Abstract
A one-step, single tube, real-time accelerated reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed for detecting the envelope gene of West Nile (WN) virus. The RT-LAMP assay is a novel method of gene amplification that amplifies nucleic acid with high specificity, efficiency, and rapidity under isothermal conditions with a set of six specially designed primers that recognize eight distinct sequences of the target. The whole procedure is very simple and rapid, and amplification can be obtained in less than 1 h by incubating all of the reagents in a single tube with reverse transcriptase and Bst DNA polymerase at 63°C. Detection of gene amplification could be accomplished by agarose gel electrophoresis, as well as by real-time monitoring in an inexpensive turbidimeter. When the sensitivity of the RT-LAMP assay was compared to that of conventional RT-PCR, it was found that the RT-LAMP assay demonstrated 10-fold higher sensitivity compared to RT-PCR, with a detection limit of 0.1 PFU of virus. By using real-time monitoring, 104 PFU of virus could be detected in as little as 17 min. The specificity of the RT-LAMP assay was validated by the absence of any cross-reaction with other, closely related, members of the Flavivirus group, followed by restriction digestion and nucleotide sequencing of the amplified product. These results indicate that the RT-LAMP assay is extremely rapid, cost-effective, highly sensitive, and specific and has potential usefulness for rapid, comprehensive WN virus surveillance along with virus isolation and/or serology.
West Nile (WN) virus, an arthropod-borne virus, is a member of the Japanese encephalitis (JE) virus serocomplex of the family Flaviviridae, genus Flavivirus. The virus has a positive-sense single-stranded RNA genome of approximately 11,000 nucleotides encoding three structural (capsid [C], premembrane [prM] or membrane [M], and envelope [E]) proteins and seven nonstructural (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) proteins (19, 21). The virus is maintained in nature through a transmission cycle involving primarily Culex species mosquitoes and birds. Humans and other mammals are incidental hosts. In areas where WN is endemic, in addition to human infections, the virus can also cause mortality among equines, as well as among certain domestic and wild birds (12).
Historically, WN virus has circulated primarily in Africa, the Middle East, Southern Europe, Australia, Russia, India, and Indonesia, causing epidemics from time to time (1, 3, 6). However, the recent outbreak of WN virus in North America is a global public health concern. Surveillance of the activity of the virus is therefore critical in implementing proper mosquito control measures that could prevent transmission to and disease among humans (12, 13). This has necessitated the development of rapid and early detection of virus activity in humans and other hosts for the prediction and prevention of large-scale epidemics.
Routine laboratory diagnosis of WN virus infection is primarily based on serodiagnosis, followed by virus isolation and identification. Serologically, WN virus infection can be inferred by immunoglobulin M (IgM) and IgG capture enzyme-linked immunosorbent assay (ELISA). However, confirmation and typing of the virus are based on the demonstration of a fourfold or greater increase in the virus-specific neutralizing antibody titer by plaque reduction neutralization (PRNT) assay with several flaviviruses (9, 11, 18). Virus isolation in cell culture from both clinical and surveillance samples has generally been unsuccessful owing to the low level of transient viremia associated with the disease process and also requires viable virus in samples. Both virus isolation and PRNT assays are time-consuming and tedious, requiring more than a week for completion. The ability to rapidly detect WN virus is therefore significant, given the nonspecificity of the ELISA and the time required for serological confirmation by PRNT assay.
Recently, several investigators have reported PCR-based detection systems for rapid detection of WN virus infection in clinical specimens that were negative for virus isolation, suggesting that nucleic acid-based assays hold greater promise for detection of WN virus infection. In addition to traditional reverse transcription-PCR (RT-PCR), more rapid and sensitive real-time PCR-based assays, such as TaqMan RT-PCR and nucleic acid sequence-based amplification (NASBA) and branched-DNA methods, have been reported and are currently under extensive evaluation with human and field mosquito samples (14, 15, 25). However, all of these nucleic acid amplification methods have the intrinsic disadvantage of requiring either a high-precision instrument for amplification or an elaborate complicated method for detection of amplified products (4, 5, 7, 8, 15, 16, 17). Owing to the problems associated with the current screening systems, it is widely accepted that test results should be confirmed by more than one type of assay. More technologies are therefore needed to complement those already available.
