Abstract
Mice homozygous for a mutation (BrdtΔBD1/ΔBD1) lacking the first bromodomain of Brdt, a testis-specific member of the BET family of double-bromodomain containing proteins, are sterile and exhibit profound defects in chromatin remodeling during spermiogenesis. We have now observed that a prominent feature of the aberrant spermatid nuclei is a fragmented chromocenter, a structure comprised of peri-centromeric heterochromatin. There was a concomitant increase in the levels of heterochromatin protein 1 alpha (Hp1α), suggesting that the presence of multiple chromocenters was correlated with a spread of heterochromatin beyond the normal centromeric region. Brdt protein was normally present throughout the nucleus but was excluded from the chromocenter. A more densely staining region of Brdt protein appeared to separate sirtuin 1 (Sirt1) protein from contact with the chromocenter. Although still nuclear, this unique localization of Brdt protein was lost in BrdtΔBD1/ΔBD1 mutant spermatids and Brdt and Sirt1 overlapped around the chromocenters. There was also ectopic localization of the H1 histone family, member N, testis-specific (H1fnt) protein in BrdtΔBD1/ΔBD1 round spermatids, which may be linked to the previously reported loss of polarized localization of peri-nuclear heterochromatin foci. The extent of chromocenter fragmentation was more severe and penetrant in mutant testes on a pure 129Sv/Ev as compared to a pure C57Bl/6 background. Indeed, all aspects of the mutant phenotype were more severe on the 129Sv/Ev background. Contrary to previous studies in genetic models where fragmented chromocenters were observed in spermatids, the BrdtΔBD1/ΔBD1 mutant spermatids do not undergo apoptosis (on either background). These observations suggest that the first bromodomain of Brdt is critical in the formation and/or maintenance of an intact chromocenter and implicate this structure in proper remodeling of the chromatin architecture of the sperm head.
Keywords: Brdt, bromodomain, chromocenter, spermiogenesis, Sirt1
Introduction
Spermiogenesis is a unique developmental process in which the male genome must be properly packaged in order to retain its integrity once exiting the body. During spermiogenesis dramatic changes occur in both chromatin architecture and cell morphology. The condensation and elongation of the sperm nucleus involves a highly regulated but poorly understood replacement of nucleosomes with transition proteins and then protamines (Govin et al., 2004). The first striking chromatin reorganization to occur after the completion of meiosis is the formation of the chromocenter. Starting in stage I round spermatids the chromocenter is formed by the coalescence of the centromeric heterochromatin of each chromatid in the center of the nucleus, while their telomeres are associated mostly with the nuclear envelope (Haaf and Ward, 1995; Meyer-Ficca et al., 1998; Zalensky et al., 1995). The chromocenter remains intact throughout spermiogenesis and can still be found at the center of the mature sperm nucleus (Hoyer-Fender et al., 2000; Zalensky et al., 1995). It has further been shown that the chromatin is not randomly organized around the chromocenter, but rather whole chromosome domains are arranged parallel to the chromocenter (Haaf and Ward, 1995) This has led to the hypothesis that the chromocenter creates a defined nuclear topology and acts as a starting point for the sequential reorganization of chromatin during spermiogenesis (Meyer-Ficca et al., 1998). The mechanisms for establishing and maintaining the chromocenter are not known.
The presence of multiple or ‘fragmented’ chromocenters has been shown to be correlated with mis-localization of the testis specific histone H1 family member N (H1fnt) (Catena et al., 2006). H1fnt is normally specifically localized to the apical pole of round and elongating spermatids where it suppresses the formation of perinuclear heterochromatin foci under the acrosome (Martianov et al., 2005). In two mutant mouse models, Tbpl1 -/- and Hmgb2 -/-, the presence of fragmented chromocenters is correlated with aberrant, bipolar localization of H1fnt (Catena et al., 2006). How aberrant localization of H1fnt affects latter stages of elongation and condensation is not known as the spermatids arrest and undergo apoptosis before the completion of spermiogenesis. In these two models, the process that gives rise to multiple or ‘fragmented’ chromocenters has not been investigated. Indeed, the fundamental question of whether an intact chromocenter is formed and is then fragmented or whether the individual centromeres fail to properly coalesce is not known.
