Abstract
Sirtuin 2 (SIRT2), a NAD-dependent deacetylase expressed by oligodendrocytes (OLs), the myelin-producing cells of the central nervous system (CNS), is markedly up-regulated during active myelination (Li et al. 2007; Southwood et al. 2007; Werner et al. 2007). SIRT2 is a component of the myelin proteome and is severely reduced in the Plp1 knockout mouse brain, in which both PLP and DM20 are absent (Werner et al. 2007). The mechanisms that regulate SIRT2 expression in OLs and myelin remain to be investigated. We report for the first time that the expression of SIRT2 is regulated by the QKI-dependent pathway and this effect is mediated through selective regulation of PLP. In the homozygous quakingviable (qkv/qkv) mutant mouse that harbors QKI deficiency in OLs (Bockbrader and Feng 2008; Ebersole et al. 1996; Hardy et al. 1996), PLP, but not DM20 mRNA, was selectively down-regulated and SIRT2 protein was severely reduced while SIRT2 mRNA expression was unaffected. Expression of the cytoplasmic isoform QKI6 in OLs (Zhao et al. 2006) rescued SIRT2 expression in the qkv/qkv mutant concomitantly with restoration of PLP expression. Moreover, SIRT2 protein is diminished in myelin tracts and compact myelin of the PLP-ISEdel mutant brain, in which PLP protein but not DM20 is selectively reduced (Wang et al. 2008). In contrast, SIRT2 expression and its cellular function in regulating process complexity are not affected by the absence of PLP in PLP-ISEdel non-myelinating oligodendrocytes. Collectively, our results indicate that the abundance of SIRT2 in myelin is dependent on PLP, but not DM20.
Keywords: oligodendrocytes, Sirtuin2, PLP, myelin, development, quaking
INTRODUCTION
Sirtuins (SIRT1-7) are NAD-dependent deacetylases which regulate a wide range of cellular functions by controlling acetylation of target proteins (Michan and Sinclair 2007). Sirtuin 2 (SIRT2) is expressed predominantly by oligodendrocytes (OLs), the myelin-producing cells of the central nervous system (CNS), is markedly up-regulated during active myelination and has emerged as a major component of the myelin proteome (Li et al. 2007; Southwood et al. 2007; Werner et al. 2007). Functionally, SIRT2 controls process arborization of differentiating OLs in vitro by regulating α-tubulin acetylation (Li et al. 2007). In addition, the alternatively spliced SIRT2 variant 2 (v2) is a component of the myelin proteome and is localized in the paranodal and compact myelin in proximity of the proteolipid protein (PLP) and the alternatively spliced isoform DM20 encoded by the Plp1 gene (Li et al. 2007; Southwood et al. 2007; Werner et al. 2007). SIRT2v2 protein, but not its message, is severely reduced in the Plp1 knockout mouse brain, in which both PLP and DM20 are absent. Furthermore, SIRT2v2 protein is nearly absent in the myelin proteome, suggesting that PLP/DM20 regulate SIRT2 transport into myelin in which SIRT2 is stabilized (Werner et al. 2007).
These discoveries have left open the question of the mechanisms that regulate SIRT2 expression in developing OLs. This is an important question, since process arborization, which is regulated by SIRT2 in OLs in vitro, is critical for proper myelination of axons in vivo. Furthermore, it is unknown whether SIRT2 transport and abundance in the myelin compartment is regulated by PLP only or both PLP and DM20. PLP serves functions that are not shared by DM20, especially in myelin-axon integrity (Edgar et al. 2004; Garbern et al. 2001; Gudz et al. 2002; Klugmann et al. 1997; Stecca et al. 2000). Importantly, SIRT2 is positioned at the paranodal myelin in close proximity of axons, where it may play a role in axo-glial interactions (Li et al. 2007; Southwood et al. 2007). In this study, we sought to investigate the regulatory pathways that control OL expression of SIRT2 during myelinogenesis and to determine whether PLP only or both PLP and DM20 regulate SIRT2 expression in OLs and SIRT2 abundance in myelin.
To investigate regulatory pathways that control SIRT2 expression, we have examined the role of the quaking proteins (QKI), RNA binding factors that play a critical role in myelinogenesis through post-transcriptional regulation of myelin specific genes (Li et al. 2000). Three QKI isoforms, QKI5, QKI6 and QKI7, are expressed in developing OLs, among which QKI5 is nuclear whereas QKI6 and QKI7 are predominantly cytoplasmic and are up-regulated as OLs differentiate (Hardy et al. 1996). In the homozygous quakingviable (qkv/qkv, abbreviated as q/q) mutant mouse that harbors QKI deficiency in OLs, myelinogenesis is severely impaired (Bockbrader and Feng 2008; Ebersole et al. 1996; Hardy et al. 1996) and re-expression of the cytoplasmic QKI6 isoform is sufficient to rescue most of the QKI targets and correct the hypomyelinating phenotype of the quakingviable mouse (Zhao et al. 2006).
To investigate the role of PLP in controlling SIRT2 expression in OLs and SIRT2 transport to myelin, we examined the PLP-ISEdel mouse, in which PLP is selectively and severely reduced, due to the absence of a critical splicing enhancer (ISE) of PLP splicing (Wang et al. 2008). Since DM20 splicing is not affected, this mouse represents an ideal model in which to experimentally assess the role of PLP in SIRT2 regulation in OLs and myelin. Importantly, this mouse develops age-dependent neurological deficits accompanied by myelin abnormalities (Wang et al. 2008).
Here, we present a comprehensive analysis of SIRT2 expression at the protein and RNA levels in the quakingviable and PLP-ISEdel mouse brains with in vivo and in vitro studies. We show that SIRT2 is regulated by a novel posttranscriptional pathway under the RNA binding protein, QKI6 through its preferential target mRNA that encodes PLP. We also show that PLP, but not DM20 selectively regulates SIRT2 transport and abundance in the myelin compartment, while it has no effect on SIRT2 expression in non myelinating OLs.
MATERIALS AND METHODS
Preparation of cell suspensions and FACS analysis
EGFP+ cells were isolated by Fluorescent Activated Cell Sorting (FACS) from post-natal day 1~3, 8~10, and 19~21 brain hemispheres dissected from CNP-EGFP mice, as previously described (Yuan et al. 2002). Briefly, cell suspensions were prepared from finely minced brain tissues by enzymatic digestion with papain in HBSS (15U/ml) at 37°C for 15′ followed by sequential passages through gauge 18, 21 and 23 needles. Dissociated cell suspensions were sorted on a MoFlo (Beckman Coulter) using a 100 micro nozzle tip at 30 psi. The FACS sorting scatter profile was Forward Scatter for relative size versus Side Scatter for granularity. Apoptotic cells appeared as smaller and more granular and were excluded from further sorting process. The GFP+ cells were sorted based on fluorescence intensity, the yield usually ranges from 1% to 3%. FACS sorted cell pellets were stored at −80°C.
Primary mouse oligodendrocyte progenitor cell culture
Mixed glial cultures and immunoselected OPC were prepared from PLP-ISEdel and C57BL/6 brains at post-natal day 1, as previously described (Huang et al. 2002) with some modifications. To enrich for OPC, mixed glial cultures were grown in chemically defined medium containing 10 ng/ml biotin, 5 ug/ml insulin, 2 mM glutamine, 100 units/ml penicillin and 100 units/ml streptomycin, 1X B27, and 5% Hyclone FBS in DMEM and 30 % of B104 conditioned medium. Microglia was removed by differential adhesion and astrocytes by sequential immunopanning with Ran-2 antibody (Huang et al. 2002). Approximately 70–80% of immunoselected cells were OPC, the remainder were astrocytes. One million purified OPC, plated onto a 35 mm poly-L-ornithine coated cell culture dish for 24 hours, were differentiated with 45 nM T3 in differentiated medium (10ng/ml biotin, 5ug/ml insulin, 2mM glutamine, 100 units/ml penicillin and 100 units/ml streptomycin, 1X B27, and 2% Hyclone FBS in DMEM) for 24 and 72 hours. Cells were harvested for protein and RNA extraction.
Immunocytochemistry, process complexity and data analysis
Immunoselected OPC and OL differentiated for 24 and 72 hrs were stained with O4 Mab and the processes were viewed under fluorescence microscopy, using a Nikon TE2000 at 40X magnification. For process complexity analysis, cells were classified in one of four groups, as previously described (Marin-Husstege et al. 2002). O4 stained OL were classified as category I, simple, if they had 1–3 primary processes, category II, intermediate, if they had secondary processes, category III, complex, if they had tertiary processes and category IV, highly complex, if they had many tertiary processes and fused membranes. For the final analysis, cells with simple and intermediate morphology and cells with complex and highly complex morphology were grouped and data were expressed as percentage of each morphological group in PLP-ISEdel and WT OPC and OL differentiated for 24 and 72 hrs. The statistical significance of differences between the PLP-ISEdel and WT cultures was determined with student t-test.