We describe the development of a loop-mediated isothermal amplification (LAMP) assay for detection of WN virus RNA. The LAMP assay is a novel approach to nucleic acid amplification that amplifies DNA with high specificity, selectivity, and rapidity under isothermal conditions, thereby obviating the need for a thermal cycler. The LAMP assay originally described by Notomi et al. (23) is based on the principle of autocycling strand displacement DNA synthesis. The reaction is performed by a DNA polymerase with high strand displacement activity and a set of two specially designed inner primers and two outer primers (23). LAMP is highly specific for the target sequence because of the recognition of the target sequence by six independent sequences in the initial stage and by four independent sequences during the later stages of the LAMP reaction. The amplification efficiency of the LAMP method is extremely high because there is no time loss for thermal change because of its isothermal reaction. Since the amplification of DNA is directly correlated with the production of magnesium pyrophosphate leading to turbidity, real-time monitoring of the LAMP reaction is possible by real-time measurement of turbidity in an inexpensive photometer (20). Further improvements in the time kinetics and sensitivity of the LAMP reaction by the use of two additional loop primers, termed accelerated LAMP, have been reported (22). Therefore, the LAMP assay has the advantages of high specificity, selectivity, and rapidity over other nucleic acid amplification methods.
A LAMP assay for hepatitis B virus using DNA as a template has been reported (23). The LAMP assay is also useful for RNA template detection upon the use of reverse transcriptase (RTase) together with DNA polymerase. Use of the RT-coupled LAMP (RT-LAMP) assay for detection of prostate-specific antigen mRNA in K562 cells has been reported (23). However, no reports are available on application of the LAMP method for detection of RNA viruses. The present study describes the development of a one-step, single-tube real-time accelerated RT-LAMP assay for rapid detection of the E gene of WN virus. Amplification of the E gene is achieved by incubating the viral RNA with a primer mixture in the presence of RTase and Bst DNA polymerase simultaneously at a constant temperature of 63°C for 1 h. Detection of amplification is accomplished by agarose gel analysis, as well as by real-time monitoring of turbidity in a turbidimeter.
MATERIALS AND METHODS
Cells and virus strains.
The virus strains used in the present study were WN virus strains NY99 (flamingo 382-99) and Eg101, Dengue virus type 2 (DEN-2) strain ThNH7/93, JE virus strain JaOArS982, and St. Louis encephalitis (SLE) virus strain Parton. The viruses were propagated by regular passaging in Aedes albopictus clone C6/36 cells (10) and titrated by plaque assay in Vero cells in accordance with the standard protocol (2).
RNA extraction.
The genomic viral RNA was extracted from 140 μl of infected culture supernatant with known numbers of PFU of virus by using the QIAamp viral RNA mini kit (QIAGEN) in accordance with the manufacturer's protocol. The RNA was eluted from QIAspin columns in a final volume of 100 μl of elution buffer and stored at −70°C until used.
Primer design.
WN virus-specific RT-LAMP primers were designed on the basis of the published sequence of strain NY99 (GenBank accession number AF196835) with the LAMP primer design support software program (Netlaboratory). A set of six primers comprising two outer, two inner, and two loop primers was designed. The two outer primers are known as the forward outer primer (F3) and the backward outer primer (B3), which helps in strand displacement. The inner primers are known as the forward inner primer (FIP) and the backward inner primer (BIP), respectively, and each has two distinct sequences corresponding to the sense and antisense sequences of the target, one for priming in the first stage and the other for self-priming in later stages. FIP contains F1C (complementary to F1), a TTTT spacer, and the F2 sequence. BIP contains the B1C sequence (complementary to B1), a TTTT spacer, and the B2 sequence. FIP and BIP were high-performance liquid chromatography-purified primers. A further two loop primers were designed to accelerate the amplification reaction. The loop F and loop B primers are composed of sequences complementary to the sequences between the F1 and F2 and the B1 and B2 regions, respectively. The sequences of the selected primers were compared to an alignment of the E gene sequences of 14 strains of WN virus from human, mosquito, and crow origins. The details of the oligonucleotide primers used for amplification of the E gene of WN virus are given in Table 1.
TABLE 1.