Brdt is a testis-specific member of the BET family of chromatin interacting proteins, all of which contain two tandem bromodomains near the N-terminus and a C-terminal ET domain (Florence and Faller, 2001). There are two Saccharomyces cerevisiae BET genes, Bdf1 and Bdf2, of which Bdf1 is the most extensively studied of all BET genes. Bdf1 is required for meiotic division (Chua and Roeder, 1995), regulates transcription of small nuclear RNAs (snRNA) (Lygerou et al., 1994), is involved in deposition of the histone variant Htz1 (H2A.Z) (Krogan et al., 2003a; Mizuguchi et al., 2004; Raisner et al., 2005; Zhang et al., 2005), and by binding to acetyl-H4, competes with the Sir2 deacetylase to stop the spread of transcriptional silencing at constitutive heterochromatin-euchromatin boundaries (Ladurner et al., 2003; Matangkasombut and Buratowski, 2003). There are four mammalian BET genes, Brd2, Brd3, Brd4 and Brdt, each of which is expressed in the testis but in distinct patterns (Shang et al., 2004). Brd2 and Brd4 are essential genes as null mutants of either are embryonic lethal (Houzelstein et al., 2002; Shang et al., 2009). As Brdt is testis-specific, a mutation in the gene that completely removed the first bromodomain (BD1), BrdtΔBD1, did not affect viability, but rather caused complete male sterility (Shang et al., 2007).
Brdt expression is restricted to pachytene and diplotene spermatocytes and round spermatids; however, no obvious morphological defect was seen in these cells in BrdtΔBD1/ΔBD1 mutant testes (Shang et al., 2007). Rather, the first obviously visible defects were observed in stage IX spermatids which fail to properly elongate and the heterochromatin foci normally observed at the nuclear envelope were absent. The severity of the BrdtΔBD1/ΔBD1 phenotype in elongating spermatids and sperm varied between mice or even among tubules of a single testis. Some spermatids seemed to elongate fairly normally and some mice had epididymal sperm. Epididymal sperm were always grossly morphologically abnormal, with excess cytoplasm, misshapen heads, and deformed or truncated tails that often lacked the midpiece. The initial observations were made on BrdtΔBD1/ΔBD1 mutant mice that were maintained on a mixed genetic background of C57BL/6J (B6) and 129/SvEv (129), which we speculated may have contributed to this heterogeneity. We have therefore backcrossed the mutation onto genetically pure C57BL/6J and 129/SvEv mice, and investigated the phenotype in greater detail in each background. We have uncovered striking defects in the chromocenters of the mutant spermatids, increased heterochromatin, and ectopic expression of specialized histones—all of which may contribute mechanistically to the abnormalities in chromatin remodeling.
Materials and methods
Backcrossing to pure 129 and B6 backgrounds
A mixed 129Sv (129) × C57Bl6/J (B6) male heterozygous for the BrdtΔBD1 mutant allele was mated with two pure strain 129 or B6 females. A single heterozygous mutant male offspring was then mated with those same two females. This was repeated for five more generations after which two new pure strain females were used for two more generations. One final cross of a heterozygous mutant female offspring with a pure 129 or B6 male was carried out to insure that the Y-chromosome was also from the pure background. This process resulted in progeny that were ten generations backcrossed, and we considered these mice to now carry the BrdtΔBD1 mutant allele on a pure background.
Histological analysis, immunohistochemistry and immunofluorescence
Histological analysis and periodic acid Schiff (PAS) staining were carried out according to our laboratory's standard protocols as previously published (Chung and Wolgemuth, 2004; Shang et al., 2007). Bouin's fixed sections were used for Hematoxylin and Eosin staining, PAS staining and also for anti-trimethyl-histone H3 (Lys9) (H3K9me3) immunostaining. The H3K9me3 immunostaining used rabbit polyclonal primary antibody (Upstate, cat#07-523) at a concentration of 1:200 and the Vectastain ABC Kit-Rabbit IgG (Vector Laboratories, Inc.). Four percent paraformaldehyde (PFA) fixed sections were used for immunofluorescence with H1fnt, H4, and H2AZ antibodies and immunostaining with Hmgb2 antibodies. The rabbit polyclonal anti-H1fnt primary antibody (Santa Cruz, cat# sc-136700) was used at 1:200, the goat polyclonal anti-H4 primary antibody (Upstate, cat# sc-8657) was used at 1:300, the rabbit polyclonal anti-H2A.Z primary antibody (AbCam, cat# 4174-100) was used at 1:200. The rabbit polyclonal anti-Hmgb2 primary antibody (Epitomics, cat# T2134) was used at 1:200. The following secondary antibodies were used: Alexa Fluor-488 donkey anti-goat IgG (Molecular Probes, cat# A11055) at 1:300, Alexa Fluor- 488 goat anti-rabbit IgG (Molecular Probes, cat #11008) at 1:300, and Alexa Fluor-594 goat anti-rabbit IgG (Molecular Probes, cat# 11012) at 1:300. The Vectastain ABC Kit-Rabbit IgG (Vector Laboratories, Inc.) was used for immunodetection of Hmgb2.
