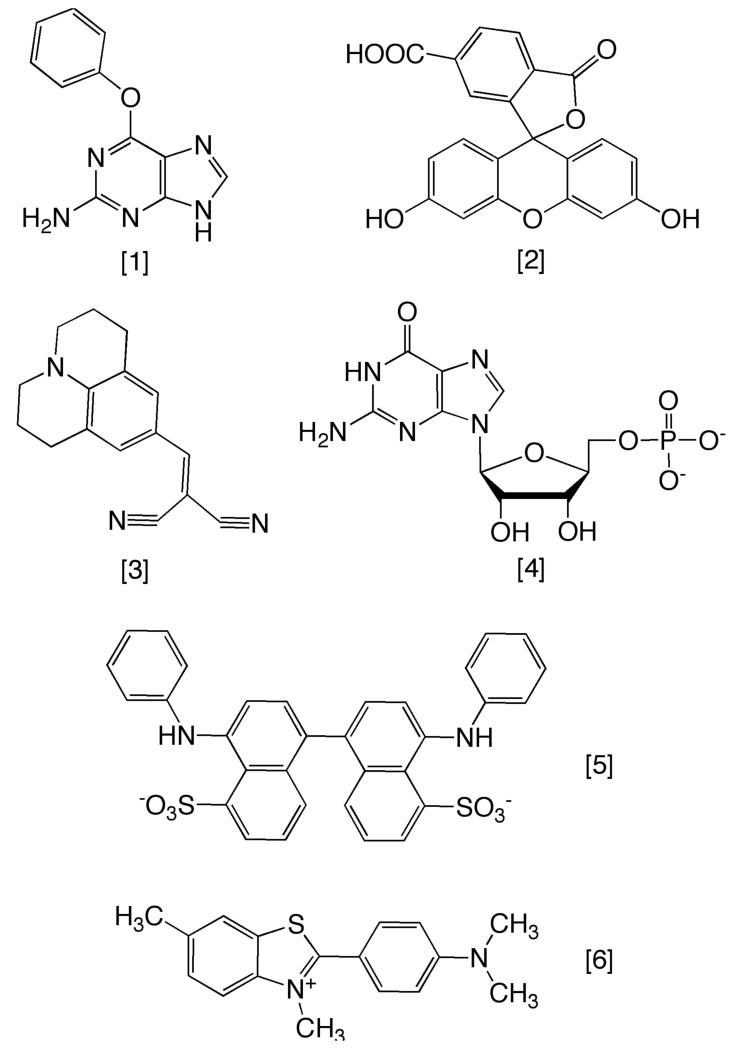
To determine whether FAM is unique in its ability to inhibit AGT’s DNA-binding and -repair activities, we tested a small number of candidate compounds (Fig. 9, Table 4). All have aromatic multi-ring structures, but they differ in size, polarity and charge. A detailed analysis of their effects will be presented elsewhere. However, in aggregate, these results show that the inhibition of DNA binding and repair is not unique to FAM and they suggest that screening of potential ligands for enhanced binding to AGT and inhibition of its activities may be practical.
Figure 9. Compounds tested for inhibition of DNA alkyltransferase activity of AGT.
Panel A. Gallery of compound structures. [1] O6-benzylguanine, [2] 6-carboxyfluorescein (FAM), [3] 9-(2,2 dicyanovinyl)julolidine (DCVJ), [4] guanosine 5′ monophosphate (5′-GMP), [5] bis-ANS, [6] thioflavin T. IC50 values for the inhibition of alkyltransferase activity by compounds 2-6 are given in Table 4.
Table 4.
Dissociation constants for DNA binding and IC50 values for DNA repair for representative compounds
| Compound | Kd, apparent (M)a | IC50 (M)b |
|---|---|---|
| FAM | 1.27 ± 0.04 × 10−5 | 6.31 ± 1.60 × 10−5 |
| Bis-ANS | 3.31 ± 0.12 × 10−5 | 4.86 ± 0.94 × 10−5 |
| DCVJ | NDc | 1.44 ± 0.77 × 10−5 |
| 5′-GMP | NDc | 3.46 ± 0.87 × 10−5 |
| ThT | NDc | 2.17 ± 0.83 × 10−4 |
From fluorescence anisotropy data.