Myelin and axogliasome membrane purification
Myelin and axogliasome membranes were prepared from brainstem and cerebral hemispheres by sucrose density gradient, as previously described (Roth et al. 2006). Briefly, brain homogenates from 90 day-old C57BL/6 and PLP-ISEdel mice (n=2) resuspended in 1.2 M sucrose solution were layered at the bottom of ultracentrifuge tubes, 0.85 M and 0.25 M sucrose buffer containing protein inhibitors were layered on top of the homogenates. After centrifugation at 100,000 g overnight at 4°C with Beckman SW28 rotor, myelin was isolated at the 0.85/0.25M interface and the axogliasomes were isolated at 1.2/0.85 M interface. The fractions were subjected to five osmotic shocks in water. Purified myelin and axogliasomes were stored at −80°C.
RNA extraction, RT-PCR and Real Time qRT-PCR
Total RNA was extracted from FACS sorted cells, OPC and OL differentiated in vitro with RNeasy mini kit (QIAGEN, Valencia, CA), and from mouse hemisphere brain tissue with TRI reagents (Sigma, St. Louis, MO). RNA was treated with DNase (Ambion, Austin, TX) to minimize genomic DNA contamination. 1ug of DNase treated RNA was reverse transcribed into cDNA using TaqMan kit (Applied Biosystems, Carlsbad, CA) with random hexamer primers. PLP and DM20 PCR products were simultaneously amplified with forward primer 5′-GGCCACTGGATTGTGTTTCT-3 ′ and reverse primer 5′-GACTGACAGGTGGTCCAGGT-3′, as previously described (Hobson et al. 2002; Wang et al. 2007). SIRT2 PCR products were amplified with forward primer 5′-TATCTCGGCCTCTTCTTGTTTC-3 ′ and reverse primer 5′-CTTCTCCAGGTTTGCATAGAGG-3′. There are three alternative spliced variants of SIRT2: v1 is the full length isoform, v2 is a shorter isoform, in which exon 2 is spliced out resulting in the utilization of a downstream start codon and a protein which is 37 amino acid shorter, and v3 in which multiple exons are deleted in frame, but the start codon is the same as in isoform 1 (NCBI, Gene ID: 64383). All three variants contain the NAD-dependent deacetylase domain (Nahhas et al. 2007). To quantify SIRT2 expression in the PLP-ISEdel mouse brain, SIRT2 transcript was amplified and normalized to GAPDH by Real Time qRT-PCR analysis using PerfeCTa SYBR Green Super Mix (Quanta Biosciences, Gaithersburg, MD). Briefly, total SIRT2 transcripts were amplified with forward primer 5′-AGGCTCAGGATTCAGACTCG-3′ and reverse primer 5′-TGTAGCGTGTCACTCCTTCG-3′. GAPDH transcript was amplified with forward primer 5′-GGTCGGTGTGAACGGATTTG-3′ and reverse primer 5′-TCGTTGATGGCAACAATCTCCACT-3′. Relative levels of SIRT2 transcript were determined by threshold cycle differences of SIRT2 versus GAPDH, as previously described (Schneeman et al. 2005). Data were expressed as fold change relative to the expression level of SIRT2 in RNA extracted from WT post-natal day 1 brain, which was set at 1. Real-time quantitative RT-PCR (qRT-PCR) of SIRT2 message in the quaking mouse brain was performed with DyNAmo SYBR Green qPCR kits (New England Biolabs, Beverly, MA). The following sets of primers were used: SIRT2 specific primers: 5′-TGGCCTGGCTGGGTGACTGT-3′ (forward) and 5′-GGCAGGCGGTGGGGACTTTC-3′ (reverse); GAPDH specific primers: 5′-GGTGAAGGTCGGTGTGAAC-3′ (forward) and 5′-CCTTGACTGTGCCGTTGAA-3′ (reverse).
RNase Protection Assays
Total RNA was extracted from qkv mouse brain stem using Trizol reagent according to the manufacturer’s protocol (Invitrogen, Carlsband, CA). The RPA probes for PLP/DM20 and GAPDH were generated as previously described (Zhao et al. 2006). Antisense riboprobes were generated by in vitro transcription in the presence of [32P] UTP (Amersham Biosciences, Piscataway, NJ) using linearized plasmids as DNA templates. Eachprobe (5 × 105 cpm) was hybridized to RNA samples as indicated in the corresponding figure legend followed by RPA analysis based on previously published procedures (Li et al. 2000).
Protein extraction and Western blot analysis
Total proteins were extracted from FACS-sorted cells, OPC and OL differentiated in vitro with RIPA lysis buffer (Millipore, Billerica, MA) supplemented with 1X Halt protease inhibitor mixture (Thermo Scientific, Rockford IL). Proteins were extracted from the PLP-ISEdel mouse brain using modified RIPA buffer (50 mM Tris–HCl, pH 7.4, 250 mM NaCl, 1% NP-40, 0.25% Deoxycholic Acid, 1 mM EDTA) supplemented with 1 mM PMSF, 5 mM DTT, 10 mM NaF, 1 mM NaVO4, 1X Halt protease inhibitor mixture. The qkv mutant and Fyn−/− brain tissues were homogenized in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.5% Triton X-100 in the presence of protease inhibitor mixtures (Roche, Indianapolis, IN), and whole cell lysates were prepared by sonication in 1X protein loading buffer. Cytoplasm and cytoskeleton associated proteins were extracted from OPC and differentiated OL, as previously described (Bargagna-Mohan et al. 2010). Briefly, after 1 × wash with PBS, cells were lysed with 50 μl of ice-cold of lysis buffer (1% NP-40 in 0.4X TBS solution) on ice for 45′. Lysates containing cytoplasmic proteins were removed and 100 ul of 2X Laemmli sample buffer (0.125 M Tris pH 6.8, 4% SDS, 20% glycerol, 0.002% bromophenol blue, 10% 2-mercaptoethanol) was added to the dish to extract cytoskeleton associated proteins. Protein concentrations were measured by Bio-Rad protein assay (Bio-Rad laboratory, Hercules, CA) and protein aliquots were stored at −80°C. Proteins were separated on either 4–20% gradient or 10% SDS-PAGE gel (Biorad, Hercules CA) and transferred to PVDF membranes. Blots were incubated with antibodies to PLP/DM20 (1:2000, AA3 hybridoma), SIRT2 (mouse specific polyclonal Ab, 1:6000, generous gift of Dr. Tainsky), CNPase (1:5000, AbCam, Cambridge, MA), caveolin-2 (1:4000, Sigma, St. Louis, MO), acetylated-α-tubulin (K40) (1:10,000, AbCam, Cambridge, MA), α-tubulin (1:10,000, Sigma, St. Louis, MO), APC/CC1 (1:200), AbCam, Cambridge, MA), β-tubulin (1:10,000, Covance), β-actin (1:10,000, Sigma, St. Louis, MO) and GAPDH (1:6000, AbCam, Cambridge, MA) at 4°C overnight, followed by HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted 1:5000~10,000 at room temperature for 1 hour and developed with Amersham ECL Plus Chemiluminescence (GE Healthcare, Piscataway, NJ). GAPDH, β-tubulin and β-actin were used as control for loading accuracy and α-tubulin as control for α-tubulin expression.
Immunohistochemistry
Mice were deeply anesthetized and transcardially perfused with heparinized saline buffer followed by ice-cold 4% paraformaldheyde in 0.01M PBS pH 7.4. The brains were fixed in 4% paraformaldheyde O/N and then cryoprotected in 20% sucrose in 0.1M PBS at 4°C O/N. Coronal sections (20 μm) were collected on gelatin-treated glass slides using Leica CM 1850 cryostat (Heidelberger, Germany). Brain slices were permeabilized and blocked with 0.2% Triton-X-100 and 0.01% Tween 20 in PBS containing 1% BSA and 2% normal gloat serum, before incubation with rabbit anti-SIRT2 (1:1,200) overnight. After three washes with PBS containing 0.1% Tween 20, FITC-labeled goat-anti-rabbit IgG (1:400, Jackson ImmunoResearch Laboratories, West Grove, PA) was used to detect SIRT2 protein. Images were captured using fluorescence microscope (IX51; Olympus, Tokyo, Japan) equipped with a RETIGA digital camera.