To investigate subcellular localization of Brdt, a single cell suspension of spermatogenic cells was made from wild-type and mutant testes by removing the tunica albuginea and placing the tubules in cold PBS. The tubules were manually sheared with scissors and by pipetting, and then passed through a 40 μm filter. The resulting suspension was dropped on slides, air dried, and then fixed in 4:1 methanol: acetone for 10 minutes. Two Brdt antibodies generated by our lab (Shang et al., 2007) were both used at a concentration of 1:300 and yielded identical results. The rabbit polyclonal anti-Sirt1 primary antibody (AbCam, cat# ab12193) was used at a concentration of 1:200. The following secondary antibodies were used: DyLight 594 Fab fragment donkey anti-rabbit IgG (Jackson ImmunoResearch cat# 711-517-003 04) at 1:300 and Alexa Fluor- 488 goat anti-rabbit IgG (Molecular Probes, cat#11008) at 1:300. The staining was done sequentially, first with anti-Sirt1, followed by DyLight 594, then anti-Brdt, then Aleza Flour-488, followed by DAPI.
Quantification of fragmented chromocenters
PFA fixed histological sections for both genotypes on both backgrounds where prepared by the histology technician and labeled numerically, without identifying genotypes or strain background. The investigators then stained the sections with anti- H3K9me3 and PAS and proceeded with the quantitation of the numbers of chromocenters per round spermatid in 100 tubules, without knowledge of the genotype or background of the sections. A cell was counted as a round spermatid if it had a visible acrosome, as having a fragmented chromocenter if there were three foci of H3K9me3 staining, and as having a severely fragmented chromocenter if there were four or more foci. Statistical significance was assessed using a standard two-tailed T-test.
Immunoblot analysis
For immunoblot analysis of components in the testicular cell chromatin our previously published procedures were followed (Shang et al., 2007). Briefly, a single cell suspension for control and mutant 129 testes were made as described above. The cells were pelleted at 2500 g for 10 minutes, and homogenized in modified RIPA buffer [50 mM Tris-Hcl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and protease inhibitor cocktail (Roche)], incubated for 30 min at 4°C, and centrifuged at 12,000 g for 10 min at 4°C. The pelleted chromatin was boiled for 5 minutes in 1X Nu PAGE loading buffer (Invitrogen). Cellular lysates from whole 129 and B6 testes were made with standard RIPA buffer and boiled for 5 minutes in 1X Nu PAGE loading buffer (Invitrogen). Chromatin preparations and lysates were separated on NuPAGE 4-12% Bis-Tris gels (Invitrogen), transferred to PVDF membranes and the blots were incubated with rabbit polyclonal anti-Fibrillarin antibody (GenScript, cat# A01462) at a concentration of 1:3000, goat polyclonal anti-Hp1α antibody (Abcam, cat# ab77256) at 1:3000, goat polyclonal anti-H4 antibody (Upstate, cat# sc-8657) at 1:3000, mouse monoclonal anti-β-actin antibody (Sigma, cat#A5316-.2ML) at 1:5000, and rabbit polyclonal anti-Hmgb2 antibody (Epitomics, cat# T2134) at 1:3000.
Quantitative real-time PCR
To preform quantitative real-time PCR, whole testes from control 129 and B6 mice were homogenized in Trizol reagent (Invitrogen) and we extracted RNA according to the manufacturer's protocol. cDNA was synthesized using random hexamers and the TaqMan Reverse Transcription Kit (Applied Biosystems) and qRT-PCR performed with the Power SYBR Green master mix (Applied Biosystems) on a ABI Prism 7500 (Applied Biosystems). Each reaction was performed in triplicate and with RNA from three different mice from each background. The values were normalized to the expression of ribosomal protein, large, PO (Rplp0) as an internal control. The following primers were used: Hmgb2 Forward: GCTCGTTATGACAGGGAGATG; Hmgb2 Reverse: TTGCCCTTGGCACGGTATG; Tbpl1 Forward: TTTGGTGCCAGACGTTTAGC; Tbpl1 Reverse: GCCACAGCCTTTACATTGGG, Rplp0 Forward: CAAAGCTGAAGCAAAGGAAGAG; Rplp0 Reverse: AATTAAGCAGGCTGACTTGGTTG.
