Abstract
Amplified fragment length polymorphism versus pulsed-field gel electrophoresis was used for fingerprinting of 85 macrolide-resistant pneumococcal isolates identified by using primarily phenotypic methods. Confirmation of identification by 16S rRNA sequencing revealed that 27 isolates were actually nonpneumococci. Amplified fragment length polymorphism but not pulsed-field gel electrophoresis offered simultaneous and accurate discrimination between pneumococci and nonpneumococcal species.
To monitor epidemiological spread of resistant pneumococci, dependable and efficient identification techniques are a prerequisite. However, accumulating data indicate that identification of Streptococcus pneumoniae by molecular biology and conventional biochemical methods leads to controversial results (2, 6, 10, 11; J. W. Mouton et al., Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. D-1690, 2003). Various fingerprinting techniques for S. pneumoniae have been described previously, including pulsed-field gel electrophoresis (PFGE) (9) and several PCR-based genomic profiling assays (5, 7). PFGE is still considered to be the “gold standard” for determination of epidemiological relationships between pneumococcal isolates. However, it is a laborious and time-inefficient method. For obvious reasons, PCR-based protocols are much more favored than the classical PFGE protocol. In this study, we explored the use of amplified fragment length polymorphism (AFLP) for epidemiological fingerprinting of macrolide-resistant S. pneumoniae isolates with emphasis on the comparison with PFGE.
We analyzed 85 erythromycin-resistant S. pneumoniae clinical isolates collected from several laboratories throughout The Netherlands between December 2001 and April 2002. Strains were stored in polypropylene vials at −70°C until testing. Strains were identified by the participating laboratories by their own standard identification techniques. All isolates were analyzed by 16S rRNA sequencing (described below). Strains were grown overnight on blood agar plates at 37°C under 5% CO2 conditions. AFLP, PFGE, and data analysis were performed according to previously described procedures with minor modifications (8). The identification of all S. pneumoniae strains was confirmed by sequence analysis of part of the 16S rRNA gene. Amplicons were generated under standard PCR conditions with the primers 5′-CGGCGTGCCTAATACATGC-3′ and 5′-CGTATTACCGCGGCTGCT-3′. After purification of the amplicons by High Pure chemistry (Roche Diagnostics), they were subjected to sequence analysis on an ABI3700 platform under the conditions recommended by the manufacturer (Applied Biosystems). The sequences obtained were compared to sequences in the public DNA libraries by using the web-based BLAST interface (1).
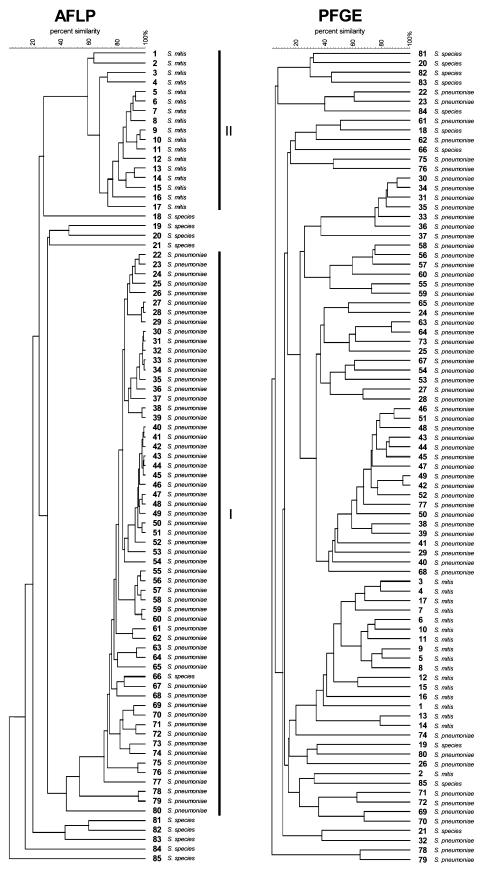
Fingerprints of the 85 strains analyzed in this study, obtained by using PFGE and AFLP, are shown in Fig. 1. As can be observed, nearly identical clusters of closely related strains can be identified by both typing methods. AFLP analysis showed, in addition to the clusters of epidemiological related strains, the formation of two clearly discernible megaclusters of strains in the dendrogram. We speculated that this collection of strains contained different species. Therefore, the identity of all strains was analyzed by 16S rRNA sequencing. All but one of the strains in AFLP cluster I (n = 58; 68% of all strains) were confirmed to be S. pneumoniae. All isolates that were not grouped in cluster I (n = 26; 31% of all strains) proved to be nonpneumococcal strains. Of these 26 isolates, 17 (65%) were Streptococcus mitis strains, which were all grouped in cluster II, and the remaining 9 (35%) were unidentifiable streptococcal species. Remarkably, one strain in the pneumococcal cluster (cluster I) in the AFLP dendrogram could not unequivocally be identified as S. pneumoniae by 16S sequencing but could only be designated as streptococcal species.
FIG. 1.