RESULTS
Expression of SIRT2 is regulated by alternative splicing in developing OLs
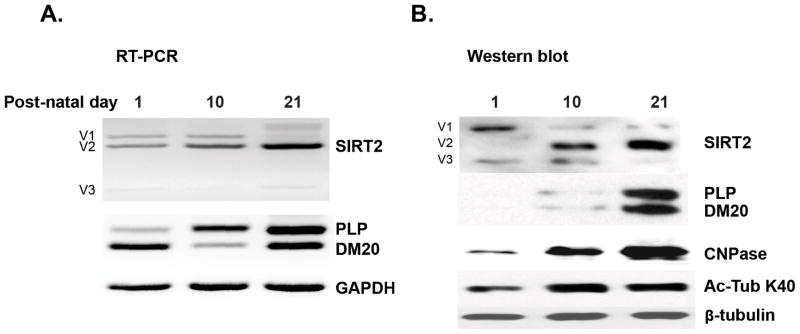
To examine the mechanism by which the differentiation-dependent increase of SIRT2v2 expression occurs in OLs in vivo, we have isolated EGFP+ OLs from the CNP-EGFP mouse brain at postnatal day 1–3 (P1-3), P10 and P21 by FACS sorting (Yuan et al. 2002). We performed RT-PCR amplification with primers spanning across the SIRT2 alternatively spliced exon in RNA extracted from OLs isolated at the aforementioned developmental time points. We detected three SIRT2 alternatively spliced transcripts (v1, v2 and v3) at all stages of development examined and show that the SIRT2v2 transcript is preferentially increased in P10 and P21 OLs, which primarily underlies the overall up-regulation of SIRT2 and coincides with the increase in the PLP/DM20 transcript ratio, consistent with the possibility that a common pathway may regulate these splicing events (Fig. 1A). Importantly, although SIRT2v2 transcript is present at all developmental time points, SIRT2v2 protein is not detected in OLs until P10, suggesting a post-transcriptional/translational regulation (Fig. 1B). The protein isoform v1 is expressed at all stages of OL development, but the highest level is in immature OL. Isoform v3 is not expressed in mature OLs (Fig. 1B). The significance of the latter findings remains to be elucidated. Interestingly, acetylation of α-tubulin at K40, a major target of SIRT2, is increased in lysates of P10 and P21 OLs, indicating that the overall degree of α-tubulin acetylation increases as OLs differentiate, in keeping with in vitro data (below) and previously published data (Li et al. 2007). Together, the data show that differentiation-dependent alternative splicing regulates the enhanced expression of SIRT2v2 transcript and protein in the developing OLs in vivo. While the greater expression of SIRT2 in differentiated OLs would be expected to result in reduced α-tubulin acetylation, an increase in acetylated α-tubulin is instead detected. These data suggest that SIRT2 does not control the overall degree of α-tubulin acetylation, but might dynamically regulate α-tubulin acetylation in discrete cytoskeleton domains [(Li et al. 2007) and see Fig. 8].
Figure 1. Differentiation-dependent regulation of SIRT2 expression and alternative splicing in developing OL in vivo.
A. Representative RT-PCR analysis of SIRT2 and PLP/DM20 transcripts in RNA extracted from EGFP+ OL isolated from CNP-EGFP mouse brain at the indicated post-natal days (n=2). The three isoforms, SIRT2v1, v2 and v3 are amplified. SIRT2v2 expression increases at post-natal day 10 concomitantly with the increase in PLP/DM20 ratio. GAPDH is amplified as control of RNA input. B. Representative Western analysis of SIRT2 and PLP/DM20 protein expression and acetylated α-tubulin in lysates prepared from EGFP+ OL isolated from CNP-EGFP mouse brain at the indicated post-natal days (n=2). SIRT2v2 begins to be expressed at p10 and increases at p20 as OL mature concomitantly with the expression of PLP/DM20. The expression of SIRT2v1 and v3 decreases. Acetylated α-tubulin (K40) is higher in lysates of OL isolated at P10 and P20 compared to day 1. CNPase is used as an additional control of differentiation.
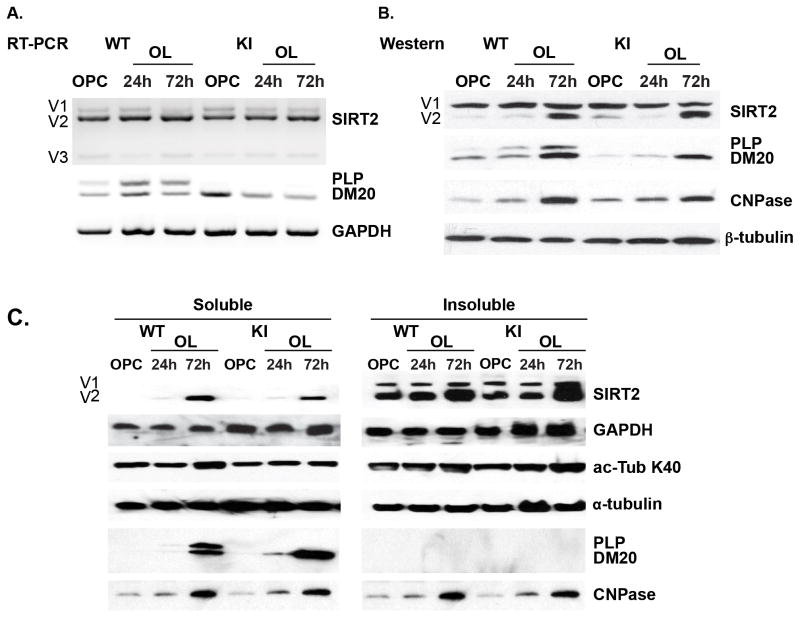
Figure 8. Oligodendrocytic SIRT2 expression is not regulated by PLP.
A. Representative RT-PCR analysis of SIRT2 and PLP/DM20 transcripts amplified in RNA extracted from WT and KI OPC and OLs differentiated for 24 and 72 hrs in vitro (n=3). SIRT2 v1, v2 and v3 transcript levels are similar in the WT and KI. PLP/DM20 ratio is greatly increased in WT differentiated OLs, while only DM20 is detected in the KI OPC and OLs. B. Representative Western blot analysis of SIRT2 and PLP/DM20 expression in lysates of WT and KI OPC and OLs differentiated for 24 and 72 hrs in vitro (n=3). SIRT2v2 is dramatically increased in WT and KI OL differentiated for 72 hrs compared to 24 hrs and OPC. PLP is detected in OLs differentiated for 24 hrs and increases at 72 hrs. DM20 only is detected in KI OPC and increases in differentiated OL. CNPase is used as an additional control of OL differentiation. β-tubulin is used as a control for accuracy of protein loading. C. Representative Western blot analysis of SIRT2, PLP/DM20 expression and α-tubulin K40 acetylation in soluble and insoluble fractions prepared from WT and KI OPC and OL differentiated for 24 and 72 hrs (n=3). SIRT2 v1 and v2 are mostly present in the insoluble fraction. SIRT2v2 is higher in the insoluble fraction of OL differentiated for 72 hrs and is detected in the soluble fraction only at this time point. Either PLP/DM20 or DM20 only is detected in the soluble fraction of differentiated WT and KI OL, respectively. Acetylated α-tubulin K40 in both soluble and insoluble fractions is comparable in WT and KI. The total amount of α-tubulin is similar in all extracts. CNPase is used as control for OL differentiation. GAPDH is used as control for accuracy of protein loading.
SIRT2 expression is severely reduced in the qkv/qkv brain and is restored by QKI-6-dependent PLP expression
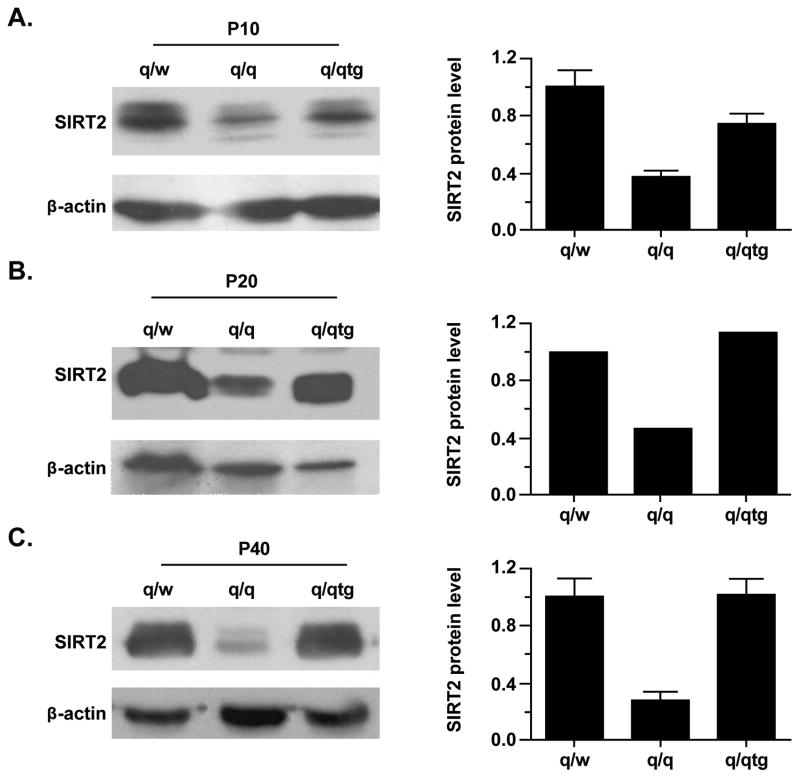
The temporal co-regulation of SIRT2 and PLP/DM20 alternative splicing in OLs suggests that these events may be regulated by the same developmental signals. To identify regulatory proteins that might be involved in these RNA processing events, we postulated that the selective RNA binding protein QKI that governs post-transcriptional expression of genes crucial for myelinogenesis, including PLP/DM20 (Li et al. 2000), may be involved in the developmental regulation of SIRT2. Three QKI isoform proteins derived from alternative splicing, named QKI5, QKI6 and QKI7, are expressed in normal OLs but diminished in q/q OLs, leading to severe hypomyelination (Hardy et al. 1996; Zhao et al. 2006). QKI6 is the predominant isoform in normal brain and is preferentially affected in the q/q mutant (Lu et al. 2003). We examined the expression of SIRT2 in lysates prepared from brainstems derived from the q/q mutant and the heterozygous non-phenotypic littermates (q/w) during myelinogenesis (P10 and P20, Fig. 2A and 2B) and young adult when myelination is largely completed (P40, Fig. 2C). SIRT2 protein is severely reduced in brainstems of q/q mutant at P10 and P20 (~60%) and P40 (>60%) compared to q/w controls. Introducing the Flag-tagged QKI6 transgene specifically expressed in the cytoplasm of q/q OLs can rescue most of the QKI targets and correct the hypomyelinating phenotype (Zhao et al. 2006). Interestingly, Flag-QKI6 restored SIRT2 protein expression in the q/q mutant by approximately 50% at P10 (Fig. 2A) and completely restored SIRT2 to normal levels starting from P20 (Fig. 2B). Furthermore, the restored expression of SIRT2 is stable over time and SIRT2 levels are similar in the q/qtg and q/w at P40 (Fig. 2C).