Sperm counts
Sperm were collected into non-capacitation medium (10 mM HEPES, 95 mM NaCl, 4.8 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 20 mM Sodium Lactate, 5 mM Sucrose, pH 7.4) at 37° from both caudal epididymides of individual animals as per our laboratory's standard protocol as previously published (Chung et al., 2011; Travis et al., 2001). Briefly, cauda were manually sheared with scissors and forceps. The solution was then passed through a 100 μm filter and medium was added up to 1ml. For sperm suspensions from control and heterozygous mice from 129 animals and all cauda from B6 animals the sperm solution was then diluted 1:10 in medium and counted on a hemicytometer under an inverted light microscope The 1:10 dilution was not needed to count the 129 mutant sperm. The individual counting the sperm did not know the genotype or background of the sperm to avoid bias.
Results
Mutant round spermatids exhibit fragmented chromocenters
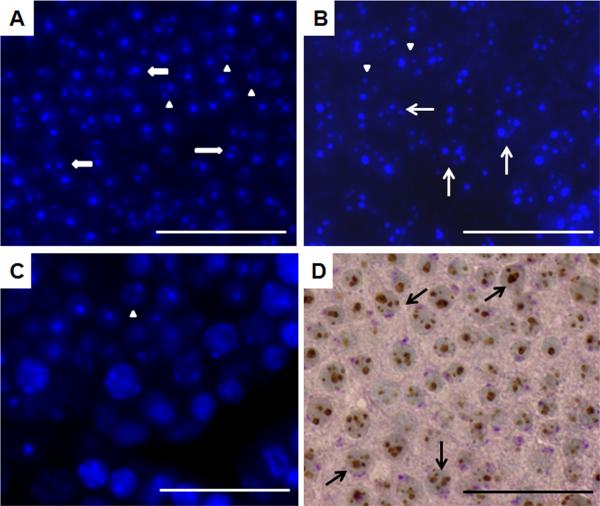
In our initial characterization of the phenotype of BrdtΔBD1/ΔBD1 mutant mice on a mixed 129/B6 background, heterogeneity was noted in the testicular morphology between mutant mice, as well as among tubules in a single testis. To assess if genetic background contributed to this heterogeneity, we backcrossed the mutation onto pure 129 and B6 backgrounds. Histological analysis of the mutant testes on the 129 background revealed a heretofore unreported component of the mutant phenotype--a striking increase in the presence of fragmented chromocenters in round spermatids. Since the earliest defect we had previously observed in the BrdtΔBD1/ΔBD1 mutant testis occurred in stage IX elongating spermatids, we initially examined the testes of 24 day-old animals, as at this age no tubules would have developed beyond stage VIII. DAPI-stained control 129 testes revealed that the majority of spermatids contain a single focus of dense heterochromatin or one large focus (the chromocenter) and a smaller lighter secondary focus (the nucleolus) (Fig. 1A, wide arrows), along with a low basal level of fragmented chromocenters--defined as those spermatids that had three or more foci (Fig 1A, arrowheads; Table 1). In striking contrast, the majority of round spermatids in mutant 129 testes contain severely fragmented chromocenters, with most spermatids exhibiting at least three and frequently four or more foci of heterochromatin (Fig. 1B, arrows; Table 1).
Figure 1. BrdtΔBD1/ΔBD1 round spermatids exhibit severe fragmentation of the chromocenter.
(A-C) DAPI staining of histological sections from post-natal day 24 testes (A) 129 control, (B) mutant and (C) B6 mutant. Wide arrows: two foci of heterochromatin, normal. Arrowheads: three foci, fragmented chromocenter. Arrows: four or more foci, severely fragmented chromocenter. The number of spermatids with multiple chromocenters seen in the mutant 129 testes is greater than that seen in control or in the B6 mutant (also see Table 1). (D) Immunostaining of histone 3 trimethylated on lysine 9 and periodic acid Schiff (PAS) staining of the acrosome of day 24 mutant 129 testis. The cells with severe fragmentation also have acrosomes and thus are round spermatids (arrows). Scale bar, 50 μm.
Table 1. Frequency and severity of fragmented chromocenters in BrdtΔBD1/BD1 mutant round spermatids is background dependent.