AFLP and PFGE dendrograms from 85 presumptive pneumococcal isolates. Two clearly discernible megaclusters containing either S. pneumoniae or S. mitis (clusters I and II, respectively) are present in the AFLP dendrogram. The identification results by 16S rRNA sequencing for all strains are also shown. Strains are sequentially numbered in the AFLP dendrogram; corresponding numbers appear in the PFGE dendrogram. The scale bar indicates the percentage of similarity.
The emergence of penicillin- and multiresistant pneumococcal isolates worldwide (4) necessitates continuous monitoring of the epidemiological spread of such strains. For this purpose, time-efficient and dependable fingerprinting techniques are essential. In the present study, we used AFLP versus PFGE for molecular typing of macrolide-resistant S. pneumoniae strains. Establishment of epidemiological relationships between pneumococcal strains was more easily established by AFLP than by PFGE. Moreover, AFLP analysis showed, in contrast to PFGE, the formation of clusters on a species level, allowing simultaneous discrimination between pneumococci and closely related species like S. mitis. Highly specific molecular biological methods like 16S sequencing provide new possibilities for more definite identification of S. pneumoniae. However, naturally occurring sequence variations within the 16S rRNA gene may complicate identification procedures, and it is not always clear where to draw the line between S. pneumoniae and genotypically similar species like S. mitis (6, 11). Clearly, this is not restricted to 16S analysis but holds true for any other single-gene method. In contrast, AFLP is a genome-wide analysis technique, much less influenced by naturally occurring minor sequence variations. In our study, one strain in the pneumococcal cluster (cluster I) in the AFLP dendrogram could not unequivocally be identified as S. pneumoniae by 16S sequencing but was only designated to be a streptococcus species. However, AFLP analysis showed that this isolate was closely related to the other pneumococci in cluster I. Therefore, this particular isolate should be designated as S. pneumoniae. AFLP analysis, showing a clear separation between pneumococcal and nonpneumococcal clusters in the dendrogram, can be used as an alternative method to 16S rRNA sequencing for a more definite identification of streptococcal isolates. Phylogenetically related species like S. mitis have reduced antimicrobial susceptibility patterns compared to S. pneumoniae, and failure to differentiate between these two species will significantly influence pneumococcal resistance rates (3, 10). This underscores the need for specific and dependable techniques for identification of S. pneumoniae in epidemiological studies. In contrast to PFGE, AFLP offers fully computerized data acquisition, which allows large numbers of isolates to be processed in a relatively short period of time. This makes this technique an efficient and dependable method for epidemiological fingerprinting of pneumococci. In summary, AFLP is an efficient alternative to PFGE for assessment of epidemiological relationships between pneumococcal isolates and is, in contrast to PFGE, effective in distinguishing S. pneumoniae from phylogenetically related species like S. mitis. In our opinion, AFLP analysis should be the preferred method for epidemiological fingerprinting of pneumococci.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler, L. J., B. S. Reisner, G. L. Woods, and A. K. Jafri. 2000. Comparison of four methods for identifying Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 37:285-287. [DOI] [PubMed] [Google Scholar]
- 3.Dobay, O., F. Rozgonyi, E. Hajdu, E. Nagy, M. Knausz, and S. G. Amyes. 2003. Antibiotic susceptibility and serotypes of Streptococcus pneumoniae isolates from Hungary. J. Antimicrob. Chemother. 51:887-893. [DOI] [PubMed] [Google Scholar]
- 4.Farrell, D. J., I. Morrissey, S. Bakker, and D. Felmingham. 2002. Molecular characterization of macrolide resistance mechanisms among Streptococcus pneumoniae and Streptococcus pyogenes isolated from the PROTEKT 1999-2000 study. J. Antimicrob. Chemother. 50(Suppl. 1):39-47. [DOI] [PubMed] [Google Scholar]
- 5.Hermans, P. W., M. Sluijter, T. Hoogenboezem, H. Heersma, A. van Belkum, and R. de Groot. 1995. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J. Clin. Microbiol. 33:1606-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaijalainen, T., S. Rintamaki, E. Herva, and M. Leinonen. 2002. Evaluation of gene-technological and conventional methods in the identification of Streptococcus pneumoniae. J. Microbiol. Methods 51:111-118. [DOI] [PubMed] [Google Scholar]
- 7.Kell, C. M., J. Z. Jordens, M. Daniels, T. J. Coffey, J. Bates, J. Paul, C. Gilks, and B. G. Spratt. 1993. Molecular epidemiology of penicillin-resistant pneumococci isolated in Nairobi, Kenya. Infect. Immun. 61:4382-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klaassen, C. H. W., H. A. van Haren, and A. M. Horrevorts. 2002. Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism. J. Clin. Microbiol. 40:101-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wester, C. W., D. Ariga, C. Nathan, T. W. Rice, J. Pulvirenti, R. Patel, F. Kocka, J. Ortiz, and R. A. Weinstein. 2002. Possible overestimation of penicillin resistant Streptococcus pneumoniae colonization rates due to misidentification of oropharyngeal streptococci. Diagn. Microbiol. Infect. Dis. 42:263-268. [DOI] [PubMed] [Google Scholar]
- 11.Whatmore, A. M., A. Efstratiou, A. P. Pickerill, K. Broughton, G. Woodard, D. Sturgeon, R. George, and C. G. Dowson. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Immun. 68:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]