Details of oligonucleotide primers used for RT-LAMP amplification of E gene of WN virus
| Primer name | Type | Length(s) | Genome positiona | Sequence (5′-3′) |
|---|---|---|---|---|
| F3 | Forward outer | 19-mer | 1028-1046 | TGGATTTGGTTCTCGAAGG |
| B3 | Reverse outer | 19-mer | 1228-1210 | GGTCAGCACGTTTGTCATT |
| FIP | Forward inner (F1C + TTTT + F2) | 46-mer; F1C, 22-mer; F2, 20-mer | F1C, 1121-1100; F2, 1050-1069 | TTGGCCGCCTCCATATTCATCATTTTCAGCTGCGTGACTATCATGT |
| BIP | Reverse inner (B1C + TTTT + B2) | 45-mer (B1C, 22-mer; B2, 19-mer) | B1C, 1144-1165; B2, 1208-1190 | TGCTATTTGGCTACCGTCAGCGTTTTTGAGCTTCTCCCATGGTCG |
| Loop F | Forward loop | 19-mer | 1093-1075 | CATCGATGGTAGGCTTGTC |
| Loop B | Reverse loop | 18-mer | 1169-1186 | TCTCCACCAAAGCTGCGT |
Genome position according to the WN virus strain NY99 (flamingo 382-99) complete genome sequence (GenBank accession number AF196835).
RT-PCR.
For comparison with the sensitivity of the RT-LAMP method, RT-PCR was performed by using the two outer primers, F3 and B3, used for the LAMP amplification as forward and reverse primers, respectively, in accordance with the standard protocol (24). Following cDNA synthesis with reverse primer B3 at 42°C for 30 min, PCR amplification was carried out with the TaKaRa LA Taq PCR kit (TAKARA BIO Inc.) by using 5 μl of cDNA and 50 pmol of each primer in a 50-μl total reaction volume by following the manufacturer's protocol with the following cycling times and temperatures: 94°C for 2 min and 30 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min. After the RT-PCR was performed, a 10-μl portion was analyzed by agarose gel electrophoresis on a 3% NuSieve 3:1 agarose gel (BMA, Rockland, Maine) and the DNA was visualized by ethidium bromide staining.
RT-LAMP.
The RT-LAMP reaction was carried out in a 25-μl total reaction mixture volume with a Loopamp DNA amplification kit (Eiken Chemical Co. Ltd.) containing 50 pmol each of inner primers FIP and BIP, 5 pmol each of outer primers F3 and B3, 25 pmol each of loop primers loop F and loop B, 1,400 μM each deoxynucleoside triphosphate, 0.6 M betaine (Sigma Chemical Co., St. Louis, Mo.), 40 mM Tris-HCl (pH 8.8), 20 mM KCl, 20 mM (NH4)2SO4, 8 mM MgSO4, 0.1% Triton X-100, 0.125 U of avian myeloblastosis virus (AMV) RTase (Invitrogen), 8 U of Bst DNA polymerase (large fragment; New England Biolabs), and the specified amounts of target RNA. The mixture was incubated at 63°C for 60 min in a heating block and then heated at 80°C for 2 min to terminate the reaction. For real-time monitoring of the RT-LAMP reaction, the reaction mixture was incubated at 63°C for 60 min in a Loopamp real-time turbidimeter (LA-200; Teramecs).
Analysis of RT-LAMP product.
Following amplification by the RT-LAMP method, 10 μl of the amplified product was analyzed by agarose gel electrophoresis with a 3% NuSieve 3:1 agarose gel (BMA) in Tris acetate-EDTA buffer (0.04 M Tris acetate, 1 mM EDTA), stained with ethidium bromide, and visualized on a UV transilluminator at 302 nm. Real-time monitoring of RT-LAMP amplification was done through spectrophotometric analysis by recording the optical density at 400 nm with a Loopamp real-time turbidimeter (LA-200; Teramecs) every 6 s. Positive real-time RT-LAMP assay results were determined by taking into account the time to positivity (Tp; in minutes), at which the turbidity increased above the threshold value fixed at 0.1, which is two times the average turbidity value of the negative controls of several replicates. None of the positive samples tested at multiple times showed positivity in terms of increased turbidity after 60 min. Therefore, a sample having Tp values of ≤60 min and turbidity above the threshold value of ≥0.1 was considered positive. The specificity of the RT-LAMP-amplified product was analyzed by restriction digestion with the AluI enzyme, as well as by nucleotide sequencing of both digested and undigested products with two outer and two inner primers.