From alkyltransferase assay.
Not determined
4.0 DISCUSSION
As shown above, AGT binds 6-carboxyfluorescein (FAM), when the dye is conjugated to DNA or free in solution. Together, several considerations support the idea that the FAM residue is bound within or near the enzyme’s active site cleft. Docking simulations (Fig. 8) predict that the highest affinity FAM-binding site is in the active site cleft. Changes in fluorescence emission on binding (Fig. 1B), and the inaccessibility of the bound state to a small polar quencher (acrylamide, Fig. 3) indicate that the binding site is a pocket with an environment that differs from the bulk solution in polarity and access of small solutes. The inhibition of FAM binding by benzylation of the active site Cys145 suggests that the binding site is near the active site or is allosterically coupled to it (Fig. 7), as does the binding competition between unliganded FAM and DNA (Fig. 5). AGT is a small enzyme (Mr = 21,519); crystal structures of AGT reveal only one pocket that is large enough to accommodate O6 alkylguanines with bulky substituents, including fluorescein [4,48]. That such bulky groups can be accommodated in the active site is shown by our simulation (Fig. 8) and by the formation of covalent adducts with the active site cysteine (C145 in the human enzyme); this is the basis of the popular “SNAP-tag” strategy for fluorescent labeling of chimeric proteins [49,50]. Finally, while the observations that FAM inhibits DNA-binding and -repair activities do not rule out the possibility that it acts by binding far from the active site, these inhibitory effects are most simply explained by models in which the FAM residue occupies and blocks the active site cleft.
The attachment of FAM to either the 5′ or the 3′ terminal residue of a short DNA molecule results in stoichiometries and equilibrium constants for AGT binding that differ significantly from values determined with unlabeled DNA homologues. This limits the usefulness of FAM-DNA derivatives in experiments to characterize native AGT-DNA interactions. However, these data do not indicate whether FAM binding is unique to AGT or generalizable to related systems. If a cleft capable of binding an extrahelical DNA base is sufficient for binding FAM or other fluorophores, then a large family of mechanistically-related enzymes may be prone to interactions with these residues. Related enzymes include the human and bacterial O6-alkylguanine alkyltransferases (AGTs), the yeast and bacterial alkyltransferase-like proteins (ATLs), human alkyladenine glycosylase (hAAG), 8-oxoguanine DNA glycosylase (hOGG), human and bacterial uracil-DNA glycosylases (UDG or UNG), oxidative DNA/RNA dealkylases such as E. coli AlkB and its human homologue ABH2, and a large number of bacterial host-restriction DNA methyltransferases such as EcoRI methylase [12,13,51-53].
The inhibition of DNA binding by FAM (Fig. 5) calls attention to the potential perturbing effects of dyes routinely used in native electrophoresis assays (EMSA) of protein-nucleic acid interaction [39,54]. Ethidium bromide and pyronin Y, in particular, share structural features with FAM, including planar triplets of fused aromatic rings and similar overall dimensions [55,56]. While we have not yet tested ethidium or related dyes, we anticipate that they will be found to bind AGT like FAM does and if so, they will be found to inhibit DNA binding. Other dyes used in electrophoresis, such as bromphenol blue and xylene cyanol FF, while less similar to FAM in over-all geometry, are charged aromatic compounds that are not-dissimilar in size from nucleotide cofactors or extrahelical nucleic acid bases that might be bound by a protein of interest [57]. While use of these dyes makes EMSA more convenient, they are not essential to the method. Performed as a control, an experiment like the one shown in Fig. 5 will reveal whether a particular dye interferes with binding activities detectable by native gel assay.