Figure 2. SIRT2 protein is severely reduced in the quakinviable brain and is rescued by transgenic expression of QKI6.
Representative Western blot analysis of SIRT2 expression in brainstem homogenates prepared from non-phenotypic heterozygous control (q/w), quakingviable mutant (q/q), and transgenic rescue mouse (q/qtg) at P10 (A), P20 (B) and P40 (C). SIRT2 is severely reduced at P10, P20 and P40 in the q/q brainstem as compared to that in q/w. Transgenic expression of QKI6 partially rescues the expression of SIRT2 at P10 (n=3) and completely restores it at P20 (n=1) and P40 (n=3). The bar graph shows the quantification of SIRT2 signals normalized to that of β-actin (mean±SD). The reduction of SIRT2 in q/q in comparison with that in q/w is significant in both P10 and P40 brain homogenates (n=3, p<0.01). The expression level of SIRT2 in q/qtg compared with that in q/q at P10 does not reach statistical significance, but is highly significant at P40 (p<0.01).
However, unlike many other myelin gene transcripts that are destabilized in the q/q OLs (Li et al. 2000; Zhao et al. 2006), real time qRT-PCR revealed no difference in SIRT2 message levels in the q/q mutant as compared to q/w (Fig. S1), suggesting that QKI does not directly regulate SIRT2 mRNA, but may indirectly regulate SIRT2 through other known QKI targets. In support of this interpretation, bioinformatics analysis of the SIRT2 transcript has not shown the presence of QKI recognition elements (QRE).
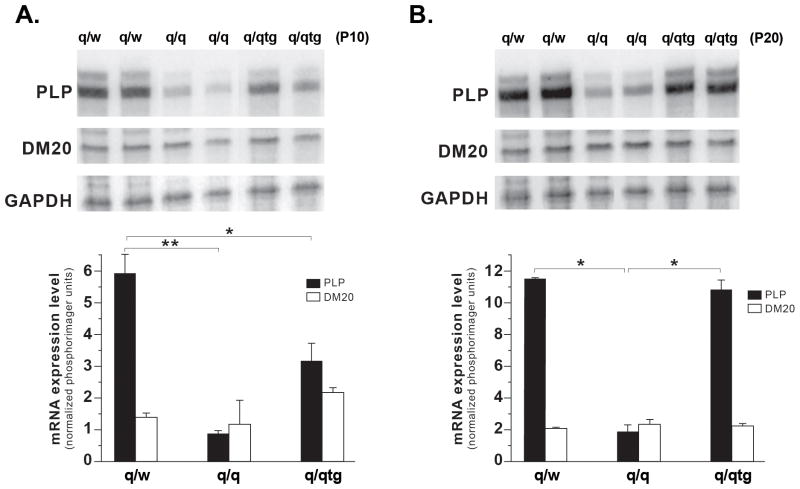
Since SIRT2 is severely reduced in the Plp1 null mouse brain (Werner et al. 2007) and PLP mRNA is a known target of QKI (Li et al. 2000; Zhao et al. 2006), we reasoned that reduced SIRT2 in the q/q brain may result from reduction in PLP caused by QKI deficiency (Zhao et al. 2006). To test this hypothesis, we developed an RNase Protection Assay (RPA) in which both PLP and DM20 transcripts are recognized by the same probe but generate protected fragments of different sizes. As shown in Figure 3, the PLP message is severely reduced in the q/q mutant brainstems at both P10 and P20, while the DM20 message is unchanged. Interestingly, the Flag-QKI6 transgene only partially restores the PLP message at P10 (Fig. 3A) and fully rescues the PLP message later at P20 (Fig. 3B). Complete rescue of SIRT2 protein expression by the Flag-QKI6 transgene (Fig. 2B) is achieved concomitantly with restoration of PLP expression (Fig. 3B). In contrast, DM20 is unlikely to be involved.
Figure 3. PLP message, but not DM20, is severely reduced in the quakingviable mouse brain and is fully restored by transgenic expression of QKI6.
Representative RNAse protection Assay (RPA) in RNA extracted from two separate non-phenotypic heterozygous control (q/w), quakingviable mutant (q/q), and transgenic rescue mouse (q/qtg) brainstem samples at P10 (A) and P20 (B). The probe used in the RPA allows protection of different size bands for PLP and DM20 (Methods). The PLP message is severely reduced, while the DM20 message is at nearly identical levels in the q/q and q/w brainstem at both P10 and P20. Transgenic expression of QKI6 partially rescued PLP expression in the P10 q/qtg brainstem and fully restored PLP message levels in P20 q/qtg brainstem. The bar graph shows the quantitation of the expression levels (mean±SD) and the statistical significance of the changes (n=3). The reduction of PLP in q/q vs. q/w is significant at both P10 and P20 (** p<0.01). The increase in PLP in the q/tg vs. q/q at P10 and P20 is significant, *p<0.05, **p<0.01 respectively.
To exclude the possibility that deficiency of SIRT2 in the q/q mutant and rescued SIRT2 expression by the QKI-6 transgene is simply represented by the ability to form myelin, we investigated SIRT2 expression in the Fyn−/− hypomyelinating mutant (Sperber et al. 2001). Importantly, unlike the majority of known hypomyelination mutant that harbor PLP deficiency, DM20 transcript is preferentially decreased in the Fyn−/− brains, whereas PLP transcript is only slightly reduced without statistical significance as shown in our previous report (Lu et al. 2005). Thus, this mouse model allows us to specifically determine whether hypomyelination per se causes loss of SIRT2. As shown in Fig. 4, SIRT2 expression is not affected in the Fyn−/− brain. PLP and DM20 protein expression was not significantly different in the Fyn−/− vs. control brain (Fig. 4A). This data clearly indicates that hypomyelination does not necessarily lead to SIRT2 reduction. Collectively, the data suggest that selective stabilization of the PLP mRNA by QKI6 forms a novel posttranscriptional pathway to control SIRT2 protein expression during CNS myelin development.
Figure 4. The abundance of SIRT2v2 protein is not affected in the Fyn −/− mouse brain.
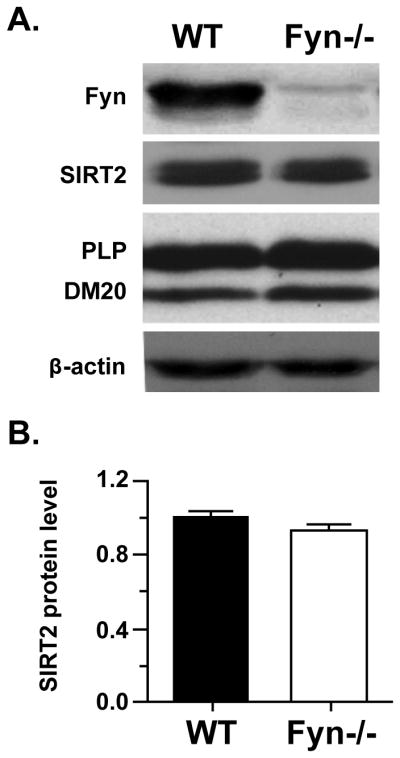
A. Representative Western blot analysis of Fyn, SIRT2 and PLP/DM20 protein expression in the cerebral cortex of Fyn−/− and wild type (WT) control mice at P20. βactin was used as a loading control. B. The bar graph shows the quantitation of SIRT2 normalized to the loading control β-actin (mean±SD) (n = 4).