Evaluation of RT-LAMP.
The feasibility of using the RT-LAMP assay for detection of WN virus-specific RNA in clinical specimens was evaluated with apparently healthy human serum samples spiked with known numbers of PFU of WN virus strain NY99 in the absence of real human or field-collected mosquito specimens. Prior to spiking, all of the healthy human volunteer serum samples were screened by IgM and IgG capture ELISA, as well as by RT-PCR assay for the presence of anti-flavivirus antibodies and WN virus RNA, respectively. A panel of 10 negative serum samples was selected for further evaluation by RT-LAMP assay. Of 10 serum samples, 4 were spiked with known numbers of PFU of virus ranging from 102 to 0.1 PFU in duplicate. Following spiking, all of the 14 (8 spiked and 6 negative) serum samples were processed for RNA extraction with a QIAamp viral RNA mini kit (QIAGEN) and screened by RT-LAMP and RT-PCR simultaneously for detection of viral RNA as described above.
RESULTS
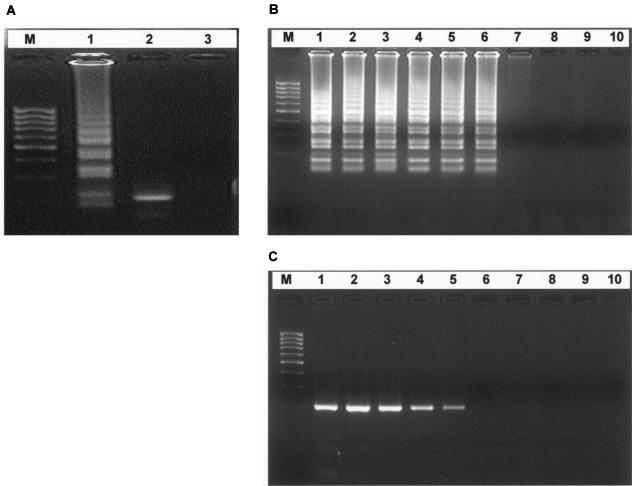
The initial standardization and optimization of the RT-LAMP assay were carried out by using a set of four primers comprising two outer and two inner primers as described in Materials and Methods. Following optimization of RT-LAMP, an accelerated RT-LAMP assay was performed by using two additional loop primers to enhance the amplification reaction so as to reduce the time of detection. Detection of gene amplification is accomplished by agarose gel analysis, as well as by real-time monitoring of turbidity. The RT-LAMP assay could amplify the 201-bp target sequence of the E gene of WN virus at 63°C in 60 min, as observed by agarose gel electrophoresis. Amplification was observed as a ladder-like pattern on the gel due to the formation of a mixture of stem-loop DNAs with various stem lengths and cauliflower-like structures with multiple loops formed by annealing between alternately inverted repeats of the target sequence in the same strand (Fig 1A, lane 1).
FIG. 1.
(A) Agarose gel electrophoresis and restriction analysis of the RT-LAMP product of the E gene of WN virus. Lanes: M, 100-bp DNA ladder (Sigma Genosys); 1, RT-LAMP with WN virus strain NY99; 2, RT-LAMP products digested with AluI (175 bp); 3, RT-LAMP without target RNA. (B and C) Comparative sensitivities of RT-LAMP and RT-PCR for detection of WN virus strain NY99 RNA. The amplification by RT LAMP (B) shows a ladder-like pattern, whereas the RT-PCR (C) shows a 201-bp amplification product. Lanes: M, 100-bp DNA ladder (Sigma Genosys); 1, 10,000 PFU; 2, 1,000 PFU; 3, 100 PFU; 4, 10 PFU; 5, 1 PFU; 6, 0.1 PFU; 7, 0.01 PFU; 8, 0.001 PFU; 9, 0.0001 PFU; 10, 0 PFU (negative control without target RNA).