Our experiments on free FAM show that the covalent linkage of dye to DNA is not required for AGT binding or for the inhibition of its DNA repair activities. These results raise the possibility that the cellular functions of AGT might be modulated by low molecular weight metabolites that are abundant in the nucleus. Potential candidates include purine and pyrimidine nucleotides and nucleosides (found in ~10−4M concentrations in cell extracts [58]). The binding of O6-alkylguanines (the basis of inhibition by chemotherapeutic enhancers O6-benzyl guanine and O6-(4-Bromothenyl)guanine [59,60]) and our observation that GMP inhibits alkyltransferase activity (Table 4) show that such interactions are possible and might serve to coordinate AGT functions with other metabolic activities of the cell.
Is FAM a prototype drug? The DNA-alkyltransferase activity of AGT protects tumor cells from toxic effects of DNA-alkylating chemotherapeutic drugs [8,44,59]. Pseudosubstrate drugs that alkylate the active site cysteine have been shown to be effective in reducing cellular resistance to DNA-alkylating chemotherapy and two are in clinical trial [7-9]. However, FAM appears to inhibit alkyltransferase activity by a different mechanism. The inhibition is reversible and concentration-dependent (rather than stoichiometric as with reactive pseudosubstrates) and it does not require that the inhibitory group be presented as an O6-adduct of guanine or an O4-adduct of thymine (a minor substrate of human AGT [12,61]). Although FAM is a poor inhibitor of DNA alkyltransferase activity (IC50 ~ 63 μM under our standard conditions), the good solubility and low toxicity of this class of compounds suggests that a related molecule, bound with higher affinity, might provide an alternative or an adjunct to current methods for the therapeutic inhibition of DNA alkyltransferases. Binding tests with FAM and Bis ANS (Figs 5, 6 and Table 4) and alkyltransferase inhibition assays carried out on representative compounds (Table 4) provide a template for screening work that may discover better inhibitors of AGT.
Highlights.
-
>
Binding and repair by O6-alkylguanine DNA alkyltransferase (AGT) were studied.
-
>
Higher stoichiometries were found with FAM-labeled DNAs than with unlabeled DNAs.
-
>
Fluorescence and activity assays suggest FAM occupies the AGT active site.
-
>
Unconjugated FAM inhibits DNA binding and repair.
-
>
FAM-like molecules belong to a new class of AGT-inhibitors.
Table 3.
Hill coefficients and apparent association constants for AGT binding single-stranded DNAs.
| Hill coefficients (n) |
|||
|---|---|---|---|
| DNA (nt) | From intensity measurements |
From anisotropy measurements |
Ka (M−1) From anisotropy measurementsx |
| 3′-FAM-16-mer | 1.74 ± 0.30 | 1.36 ± 0.05 | 2.82 ± 0.08 × 105 |
| 5′-FAM-16-mer | 2.55 ± 0.70 | 1.74 ± 0.04 | 2.95 ± 0.04 × 105 |
| 3′-FAM-26-mer | 3.33 ± 0.27 | 2.13 ± 0.14 | 3.30 ± 0.12 × 105 |
| 5′-FAM-26-mer | 2.92 ± 0.62 | 2.02 ± 0.17 | 3.33 ± 0.15 × 105 |
5.0 ACKNOWLEDGMENTS
We thank the reviewers for their thoughtful suggestions. This work was supported by NIH grants NS-38041 and P20 RR-20171 to D.W.R and GM-070662 to M.G.F.