PLP, but not DM20, is necessary to control the abundance of SIRT2 protein in myelin compartments
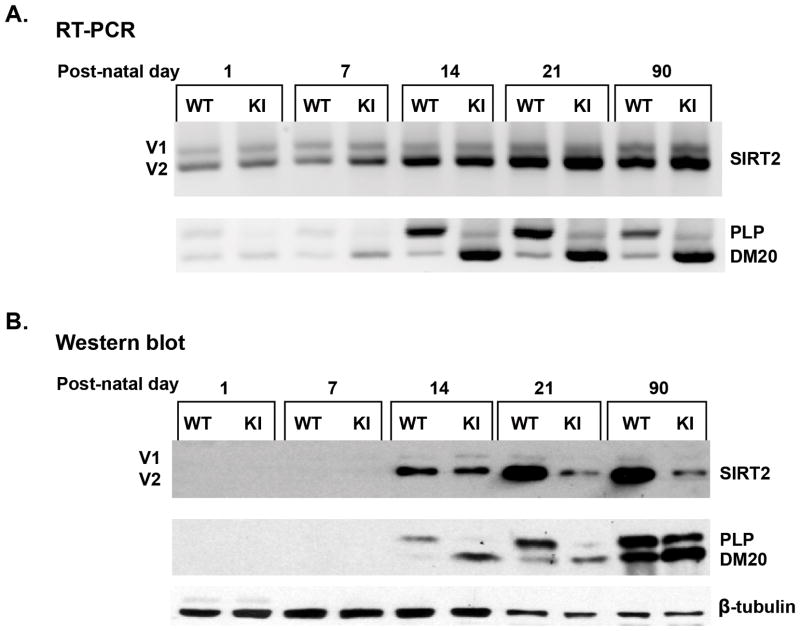
To directly demonstrate that PLP, but not DM20, regulates SIRT2 abundance, we examined the expression of SIRT2 in the developing post-natal PLP-ISEdel mouse brain, in which PLP is selectively decreased, while DM20 is increased [Fig. 5 and (Wang et al. 2008)]. Homogenates were prepared from PLP-ISEdel and control littermate cerebral hemispheres at postnatal days (P) 1, 7, 14, 21 and 90. Expression of SIRT2v2 protein is detected at P14, increases at P21 and parallels the selective increase in PLP protein at the same time points, SIRT2v1 expression is essentially unchanged (Fig. 5B). SIRT2v2 levels remain stable at post-natal day 90 (Fig. 5B). The developmental increase in SIRT2v2 protein does not take place in the PLP-ISEdel brain and SIRT2 expression is lower than in WT littermates at all postnatal time points (Fig. 5B). Similar results were obtained by analyzing brainstem tissues (data not shown). We examined whether decreased SIRT2 abundance results in an increase in α-tubulin acetylation in the PLP-ISEdel post-natal brain lysates. Acetylation of α-tubulin K40 in the developing and adult PLP-ISEdel mouse brain was comparable to that of the WT (data not shown). RT-PCR with primers that span across SIRT2 alternative spliced exon detected SIRT2v2 message increase similarly in the control and PLP-ISEdel brain at P14, suggesting that alternative splicing of SIRT2 proceeds normally in the PLP-ISEdel mouse brain (Fig. 5A). PLP message is severely reduced due to the absence of a critical intronic splicing enhancer of PLP splicing (Fig. 5A and (Wang et al. 2008)). No differences in total SIRT2 message abundance were detected by qRT-PCR at all postnatal days (data not shown). These results indicate that the decrease of SIRT2v2 protein in the PLP-ISEdel mouse is not due to defects in either alternative splicing or accumulation of SIRTv2 message, supporting the conclusion that PLP regulates the abundance of SIRT2v2 protein by posttranscriptional mechanisms.
Figure 5. SIRT2v2 protein is severely reduced in the PLP-ISEdel mouse brain.
A. Representative RT-PCR analysis of SIRT2 and PLP/DM20 transcripts amplified in RNA extracted from control (WT) and PLP-ISEdel (KI) brains at the indicated post-natal days (n=3). SIRT2v2 transcript increases during development concomitantly with the developmental switch in PLP/DM20 ratio. SIRT2 alternative splicing regulation is similar in the KI and WT mouse brain. B. Representative Western blot analysis of SIRT2 and PLP/DM20 abundance in WT and KI brain homogenates prepared at the indicated post-natal days (n=3). SIRT2v2 protein is severely reduced in the KI mouse brain in association with the reduction in PLP. SIRT2v1 level is essentially unchanged. 30 μg of protein of day 1, 7, 14 brain homogenates and 15 μg of protein of day 21 and 90 homogenates were loaded to adjust for the greater levels of expression of PLP/DM20 and SIRT2 at later time points.
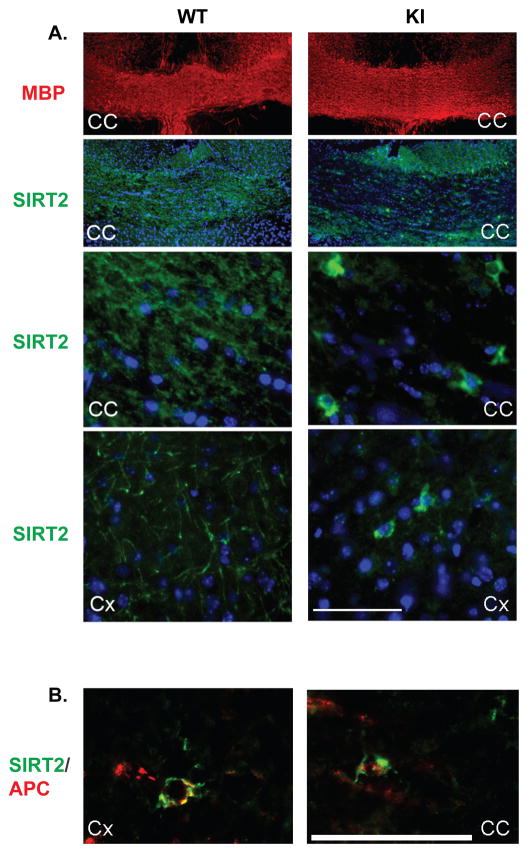
Next, we examined the presence of SIRT2 in OL soma, processes and myelin fibers by immunohistochemistry in PLP-ISEdel and WT brain coronal cryosections containing the corpus callosum and cortex (Fig. 6A). SIRT2 is severely reduced in myelin tracts of the PLP-ISEdel corpus callosum and cortex (Fig. 6A), but appears to be over-concentrated in the OL soma and proximal processes and co-localizes with the oligodendrocyte-specific marker APC/CC1 (Fig. 6A and B). In contrast, myelin and OL cell bodies and processes are uniformly stained for SIRT2 in the WT corpus callosum and cortex (Fig. 6A). Unlike SIRT2, comparable intensity immunofluorescent signals of MBP were detected in both WT and PLP-ISEdel corpus callosum (Fig. 6A), consistent with normal myelin formation in the PLP-ISEdel brain (Wang et al. 2008). These data clearly indicate that SIRT2 is expressed by the PLP-ISEdel OLs, but its transport to myelin is severely impaired by the reduced abundance of PLP, resulting in “trapping” of SIRT2 in the OL soma and processes.
Figure 6. SIRT2 is absent in the PLP-ISEdel myelin but present in the OL cell bodies.
A. SIRT2 and MBP were immunostained in the corpus callosum (CC) and cortex (Cx) of the PLPISEdel (KI) and control littermates (WT). Cross-sections were stained with anti-MBP antibody (red) and anti-SIRT2 antibody (green). A. Low magnification micrographs show equal staining of WT and KI CC myelin with MBP, SIRT2 is essentially absent in the KI CC myelin and present in the WT CC myelin (top two panels). High magnification image of the CC and the Cx show SIRT2 staining in the OL soma and entire process arbor as well as myelin tracts in WT. In contrast, SIRT2 staining was over concentrated in the OL soma but nearly absent in the myelin tracts in the KI mutant brain regions (bottom two panels). B. Sections of the KI cortex (Cx) and corpus callosum (CC) were immunostained with anti-SIRT2 (green) and the oligodendrocyte-specific APC/CC1 (red) antibody. SIRT2 is accumulated in the cell bodies of APC+ OL. The scale bars for both A and B are shown.