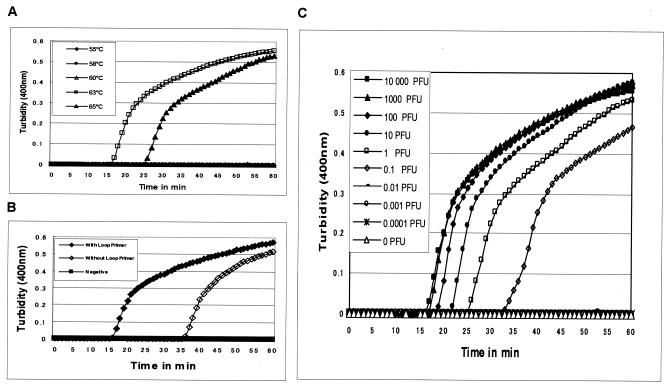
Amplification of RNA by RT-LAMP assay requires both RTase and Bst DNA polymerase since no amplification was observed when one of these two enzymes was omitted from the reaction mixture. The effect of two different types of RTases, such as AMV RTase and Moloney murine leukemia virus RTase, on the kinetics of the RT-LAMP reaction was also studied with a fixed amount of target RNA. AMV RTase was found to be more rapid in generating the amplification signal in the form of turbidity, as well as improved sensitivity, compared to Moloney murine leukemia virus RTase (data not shown). The optimum temperature required for efficient amplification by the RT-LAMP assay was also studied by incubating the reaction mixture at temperatures ranging from 55 to 65°C. The results indicate that the optimum temperature for the RT-LAMP reaction is 63°C, which is optimum for the activity of Bst DNA polymerase (Fig 2A).
FIG. 2.
(A) Effect of temperature on the time kinetics of the RT-LAMP reaction of WN virus strain NY99 as monitored by measurement of turbidity in a Loopamp real-time turbidimeter (LA-200; Teramecs). (B) Kinetics of RT-LAMP amplification of WN virus strain NY99 RNA with and without the loop primers as monitored by real-time measurement of turbidity in a Loopamp real-time turbidimeter (LA-200; Teramecs). (C) Sensitivity of the RT-LAMP assay for detection of WN virus RNA as monitored by real-time measurement of turbidity (LA-200, Teramecs). Serial 10-fold dilutions of virus strain NY99 ranging from 10,000 plaque-forming units to 0.0001 plaque-forming unit were tested.
The real-time kinetics of the RT-LAMP reaction with or without loop primers was studied at 63°C by monitoring turbidity as described in Materials and Methods. The results indicate that the time required for initiation of amplification was 17 or 35 min with or without loop primers, respectively (Fig. 2B). This finding suggests that the use of loop primers accelerated the amplification by reducing the time of detection to half compared to RT-LAMP without loop primers. It was also observed that there is continuous amplification of the target sequence in a stepwise gradient manner, as observed by increased turbidity compared to that of the negative control having no template, where the turbidity gets fixed at around 0.01 below the threshold value (Fig. 2B).
Following standardization, optimization of the RT-LAMP assay was also carried out with regard to the effect of primer and template concentrations on reaction kinetics and sensitivity. The optimal ratio of primer (inner-outer-loop) concentrations for RT-LAMP reaction was found to be 10:1:5 with 50, 5, and 25 pmol, respectively. The effect of higher concentrations of primers and RNA template on the RT-LAMP reaction kinetics was also studied by real-time monitoring. However, no significant improvements could be observed with regard to reaction kinetics when either primer or template concentrations were increased.
Sensitivity and specificity of RT-LAMP
The sensitivity of the accelerated RT-LAMP assay for detection of WN virus RNA was determined by testing serial 10-fold dilutions of virus that had previously been quantified by plaque assay and compared with that of conventional RT-PCR. The accelerated RT-LAMP assay was able to amplify the 104 PFU of virus in 17 min, having a detection limit of 0.1 PFU of virus (Fig. 2C). The comparative sensitivities of the RT-LAMP and RT-PCR assays revealed that the RT-LAMP assay was 10-fold more sensitive than the RT-PCR assay, which has a detection limit of 1 PFU of virus, as indicated by the presence of a 201-bp amplicon (Fig. 1B and C). The sensitivity of the RT-LAMP assay was found to be same for both the NY99 and Eg101 strains of WN virus tested in this study.