ABBREVIATIONS
- AGT
O6 alkylguanine-DNA alkyltransferase
- FAM
6-carboxyfluorescein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6.0 REFERENCES
- [1].Pegg AE, Perry W. Alkylation of nucleic acids and metabolism of small doses of dimethylnitrosamine in the rat. Cancer Res. 1981;41:3128–3132. [PubMed] [Google Scholar]
- [2].Maher VM, Domoradzki J, Bhattacharyya NP, Tsujimura T, Corner RC, McCormick JJ. Alkylation damage, DNA repair and mutagenesis in human cells. Mutation Res. 1990;233:235–245. doi: 10.1016/0027-5107(90)90166-2. [DOI] [PubMed] [Google Scholar]
- [3].Daniels DS, Tainer JA. Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O6-alkylguanine-DNA alkyltransferase. Mutation Res. 2000;460:151–163. doi: 10.1016/s0921-8777(00)00024-0. [DOI] [PubMed] [Google Scholar]
- [4].Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- [5].Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23:823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- [6].Liu L, Xu-Welliver M, Kanugula S, Pegg AE. Inactivation and degradation of O6-alkylguanine-DNA alkyltransferase after reaction with nitric oxide. Cancer Res. 2002;62:3037–3043. [PubMed] [Google Scholar]
- [7].Quinn JA, Jiang SX, Carter J, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Bigner DD, Sampson JH, McLendon RE, Herndon JE, 2nd, Threatt S, Friedman HS. Phase II trial of Gliadel plus O6-benzylguanine in adults with recurrent glioblastoma multiforme. Clin Cancer Res. 2009;15:1064–1068. doi: 10.1158/1078-0432.CCR-08-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Watson AJ, Middleton MR, McGown G, Thorncroft M, Ranson M, Hersey P, McArthur G, Davis ID, Thomson D, Beith J, Haydon A, Kefford R, Lorigan P, Mortimer P, Sabharwal A, Hayward O, Margison GP. O6-methylguanine-DNA methyltransferase depletion and DNA damage in patients with melanoma treated with temozolomide alone or with lomeguatrib. Br J Cancer. 2009;100:1250–1256. doi: 10.1038/sj.bjc.6605015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kefford RF, Thomas NP, Corrie PG, Palmer C, Abdi E, Kotasek D, Beith J, Ranson M, Mortimer P, Watson AJ, Margison GP, Middleton MR. A phase I study of extended dosing with lomeguatrib with temozolomide in patients with advanced melanoma. Br J Cancer. 2009;100:1245–1249. doi: 10.1038/sj.bjc.6605016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pegg AE, Xu-Welliver M, Loktionova NA. The DNA repair protein O6-alkylguanine-DNA alkyltransferase as a target for cancer chemotherapy. In: Ehrlich EM, editor. DNA alterations in cancer: genetic and epigenetic changes. Eaton Publishing; Natick, MA: 2000. pp. 471–488. [Google Scholar]
- [11].Margison GP, Santibáñez-Koref MF. O6-Alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. BioEssays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- [12].Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 2007;6:1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roberts RJ, Cheng X. Base Flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- [14].Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol. 2006;301:203–225. doi: 10.1007/3-540-31390-7_7. [DOI] [PubMed] [Google Scholar]
- [15].Rasimas JJ, Kar SR, Pegg AE, Fried MG. Interactions Of Human O6-Alkylguanine-DNA Alkyltransferase (AGT) With Short Single-Stranded DNAs. J. Biol. Chem. 2007;282:3357–3366. doi: 10.1074/jbc.M608876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Melikishvili M, Rasimas JJ, Pegg AE, Fried MG. Interactions of human O6-alkylguanine-DNA alkyltransferase (AGT) with short double-stranded DNAs. Biochemistry. 2008;47:13754–13763. doi: 10.1021/bi801666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pegg AE. Properties of mammalian O6-alkylguanine-DNA alkyltransferases. Mutation Res. 1990;233:165–175. doi: 10.1016/0027-5107(90)90160-6. [DOI] [PubMed] [Google Scholar]
- [18].Luu KX, Kanugula S, Pegg AE, Pauly GT, Moschel RC. Repair of oligodeoxyribonucleotides by O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2002;41:8689–8697. doi: 10.1021/bi025857i. [DOI] [PubMed] [Google Scholar]