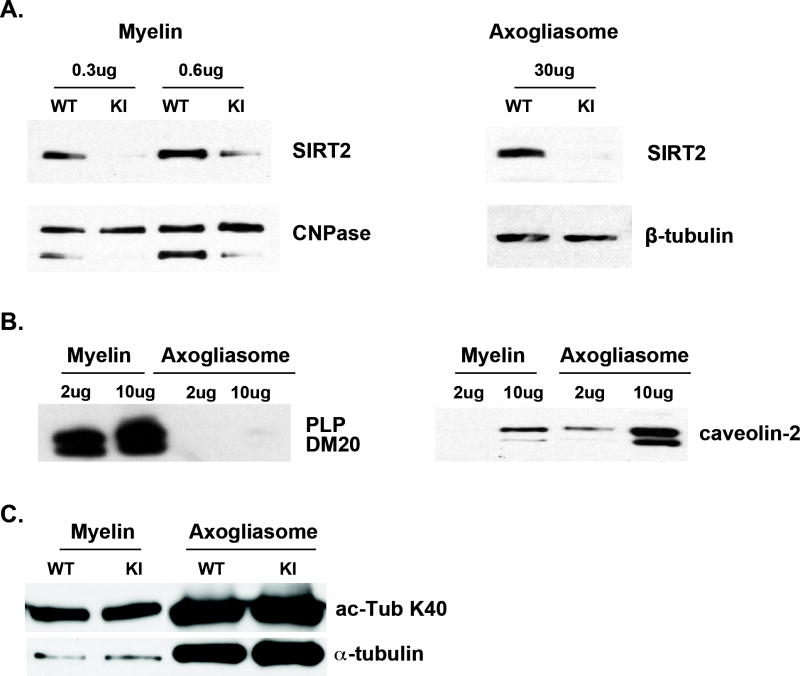
Lastly, we examined whether SIRT2 is reduced to the same extent in the PLP-ISEdel compact myelin and axogliasomes, which comprise the node and flanking paranodal domains, by Western blot analysis of these fractions prepared from adult PLP-ISEdel and WT brains by sucrose density gradients (Roth et al. 2006). As shown in Figure 7, SIRT2 is hardly detectable in the PLP-ISEdel myelin and axogliasomes compared to the WT. The purity of each fraction was verified by the enrichment of PLP/DM20 in the myelin fraction and caveolin, a protein present in the membrane lipid rafts, in the axogliasomes [Fig. 7B, (Roth et al. 2006)]. PLP/DM20 is detected exclusively in myelin, but not in axogliasomes, while caveolin was highly enriched in the axogliasome fraction (Fig. 7B). Finally, α-tubulin K40 acetylation in the PLP-ISEdel axogliasome and myelin fractions was similar to that of the WT (Fig. 7C). These data indicate that SIRT2v2 fails to be transported to the myelin membranes and the axogliasomes in the PLP-ISEdel mouse brain, suggesting that SIRT2v2 is specifically targeted by PLP. Despite the marked reduction of SIRT2 in the PLP-ISEdel myelin and axogliasome compartments there is no difference in α-tubulin acetylation as compared to WT, in keeping with the data in brain lysates. Collectively, the data support the interpretation that PLP is solely responsible for SIRT2v2 protein abundance in myelin compartments
Figure 7. SIRT2 is reduced in PLP-ISEdel compact myelin and axogliasomes.
A. Representative Western blot analysis of SIRT2 expression in myelin and axogliasome fractions prepared from 90-day old WT and KI mouse brains (n=2). Myelin proteins (0.3 and 0.6 μg) and axogliasome proteins (30 μg) were separated by SDS-PAGE and reacted with an antibody specific for mouse SIRT2. SIRT2 abundance is severely reduced in the KI myelin and axogliasomes compared to the WT. B. The purity of the membrane preparations is shown. Myelin and axogliasome proteins (2 and 10 μg) were separated by SDS-PAGE and reacted with an antibody to PLP/DM20 (AA3) and to caveolin. PLP/DM20 is not detected in the axogliasomes, and caveolin is highly enriched in axogliasomes and detected in very low amount only when 10 μg myelin protein were loaded. C. Representative Western blot of α-tubulin and acetylated K40 α-tubulin in myelin and axogliasome fractions prepared from WT and KI mouse brains (n=2). There is no difference in acetylated K40 in the KI vs. WT.
SIRT2 expression is not regulated by PLP in pre-myelinating OLs
Since PLP and SIRT2 are co-regulated in developing OLs in vivo (Fig. 1), we investigated whether PLP is required for the expression of SIRT2 in differentiating OLs. To this end, we have examined the expression and subcellular distribution of SIRT2 in non-myelinating PLP-ISEdel and WT OLs differentiated in vitro. SIRT2v2 protein was highly expressed in both WT and PLP-ISEdel OLs undergone 72 hrs differentiation, while SIRT2v1 remained constant through lineage progression in vitro (Fig. 8B). SIRT2v2 transcript increases in both WT and PLP-ISEdel OLs differentiated for 72 hrs, while transcripts of SIRT2v1 and SIRT2v3 remain constant (Fig. 8A), consistent with the results in developing EGFP+ OL (Fig. 1) and postnatal brain (Fig. 5A). However, unlike EGFP+ OLs, a protein product for SIRT2 v3 was not detected in cultured OLs. PLP was expressed in WT OLs differentiated for 24 and 72 hrs, only DM20 was detected in PLP-ISEdel OLs (Fig. 8A and B). These data indicate that the developmental regulation of SIRT2 expression and alternative splicing proceeds normally in PLP deficient OL in vitro before myelination.
Since SIRT2 is distributed in the cytoplasm and cytoskeleton of OLs, in which it controls acetylation of α-tubulin (Li et al. 2007; Southwood et al. 2007), we investigated whether deficiency of PLP reduces the cytoskeletal-associated SIRT2 and results in increased α-tubulin acetylation in the PLP-ISEdel OLs in vitro. Soluble and insoluble fractions (Bargagna-Mohan et al. 2010) from WT and PLP-ISEdel OPCs and OLs undergone 24 and 72 hrs of differentiation in vitro were examined by Western blot analysis (Fig. 8C). Both SIRT2 v1 and v2 are mostly present in the insoluble fraction, in which SIRT2v2 is similarly increased in both WT and PLP-ISEdel OLs differentiated for 72 hrs (Fig. 8C). In addition, a small amount of SIRT2v2 is also detected in the soluble fraction extracted from WT and PLP-ISEdel OLs differentiated for 72 hrs (Fig. 8C). There was no difference in the extent of α-tubulin K40 acetylation in the soluble and insoluble fraction of PLP-ISEdel OPC and differentiated OLs as compared to WT (Fig. 8C). In keeping with these data, morphological differentiation of PLP-ISEdel OLs in vitro proceeds similarly to WT OLs. The number of cells with simple, intermediate, complex and highly complex processes was counted in cultures of PLP-ISEdel and WT OPC and OL differentiated for 24 and 72 hrs was similar (Li et al. 2007; Marin-Husstege et al. 2002) (data not shown). Furthermore, SIRT2 is present in the cell body, processes and myelin sheaths of the PLP-ISEdel OL differentiated in vitro for 72 hrs (Fig. S2). Together, our results indicate that comparable SIRT2 is expressed and associated with cytoskeleton in pre-myelinating PLP-ISEdel compared to WT OLs during early differentiation, thus accounting for normal OL morphological differentiation. The data further support the conclusion that only the abundance of SIRT2 in myelin is regulated by PLP, while cell restricted SIRT2 and morphological differentiation are not affected.
DISCUSSION
In this study, we report for the first time that the expression of SIRT2 is regulated by the QKI-6-dependent pathway in OLs and this effect is mediated through selective regulation of PLP. Using two genetic mouse model systems, the quakingviable mutant and the PLP-ISEdel mutant, we demonstrate that the abundance of SIRT2 in myelin is dependent on the full dosage of PLP, but not DM20. Furthermore, the expression of SIRT2 in pre-myelinating oligodendrocytes is independent of the abundance of PLP and the cellular function of SIRT2 in regulating process complexity is not affected by the absence of PLP.
SIRT2 is a major component of the myelin proteome and an OL-specific deacetylase co-regulated with the program of OL differentiation and the activation of myelination (Li et al. 2007). In the present study, we show that the three alternatively spliced variants of SIRT2 are developmentally regulated at the transcript and protein levels in OLs in vivo and in vitro. SIRT2v3 was exclusively detected in OLs either isolated by sorting or cultured in vitro, while it was not detected in the developing brain. The functional relevance of this isoform and the significance of this finding remain to be investigated. Importantly, SIRTv2 is preferentially up-regulated and becomes the major functional isoform in differentiated OL and in mature brain. The increase in SIRT2v2 correlates temporally with the program of myelin gene activation and myelin protein expression, likely due to enhanced transcription as well as splicing of the primary SIRT2 transcripts under regulatory pathways that control OL differentiation and myelin gene expression.