The specificity of the RT-LAMP primers for the E gene of WN virus, as shown in Table 1, was established by checking the reactivity with another strain of the WN virus (Eg101), as well as the cross-reactivity with other serologically related members of the flavivirus group, such as DEN-2 (ThNH7/93 strain), JE virus (JaOArS982 strain), and SLE virus (Parton strain). The WN virus-specific RT-LAMP primers demonstrated a high degree of specificity for WN virus by amplifying WN virus strains NY99 and Eg101 but yielding negative results with all of the other viruses tested. (Table 2). The specificity of the amplification was confirmed by restriction endonuclease digestion with the AluI enzyme, which resulted in a 175-bp product, which is in good agreement with the predicted size (Fig. 1A, lane 2). Further confirmation of the structures of the amplified products was also carried out by nucleotide sequencing with inner primers wherein the sequences obtained perfectly matched the expected nucleotide sequences (data not shown).
TABLE 2.
Comparative sensitivities and specificities of the RT-LAMP and RT-PCR assay systems for detection of WN virusa
| Virus | Quantity (PFU) | RT-PCRb | RT-LAMP
|
||
|---|---|---|---|---|---|
| Agarose gel analysis | Real-time monitoring of turbidity
|
||||
| Tp (min) | Interpretationb | ||||
| WN virus strain NY99 | 10,000 | Pos | Pos | 17 | Pos |
| 1,000 | Pos | Pos | 18 | Pos | |
| 100 | Pos | Pos | 19 | Pos | |
| 10 | Pos | Pos | 22 | Pos | |
| 1 | Pos | Pos | 25 | Pos | |
| 0.1 | Neg | Pos | 33 | Pos | |
| 0.01 | Neg | Neg | NA | Neg | |
| 0.001 | Neg | Neg | NA | Neg | |
| 0.0001 | Neg | Neg | NA | Neg | |
| 0 | Neg | Neg | NA | Neg | |
| WN virus strain Eg101 | 1 | Pos | Pos | 28 | Pos |
| 0.1 | Neg | Pos | 37 | Pos | |
| 0.01 | Neg | Neg | NA | Neg | |
| JE virus | 100,000 | Neg | Neg | NA | Neg |
| DEN-2 | 100,000 | Neg | Neg | NA | Neg |
| SLE virus | ND | Neg | Neg | NA | Neg |
Pos, positive; Neg, negative; NA, turbidity did not appear; ND, not determined.
Positive result by RT-PCR revealed 201-bp amplicon on 3% agarose gel analysis.
Tp of ≤60 min is interpreted as positive by the real-time RT-LAMP method.
The comparative evaluation of RT-LAMP vis-à-vis traditional RT-PCR with a limited number of spiked serum samples revealed a very good correlation in detecting viral RNA. A 100% concordance between the two test systems with regard to sensitivity and specificity was observed. The sensitivity of both the test systems with spiked human serum sample was found to be 1 PFU of virus. All of the negative serum samples were negative by both the tests.
DISCUSSION
The recent outbreaks of WN virus in the northeastern part of the American continent and other regions of the world have made it essential to develop an efficient protocol for surveillance of WN virus. The best approach to minimize the risk to humans should involve continuous monitoring of virus activity among migratory birds, mosquitoes, and equines to track down the course of infection for pre-warning of large-scale epidemics. The accumulation of data on viral infection among various species of mosquitoes and birds, as well as other vertebrates, such as bats and horses, in an expanding geographic area during the transmission season provides valuable information for the prevention and prediction of future outbreaks. This is critical for implementing proper mosquito control measures that could prevent virus transmission to and disease among humans.
The difficulty in isolating the virus from clinical specimens has also necessitated the development of rapid and reliable virus detection assay systems. Nucleic acid-based techniques, especially RT-PCR, have the advantage of sensitivity, specificity, and rapidity. Despite their simplicity and the obtainable magnitude of amplification, the requirement of a high-precision thermal cycler prevents this powerful method from being widely used in places such as private clinics as a routine diagnostic tool. In addition to RT-PCR, other sensitive and real-time nucleic acid-based assays, including the TaqMan RT-PCR, NASBA, and branched-DNA amplification methods, are also available for the detection of WN virus RNA (14, 15, 25). These methods can amplify target nucleic acids to similar magnitudes, all with a detection limit of less than 10 copies and within an hour or so, but still have shortcomings to overcome. All of these assays require either a precision instrument for amplification or an elaborate method for detection of the amplified products because of poor specificity of target sequence selection.