- [19].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [20].Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- [21].Maxam A, Gilbert WS. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U.S.A. 1977;74:560–565. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Maniatis T, Efstratiadis A. Fractionation of low molecular weight DNA or RNA in polyacrylamide gels containing 98% formamide or 7M urea. Method Enzymol. 1980;65:299–305. doi: 10.1016/s0076-6879(80)65040-x. [DOI] [PubMed] [Google Scholar]
- [23].Voigt JM, Topal MD. O6-methylguanine in place of guanine causes asymmetric single-strand cleavage of DNA by some restriction enzymes. Biochemistry. 1990;29:1632–1637. doi: 10.1021/bi00458a039. [DOI] [PubMed] [Google Scholar]
- [24].Cohn EJ, Edsall JT. Proteins, Amino Acids and Peptides as Ions and Dipolar Ions. In: Cohn EJ, Edsall JT, editors. Proteins, Amino Acids and Peptides as Ions and Dipolar Ions. Reinhold; New York: 1943. pp. 370–381.pp. 428–431. [Google Scholar]
- [25].Cohen G, Eisenberg H. Deoxyribonucleate solutions: sedimentation in a density gradient, partial specific volumes, density and refractive density increments and preferential interactions. Biopolymers. 1968;6:1077–1100. doi: 10.1002/bip.1968.360060805. [DOI] [PubMed] [Google Scholar]
- [26].Dam J, Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- [27].Schuck P. Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophysical Journal. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-Aided Interpretation of Analytical Sedimentation Data For Proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. The Royal Society of Chemistry; Cambridge, England: 1992. pp. 90–125. [Google Scholar]
- [29].Lakowicz JR. Principles of Fluorescence Spectroscopy. Plenum; N.Y.: 1983. [Google Scholar]
- [30].Eftink MR, Ghiron CA. Fluorescence quenching. Anal. Biochem. 1981;114:199–227. doi: 10.1016/0003-2697(81)90474-7. [DOI] [PubMed] [Google Scholar]
- [31].Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Heyduk T, Ma Y, Tang H, Ebright RH. Fluorescence anisotropy: rapid, quantitative assay for protein-DNA and protein-protein interaction. Methods Enzymol. 1996;274:492–503. doi: 10.1016/s0076-6879(96)74039-9. [DOI] [PubMed] [Google Scholar]
- [34].Melikishvili M, Hellman LM, Fried MG. Use of DNA length varition to detect periodicities in positively cooperative, nonspecific binding. Methods in Enzymology. 2009;466:66–82. doi: 10.1016/S0076-6879(09)66004-3. [DOI] [PubMed] [Google Scholar]
- [35].Goutelle S, Maurin M, Rougier F, Barbaut X, Bourguignon L, Ducher M, Maire P. The Hill equation: a review of its capabilities in pharmacological modelling. Fundamental & Clinical Pharmacology. 2008;22:633–648. doi: 10.1111/j.1472-8206.2008.00633.x. [DOI] [PubMed] [Google Scholar]
- [36].Weiss JN. The Hill equation revisited: uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- [37].Sjöback R, Nygren J, Kubista M. Absorption and fluorescence properties of fluorescein. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 1995;51:L7–L21. [Google Scholar]
- [38].Klonis N, Clayton AHA, Voss J, W E, Sawyer WH. Spectral Properties of Fluorescein in Solvent-Water Mixtures: Applications as a Probe of Hydrogen Bonding Environments in Biological Systems. Photochemistry and Photobiology. 1998;67:500–510. [PubMed] [Google Scholar]
- [39].Fried MG, Crothers DM. Equilibria and Kinetics of Lac Repressor-Operator Interactions by Polyacrylamide Gel Electrophoresis. Nucl. Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Adams CA, Melikishvili M, Rodgers DW, Rasimas JJ, Pegg AE, Fried MG. Topologies of complexes containing O6-alkylguanine-DNA alkyltransferase and DNA. J Mol Biol. 2009;389:248–263. doi: 10.1016/j.jmb.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Eftink MR, Ghiron CA. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- [42].Rasimas JJ, Pegg AE, Fried MG. DNA-binding mechanism of O6-alkylguanine-DNA alkyltransferase. Effects of protein and DNA alkylation on complex stability. J Biol Chem. 2003;278:7973–7980. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- [43].Pegg AE, Boosalis M, Samson L, Moschel RC, Byers TL, Swenn K, Dolan ME. Mechanism of inactivation of human O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine. Biochemistry. 1993;32:11998–12006. doi: 10.1021/bi00096a009. [DOI] [PubMed] [Google Scholar]