The products of the qk gene, QKI5, 6 and 7, play critical roles in OL differentiation, lineage progression and myelination (Chen et al. 2007; Ebersole et al. 1996; Hardy et al. 1996; Larocque et al. 2005; Zhao et al. 2006). In the quakingviable mutant mouse, selective QKI deficiency in OLs impairs OL differentiation and myelin formation (Hardy et al. 1996; Li et al. 2000; Zhao et al. 2006). QKIs are K homology (KH) domain RNA binding proteins that regulate several aspects of RNA metabolism, including alternative splicing, message stability and translation, through a QKI recognition element (QRE) [review (Bockbrader and Feng 2008),(Galarneau and Richard 2005; Larocque and Richard 2005). This motif has been identified, based on in vitro binding, bioinformatically in >1,400 transcripts, among which many encodes proteins crucial for OL and myelin development (Galarneau and Richard 2005). QKI5, 6 and 7 are developmentally regulated during postnatal brain development. The cytoplasmic QKI6 and 7 become the predominant isoforms by the peak of myelination with a reciprocal decline of the nuclear QKI5 (Hardy et al. 1996). Notably, QKI6 is the most abundant isoform and is sufficient to restore myelin formation and rescue the deregulated myelin gene transcript levels in the hypomyelinating quakingviable mouse (Zhao et al. 2010; Zhao et al. 2006). Interestingly, SIRT2v2 protein, but not its message, is severely reduced in the quakinviable mouse brain and SIRT2 protein levels are restored by the Flag-QKI6 transgene through regulation of PLP abundance. It is important to point out that QKI6 selectively regulates the abundance of the PLP message, not affecting the DM20 transcript. In the Fyn−/− mouse brain, in which hypomyelination is not accompanied by significant reduction of PLP, SIRT2 abundance is not affected. These data show that hypomyelination per se is not sufficient to cause a reduction of SIRT2. Rather, the specific reduction of PLP in q/q mice due to QKI6 deficiency mediates the dysregulation of SIRT2. How this selective regulation of PLP is achieved still remains elusive. The QREs are identified in the 3′ UTR commonly shared by both the PLP/DM20 transcripts (Macklin et al. 1987), hence these QREs would not account for the selective regulation of PLP by QKI6. Another possibility is that the QKI pathway regulates the alternative splicing of PLP/DM20 and may control the developmentally regulated increase in the PLP transcript. Temporally, QKI6 expression increases in the developing brain at the time in which PLP alternative splicing is activated (Hardy et al. 1996; Zhao et al. 2006) Zhu and Cambi, unpublished data). Consistent with the idea, QKI6 was recently reported to regulate alternatively splicing of Myelin Associated Glycoprotein (MAG) by controlling the expression of the canonical splicing factor hnRNPA1 (Zhao et al. 2010).
In addition, we have found that over-expression of QKI6 increases PLP/DM20 ratio in Oli-neu cells, suggesting a direct role of QKI6 in PLP alternative splicing regulation (Wang and Cambi, unpublished observations). In addition, the nuclear QKI5 acts as a direct modulator of alternative splicing of MAG (Hafner et al. 2010; Wu et al. 2002), however, QKI5 does not appear to play a role in the alternative splicing of PLP. Over-expression of QKI5 in Oli-neu cells has very little effect on the PLP/DM20 ratio (Wang and Cambi, unpublished data) and the QKI-6 transgene is sufficient to rescue the PLP/DM20 ratio in the q/q mouse OLs (Fig. 2).
Although other downstream targets of QKI could be involved in regulating SIRT2, collectively our data strongly support the conclusion that PLP, but not DM20, plays a major and specific role in regulating SIRT2 expression. The abundance of SIRT2 in myelin depends on PLP full dosage, suggesting that PLP controls the transport to and/or stability of SIRT2 in the myelin compartment. Because myelin formation proceeds normally in the PLP-ISEdel mouse, the deficit in myelin SIRT2 is not caused by hypomyelination. By contrast, SIRT2v2 expression in pre-myelinating OLs is not regulated by PLP, but by differentiation-dependent signals. Consistent with the fact that the cellular expression of SIRT2v2 is unaffected in pre-myelinating OLs by the reduction of PLP, the cellular function of SIRT2 in controlling α-tubulin acetylation and process morphology was not disrupted in the PLP-ISEdel OL during morphogenesis. Furthermore, α-tubulin acetylation in the PLP-ISEdel developing brain is largely unaffected by the marked decrease in SIRT2 abundance. Our data are in keeping with data in the PLp1 null mouse (Werner et al. 2007) and suggest that either SIRT2 is not necessary for regulation of tubulin acetylation or that the other isoforms, SIRT2v1 and SIRT2v3, or other deacetylases compensate for the absence and/or reduction of SIRT2. Interestingly, SIRT2 appears to be present in greater amount in the OL cell bodies in PLP-ISEdel brain sections, suggesting that defects in transporting SIRT2 into the myelin compartment eventually results in abnormal accumulation of SIRT2 in the cell body and proximal processes. Such a finding was not observed in the non-myelinating PLP-ISEdel OL in vitro, suggesting that this is a myelinating-dependent phenotype.
The selective regulation of SIRT2v2 by PLP suggests biochemical interaction and a specific subcellular co-localization of SIRT2v2 with PLP. It remains to be determined whether PLP and SIRT2v2 interact directly or indirectly through an adaptor/partner and which domains in the two proteins mediate the interaction. It is conceivable that the PLP specific domain may participate in the interaction specifically with SIRT2v2. Since SIRT2v2 differs from SIRT2v1 in the N-terminus domain, it is reasonable to postulate that the N-terminus of SIRT2 isoforms may regulate SIRT2 conformation and its interaction with protein partners for subcellular localization. Although in other cell types, SIRT2v1 and v2 have been shown to be equivalent in their substrate and enzymatic activity (Nahhas et al. 2007), SIRT2v2 may have evolved in OLs to serve unique functions in the myelin compartment. It is tempting to speculate that myelin SIRT2 plays a role in the PLP-dependent pathway that regulates axonal integrity. Progressive axonal loss occurs in the PLp1 null mouse brain, in which PLP/DM20 and SIRT2 are absent.
The identification of a QKI6-PLP pathway that regulates SIRT2 has important implications for human disorders in which QKI and/or PLP expression are disrupted. In schizophrenia brains, there is a consistent reduction in QKI and PLP/DM20 transcripts, which correlates with white matter and myelin abnormalities associated with this disorder (Aberg et al. 2006a; Aberg et al. 2006b; Haroutunian et al. 2006; Tkachev et al. 2007; Tkachev et al. 2003). How these white matter and oligodendrocyte abnormalities may contribute to synaptic and neuronal dysregulation remains to be determined. A reduction in SIRT2 would be expected in schizophrenia brains and may represent a pathogenic link between white matter changes and neuronal dysfunction. SIRT2 is greatly enriched in paranodal myelin where it may play important functions in axo-glial interactions and mediate long-term axonal integrity. Loss/reduction in SIRT2 is likely to occur in Pelizaeus-Merzbacher disease caused by either deletion or point mutations of the PLP/DM20 gene. Recently, SIRT2 was shown to be decreased in the rumpshaker mouse mutant and its expression is restored by PLP expression, which rescues the myelinating phenotype (Barrie et al. 2010). Importantly, the rumpshaker mutation is associated with X-linked complicated spastic paraplegia in humans (Saugier-Veber et al. 1994). Since mammalian sirtuins have a wide range of metabolic and stress-related functions (Sinclair 2005), SIRT2 represents a potential target for therapeutic interventions in these disorders.
Supplementary Material
SIRT2 message levels were quantitated by Real Time qRT-PCR in RNA extracted from q/w and q/q brainstem at P40 (n=3). Bar graph is the mean±SD of data obtained from three independent determinations in three separate mice. There are not differences between q/w and q/q.
Representative immunocytochemical staining of a PLP-ISEdel OL differentiated for 72 hrs with SIRT2 (red) and the oligodendrocyte-specific surface marker recognized by the O4 antibody (green). The merged image shows that SIRT2 is localized in the cell body, processes and in the cytoplasm of the myelin sheath (arrows) (n=3).
Acknowledgments
This work was supported by NIH/NINDS (RO1NS053905) to FC and European Leukodystrophy Association (2008-013I1) to FC and EW, NIH/NINDS (R01NS056097) and NMSS (RG4010-A-2) to YF. The authors thank Dr. Tainsky for the generous gift of SIRT2 antibody, Dr. Vittorio Gallo for the generous gift of the CNPase-EGFP mouse and Ms. Jennifer Strange for the excellent technical assistance with FACS isolation of oligodendrocytes.