This report describes the development of one-step, single-tube, real-time, accelerated RT-LAMP assay for rapid detection of the E gene of WN virus. Just as in a one-step RT-PCR, cDNA synthesis and follow-up amplification can be achieved in a single tube by incubating the viral RNA with a primer mixture in the presence of RTase and Bst DNA polymerase simultaneously at a constant temperature of 63°C for 60 min in a heating block. Amplification by the RT-LAMP assay is based on the principle of autocycling strand displacement DNA synthesis. In the initial steps of the RT-LAMP reaction, all four primers are used, but later, during the cycling reaction, only the inner primers are used for strand displacement DNA synthesis. The RT-LAMP assay used one of two formats for detection of virus-specific amplification, either postamplification detection by agarose gel analysis or real-time monitoring of turbidity in a turbidimeter. The cost of the turbidimeter is $5,000, which is very low compared to that of the ABI Prism 7700 sequence detection system instrument (PE Applied Biosystems) required for the real-time TaqMan RT-PCR and NASBA assays, which is $100,000.
The accelerated RT-LAMP assay for WN virus reported here showed exceptionally higher sensitivity, specificity, and rapidity in comparison with the traditional RT-PCR assay. The RT-LAMP assay was found to be 10-fold more sensitive than the traditional RT-PCR assay, detecting 0.1 PFU of WN virus. The RT-LAMP assay also demonstrated a high degree of specificity for WN virus, having no false-positive results with any of the serologically related flaviviruses tested, indicating that it is highly specific for the target sequence (Table 2). This is attributed to the choice of primers, which were designed after analysis of the sequence alignment of 14 different strains of WN virus from diverse geographical locations like Uganda and Egypt and including recent U.S. isolates of from humans, crows, mosquitoes, and horses. The sequence alignment showed 95 to 100% homology among the different strains. All of the isolates of U.S. origin showed almost 98 to 100% homology. Only a few mismatches of two or three bases could be observed in Ugandan and Eg101 strains in some of the primer sets, but these mismatches did not show any significant difference in terms of detection of viral RNA by the LAMP assay, as reflected by the similar sensitivities obtained with both the NY99 and Eg101 strains of WN virus included in this study.
The accelerated RT-LAMP method using two additional loop primers cuts the signal generation time in half and thus requires only 17 min to detect the amplification that can be either observed visually or monitored in real time with a turbidimeter. The loop primers hybridize to the stem-loops, except for the loops that are hybridized by inner primers and thus prime strand displacement DNA synthesis (22). In addition to its higher amplification efficiency, the RT-LAMP reaction yields a large amount of the by-product pyrophosphate ion, leading to the accumulation of a white magnesium pyrophosphate precipitate in the reaction mixture. Since the increase in the turbidity of the reaction mixture as a result of this precipitate production correlates with the amount of DNA synthesized, real-time monitoring of the RT-LAMP reaction can be achieved by real-time measurement of turbidity (20).
As discussed above, execution of the RT-LAMP reaction and turbidity measurement is extremely simple compared to that of the existing real-time TaqMan RT-PCR and NASBA assays, which requires fluorogenic primers and probes, as well as expensive detection equipment, as described earlier. Since turbidity can be confirmed visually, the only device required for the LAMP reaction is a laboratory water bath or heating block to provide a constant temperature of 63°C. Particularly important is the fact that substantially less time is required for confirmation of the results of the RT-LAMP assay, i.e., less than 1 h (as little as 17 min), compared to 3 to 4 h in the case of RT-PCR. The suitability of the RT-LAMP assay for detection of viral RNA in clinical specimens was validated with a limited number of spiked human serum samples in the absence of real-world human and field-collected mosquito specimens. However, further evaluation with a more defined panel of serum, cerebrospinal fluid, mosquito pools, and avian tissue samples is warranted to establish the RT-LAMP assay as a tool for surveillance of WN virus. Taken together, the data reported here indicate that the RT-LAMP assay is extremely rapid, cost-effective, highly sensitive, and specific and could be used along with virus isolation and/or serology as a comprehensive WN virus detection system in the diagnostic laboratory.
Acknowledgments
The financial support for this study from the Ministry of Education, Culture, Sports, Science and Technology, in the form of a Monbusho scholarship [Grant-in-Aid for Scientific Research (A) 14206036], is thankfully acknowledged.