- [44].Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- [45].Kanugula S, Goodtzova K, Pegg AE. Probing of conformational changes in human O6-alkylguanine-DNA alkyltransferase protein in its alkylated and DNA bound states by limited proteolysis. Biochem. J. 1998;329:545–550. doi: 10.1042/bj3290545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rasimas JJ, Kanugula S, Dalessio PM, Ropson IJ, Fried MG, Pegg AE. Effects of zinc occupancy on human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2003;42:980–990. doi: 10.1021/bi026970b. [DOI] [PubMed] [Google Scholar]
- [47].Takashi R, Tonomura Y, Morales MF. 4,4′-Bis (1-anilinonaphthalene 8-sulfonate) (bis-ANS): a new probe of the active site of myosin. Proc Natl Acad Sci U S A. 1977;74:2334–2338. doi: 10.1073/pnas.74.6.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Duguid EM, Rice PA, He C. The Structure of the Human AGT Protein Bound to DNA and its Implications for Damage Detection. J. Mol. Biol. 2005;350:657–666. doi: 10.1016/j.jmb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- [49].Gautier A, Juillerat A, Heinis C, Corrêa IR, Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- [50].Hinner MJ, Johnsson K. How to obtain labeled proteins and what to do with them. Curr Opin Biotechnol. 2010;21:766–776. doi: 10.1016/j.copbio.2010.09.011. [DOI] [PubMed] [Google Scholar]
- [51].Neely RK, Tamulaitis G, Chen K, Kubala M, Siksnys V, Jones AC. Time-resolved fluorescence studies of nucleotide flipping by restriction enzymes. Nucleic Acids Res. 2009;37:6859–6870. doi: 10.1093/nar/gkp688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chen B, Liu H, Sun X, Yang CG. Mechanistic insight into the recognition of single-stranded and double-stranded DNA substrates by ABH2 and ABH3. Mol Biosyst. 2010;6:2143–2149. doi: 10.1039/c005148a. [DOI] [PubMed] [Google Scholar]
- [53].Hopkins BB, Reich NO. Simultaneous DNA binding, bending, and base flipping: evidence for a novel M.EcoRI methyltransferase-DNA complex. J Biol Chem. 2004;279:37049–37060. doi: 10.1074/jbc.M404573200. [DOI] [PubMed] [Google Scholar]
- [54].Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Waring MJ. Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol. 1965;13:269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- [56].Wu L, Burgess K. Fluorescent amino- and thiopyronin dyes. Org. Lett. 2008;10:1779–1782. doi: 10.1021/ol800526s. [DOI] [PubMed] [Google Scholar]
- [57].Kreft S, Kreft M. Physicochemical and physiological basis of dichromatic colour. Naturwissenschaften. 2007;94:935–939. doi: 10.1007/s00114-007-0272-9. [DOI] [PubMed] [Google Scholar]
- [58].Ng M, Blaschke TF, Arias AA, Zare RN. Analysis of free intracellular nucleotides using high-performance capillary electrophoresis. Anal. Chem. 1992;64:1682–1684. doi: 10.1021/ac00039a010. [DOI] [PubMed] [Google Scholar]
- [59].Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-Methylguanine-DNA methyltransferase inactivation and chemotherapy. Br. Med. Bull. 2008;85:17–33. doi: 10.1093/bmb/ldm036. [DOI] [PubMed] [Google Scholar]
- [60].Kaina B, Margison GP, Christmann M. Targeting O6 -methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy. Cell Mol Life Sci. 2010;67:3663–3681. doi: 10.1007/s00018-010-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fang Q, Kanugula S, Tubbs JL, T JA, Pegg AE. Repair of O4-alkylthymine by O6-alkylguanine-DNA alkyltransferases. J Biol Chem. 2010;285:8185–8195. doi: 10.1074/jbc.M109.045518. [DOI] [PMC free article] [PubMed] [Google Scholar]