References
- Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci U S A. 2006a;103:7482–7. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg K, Saetre P, Lindholm E, Ekholm B, Pettersson U, Adolfsson R, Jazin E. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am J Med Genet B Neuropsychiatr Genet. 2006b;141B:84–90. doi: 10.1002/ajmg.b.30243. [DOI] [PubMed] [Google Scholar]
- Bargagna-Mohan P, Paranthan RR, Hamza A, Dimova N, Trucchi B, Srinivasan C, Elliott GI, Zhan CG, Lau DL, Zhu H, et al. Withaferin A targets intermediate filaments glial fibrillary acidic protein and vimentin in a model of retinal gliosis. J Biol Chem. 2010;285:7657–69. doi: 10.1074/jbc.M109.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrie JA, Montague P, Karim S, Kirkham D, Nave KA, Anderson TJ, Griffiths IR, McLaughlin M. Modulation of rumpshaker phenotype with wild-type PLP/DM20 suggests several pathogenic mechanisms. J Neurosci Res. 2010;88:2135–45. doi: 10.1002/jnr.22379. [DOI] [PubMed] [Google Scholar]
- Bockbrader K, Feng Y. Essential function, sophisticated regulation and pathological impact of the selective RNA-binding protein QKI in CNS myelin development. Future Neurol. 2008;3:655–668. doi: 10.2217/14796708.3.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tian D, Ku L, Osterhout DJ, Feng Y. The selective RNA-binding protein quaking I (QKI) is necessary and sufficient for promoting oligodendroglia differentiation. J Biol Chem. 2007;282:23553–60. doi: 10.1074/jbc.M702045200. [DOI] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MJ, Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat Genet. 1996;12:260–5. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Edgar JM, McLaughlin M, Yool D, Zhang SC, Fowler JH, Montague P, Barrie JA, McCulloch MC, Duncan ID, Garbern J, et al. Oligodendroglial modulation of fast axonal transport in a mouse model of hereditary spastic paraplegia. J Cell Biol. 2004;166:121–31. doi: 10.1083/jcb.200312012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12:691–8. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- Garbern J, Shy M, Krajewski K, Kamholz J, Hobson G, Cambi F. Evidence for neuroaxonal injury in patients with proteolipid gene mutations. Neurology. 2001;57:1938–1939. doi: 10.1212/wnl.57.10.1938-a. [DOI] [PubMed] [Google Scholar]
- Gudz TI, Schneider TE, Haas TA, Macklin WB. Myelin proteolipid protein forms a complex with integrins and may participate in integrin receptor signaling in oligodendrocytes. J Neurosci. 2002;22:7398–407. doi: 10.1523/JNEUROSCI.22-17-07398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RJ, Loushin CL, Friedrich VL, Jr, Chen Q, Ebersole TA, Lazzarini RA, Artzt K. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice. J Neurosci. 1996;16:7941–9. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroutunian V, Katsel P, Dracheva S, Davis KL. The human homolog of the QKI gene affected in the severe dysmyelination “quaking” mouse phenotype: downregulated in multiple brain regions in schizophrenia. Am J Psychiatry. 2006;163:1834–7. doi: 10.1176/ajp.2006.163.10.1834. [DOI] [PubMed] [Google Scholar]
- Hobson GM, Huang Z, Sperle K, Stabley DL, Marks HG, Cambi F. A PLP splicing abnormality is associated with an unusual presentation of PMD. Ann Neurol. 2002;52:477–88. doi: 10.1002/ana.10320. [DOI] [PubMed] [Google Scholar]
- Huang Z, Tang XM, Cambi F. Down-regulation of the retinoblastoma protein (rb) is associated with rat oligodendrocyte differentiation. Mol Cell Neurosci. 2002;19:250–62. doi: 10.1006/mcne.2001.1077. [DOI] [PubMed] [Google Scholar]
- Klugmann M, Schwab MH, Pühlhofer A, Schneider A, Zimmermann F, Griffiths IR, Nave K-A. Assembly of CNS myelin in the absence of proteolipid protein. Neuron. 1997;18:59–70. doi: 10.1016/s0896-6273(01)80046-5. [DOI] [PubMed] [Google Scholar]
- Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- Larocque D, Richard S. QUAKING KH domain proteins as regulators of glial cell fate and myelination. RNA Biol. 2005;2:37–40. doi: 10.4161/rna.2.2.1603. [DOI] [PubMed] [Google Scholar]
- Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, Guo J, Ling EA, Liang F. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007;27:2606–16. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Li D, Feng Y. Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J Neurosci. 2000;20:4944–53. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Ku L, Chen Y, Feng Y. Developmental abnormalities of myelin basic protein expression in fyn knock-out brain reveal a role of Fyn in posttranscriptional regulation. J Biol Chem. 2005;280:389–95. doi: 10.1074/jbc.M405973200. [DOI] [PubMed] [Google Scholar]
- Lu Z, Zhang Y, Ku L, Wang H, Ahmadian A, Feng Y. The quakingviable mutation affects qkI mRNA expression specifically in myelin-producing cells of the nervous system. Nucleic Acids Res. 2003;31:4616–24. doi: 10.1093/nar/gkg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklin WB, Campagnoni CW, Deininger PL, Gardinier MV. Structure and expression of the mouse myelin proteolipid protein gene. Journal of Neuroscience Research. 1987;18:383–394. doi: 10.1002/jnr.490180302. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–45. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahhas F, Dryden SC, Abrams J, Tainsky MA. Mutations in SIRT2 deacetylase which regulate enzymatic activity but not its interaction with HDAC6 and tubulin. Mol Cell Biochem. 2007;303:221–30. doi: 10.1007/s11010-007-9478-6. [DOI] [PubMed] [Google Scholar]
- Roth AD, Ivanova A, Colman DR. New observations on the compact myelin proteome. Neuron Glia Biol. 2006;2:15–21. doi: 10.1017/S1740925X06000068. [DOI] [PubMed] [Google Scholar]
- Saugier-Veber P, Munnich A, Bonneau D, Rozet JM, Le Merrer M, Gil R, Boespflug-Tanguy O. X-linked spastic paraplegia and Pelizaeus-Merzbacher disease are allelic disorders at the proteolipid protein locus. Nature Genetics. 1994;6:257–262. doi: 10.1038/ng0394-257. [DOI] [PubMed] [Google Scholar]
- Schneeman TA, Bruno ME, Schjerven H, Johansen FE, Chady L, Kaetzel CS. Regulation of the polymeric Ig receptor by signaling through TLRs 3 and 4: linking innate and adaptive immune responses. J Immunol. 2005;175:376–84. doi: 10.4049/jimmunol.175.1.376. [DOI] [PubMed] [Google Scholar]
- Sinclair D. Sirtuins for healthy neurons. Nat Genet. 2005;37:339–40. doi: 10.1038/ng0405-339. [DOI] [PubMed] [Google Scholar]
- Southwood CM, Peppi M, Dryden S, Tainsky MA, Gow A. Microtubule deacetylases, SirT2 and HDAC6, in the nervous system. Neurochem Res. 2007;32:187–95. doi: 10.1007/s11064-006-9127-6. [DOI] [PubMed] [Google Scholar]
- Sperber BR, Boyle-Walsh EA, Engleka MJ, Gadue P, Peterson AC, Stein PL, Scherer SS, McMorris FA. A unique role for Fyn in CNS myelination. J Neurosci. 2001;21:2039–47. doi: 10.1523/JNEUROSCI.21-06-02039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Southwood CM, Gragerov A, Kelley KA, Friedrich VL, Jr, Gow A. The evolution of lipophilin genes from invertebrates to tetrapods: DM-20 cannot replace proteolipid protein in CNS myelin. Journal of Neuroscience. 2000;20:4002–4010. doi: 10.1523/JNEUROSCI.20-11-04002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Huffaker SJ, Ryan M, Bahn S. Further evidence for altered myelin biosynthesis and glutamatergic dysfunction in schizophrenia. Int J Neuropsychopharmacol. 2007;10:557–63. doi: 10.1017/S1461145706007334. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, Starkey M, Webster MJ, Yolken RH, Bahn S. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35:4164–78. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Dimova N, Sperle K, Huang Z, Lock L, McCulloch MC, Edgar JM, Hobson GM, Cambi F. Deletion of a splicing enhancer disrupts PLP1/DM20 ratio and myelin stability. Exp Neurol. 2008;214:322–30. doi: 10.1016/j.expneurol.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Mobius W, et al. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27:7717–30. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Reed RB, Grabowski PJ, Artzt K. Function of quaking in myelination: regulation of alternative splicing. Proc Natl Acad Sci U S A. 2002;99:4233–8. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Chittajallu R, Belachew S, Anderson S, McBain CJ, Gallo V. Expression of the green fluorescent protein in the oligodendrocyte lineage: a transgenic mouse for developmental and physiological studies. J Neurosci Res. 2002;70:529–45. doi: 10.1002/jnr.10368. [DOI] [PubMed] [Google Scholar]
- Zhao L, Mandler MD, Yi H, Feng Y. Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc Natl Acad Sci U S A. 2010;107:19061–6. doi: 10.1073/pnas.1007487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Tian D, Xia M, Macklin WB, Feng Y. Rescuing qkV dysmyelination by a single isoform of the selective RNA-binding protein QKI. J Neurosci. 2006;26:11278–86. doi: 10.1523/JNEUROSCI.2677-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SIRT2 message levels were quantitated by Real Time qRT-PCR in RNA extracted from q/w and q/q brainstem at P40 (n=3). Bar graph is the mean±SD of data obtained from three independent determinations in three separate mice. There are not differences between q/w and q/q.
Representative immunocytochemical staining of a PLP-ISEdel OL differentiated for 72 hrs with SIRT2 (red) and the oligodendrocyte-specific surface marker recognized by the O4 antibody (green). The merged image shows that SIRT2 is localized in the cell body, processes and in the cytoplasm of the myelin sheath (arrows) (n=3).