We are thankful to Basudev Pandey, Bipolo Sophie, and Afjal Hossain Khan for technical assistance and support during this study. We are also thankful to D. J. Gubler and Robert Lanciotti (CDC, Fort Collins, Colo.) for the kind supply of the WN virus NY-99 strain and to Tsugunori Notomi (Eiken Chemical Co., Ltd.) for his technical suggestions.
REFERENCES
- 1.Anderson, J. F., T. G. Andreadis, C. R. Vossbrinck, S. Tirrell, E. M. Wakem, R. A. French, A. E. Garmendia, and H. J. Van Kruiningen. 1999. Isolation of West Nile virus from mosquitoes, crows, and a Cooper's hawk in Connecticut. Science 286:2331-2333. [DOI] [PubMed] [Google Scholar]
- 2.Beaty, B. J., C. H. Calisher, and R. S. Shope. 1989. Arboviruses, p. 797-856. In N. J. Schmidt and R. W. Emmons (ed.), Diagnostic procedures for viral, rickettsial and chlamydial infections. American Public Health Association, Washington, D.C.
- 3.Centers for Disease Control and Prevention. 2000. Update: West Nile virus activity, eastern United States, 2000. Morb. Mortal. Wkly. Rep. 49:1044-1047. [PubMed] [Google Scholar]
- 4.Chan, A. B., and J. D. Fox. 1999. NASBA and other transcription-based amplification methods for research and diagnostic microbiology. Rev. Med. Microbiol. 10:185-196. [Google Scholar]
- 5.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. [DOI] [PubMed] [Google Scholar]
- 6.Hayes, C. G. 1989. West Nile fever, p. 59-88. In T. P. Monath (ed.), The arboviruses: epidemiology and ecology, vol. V. CRC Press, Inc., Boca Raton, Fla.
- 7.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reaction. Bio/Technology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 9.Hunt, A. R., R. A. Hall, A. J. Kerst, R. S. Nasci, H. M. Savage, N. A. Panella, K. L. Gottfried, K. L. Burkhalter, and J. T. Roehrig. 2002. Detection of West Nile virus antigen in mosquitoes and avian tissues by a monoclonal antibody-based capture enzyme immunoassay. J. Clin. Microbiol. 40:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igarashi, A. 1978. Isolation of a Singh's Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 40:531-544. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, A. J., D. A. Martin, N. Karabatsos, and J. T. Roehrig. 2000. Detection of antiarboviral immunoglobulin G by using a monoclonal antibody-based capture enzyme-linked immunosorbent assay. J. Clin. Microbiol. 38:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komar, N. 2000. West Nile viral encephalitis. Rev. Sci. Tech. Off. Int. Epizoot. 19:166-176. [DOI] [PubMed] [Google Scholar]
- 13.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern U.S. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 14.Lanciotti, R. S., A. J. Kerst, R. S. Nasci, M. S. Godsey, C. J. Mitchell, H. M. Savage, N. Komar, N. A. Panella, B. C. Allen, K. E. Volpe, B. S. Davis, and J. T. Roehrig. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 38:4066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanciotti, R. S., and A. J. Kerst. 2001. Nucleic acid sequence based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J. Clin. Microbiol. 39:4506-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leone, G., H. van Schijndel, B. van Gemen, F. R. Kramer, and C. D. Schoen. 1998. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 26:2150-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, D. A., D. A. Muth, T. Brown, A. J. Johnson, N. Karabatsos, and J. T. Roehrig. 2000. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J. Clin. Microbiol. 38:1823-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monath, T. P., and F. X. Heinz. 1996. Flaviviruses, p. 978-984. In B. N. Fields (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa.
- 20.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 21.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers. 1995. Virus taxonomy, classification and nomenclature of viruses. Arch. Virol. 10(Suppl.):1-586. [Google Scholar]
- 22.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 23.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28(12):i-vii. [DOI] [PMC free article] [PubMed]
- 24.Saiki, R. K. 1989. The design and optimization of the PCR, p. 7-16. In H. A. Erlich (ed.), PCR technology. Stockton Press, New York, N.Y.
- 25.Shi, P., E. B. Kauffman, P. Ren, A. Felton, J. H. Tai, A. P. Dupuis, S. A. Jones, K. A. Ngo, D. C. Nicholas, J. Maffei, G. D. Ebel, K. A. Bernard, and L. D. Kramer. 2001. High-throughput detection of West Nile virus RNA. J. Clin. Microbiol. 39:1264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]