In both backgrounds there is a statistically significant increase in the number of spermatids with fragmented chromocenters in the mutants (pValue ≤ 1.0×10-6 for B6 and ≤ 5.0×10-8 for 129). There is also a statistically significant difference in basal level of fragmented chromocenters between the two backgrounds (pValue ≤ 5.0×10-3). The difference in the number of fragmented chromocenters in the mutants of the two backgrounds is significant (pValue ≤ 5.0×10-7) as is the difference in the number of severely fragmented chromocenters (pValue ≤ 5.0×10-17).
| B6 +/+ | B6 ΔBD1/ΔBD1 | 129 +/+ | 129 ΔBD1/ΔBD1 | |
|---|---|---|---|---|
| Total nubmer of round spermatids (RS) per tubule of 100 tubules examined | 128.4±15.1 | 115.5±11.2 | 123.8±7.5 | 100.6±9.7 |
| RS with non-fragmented chromcenters (avr. per tubule of 100 tubules examined) | 119.5±13.1 | 84.3±6.9 | 104.3±5.4 | 33.5±4.0 |
| RS with fragmented chromocenters (avr. per tubule of 100 tubules examined) | 8.9±2.0 | 31.2±4.3 | 19.5±2.1 | 67.1±5.7 |
| RS with severely fragmented chromocenters (avr. per tubule of 100 tubules examined) | 0.2±0.1 | 6.4±0.8 | 2.6±0.6 | 55.1±4.6 |
| Percent of RS that have fragmented chromocenters (≥3 heterochromatin foci) | 7% | 23% | 17% | 67% |
| Percent of RS that have severely fragmented chromocenters (≥4 heterochromatin foci) | <1% | 6% | 2% | 55% |
As secondary spermatocytes also exhibit foci of heterochromatin (Cobb et al., 1999), it was critical to ensure that the cells in question were indeed round spermatids. We therefore performed PAS staining which permitted visualization of the acrosome. To concomitantly visualize the chromocenters, the sections were also stained with anti- H3K9me3 (Greaves et al., 2006). Cells with severely fragmented chromocenters did indeed have developing acrosomes and thus could only be spermatids (Fig 1D, arrows).
Brdt protein is nuclear but excluded from the chromocenter
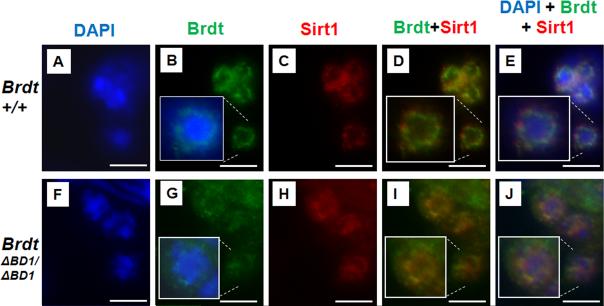
Our initial studies clearly showed that Brdt is a nuclear protein and associated with chromatin (Shang et al., 2007). Given the new observation of chromocenter abnormalities, we characterized the distribution of Brdt protein within sub-nuclear regions of spermatids using fluorescent immunostaining. Brdt protein localized throughout the nucleus as predicted, but did not extend into the dense region of the chromocenter (Fig. 2B). With these preparations, Brdt protein appeared to be more concentrated immediately surrounding the chromocenter (Fig. 2B, inset). In contrast, although the truncated protein produced in BrdtΔBD1/ΔBD1 mutant spermatids localized to the nucleus, it is more evenly distributed throughout the nucleus (Fig. 2G, inset). This observation suggests that the BD1 is involved in determining the sub-nuclear association of Brdt to specific chromatin.
Figure 2. Dense Brdt expression separates the chromocenter from Sirt1 protein.
Fluorescent staining of control (A-E) and mutant (F-J) round spermatids with Brdt and Sirt1 antibodies and the merge of the two.(A-B) Brdt is present in the nucleus of round spermatids, with regions of denser staining surrounding the chromocenter from which it is excluded (inset merge of Brdt and DAPI). (C) Sirt1 is also present in the nucleus of round spermatids, and is also excluded from the chromocenter (inset in E). (D-E) Brdt and Sirt1 protein localization overlap except for the region directly surrounding the chromocenter where Brdt is strongly detected but Sirt1 is excluded (green fluorescence only). (F-G) Truncated Brdt protein is still present in the nucleus of mutant round spermatids, but the denser staining around the chromocenter is no longer seen (inset merge of Brdt and DAPI). (H) Sirt1 expression is still nuclear and excluded from the chromocenter. (I-J) Brdt and Sirt1 protein localization overlap completely (no exclusive green fluorescence), and both are present directly adjacent to the chromocenter. While there are still some regions of dense Brdt staining, it is no longer localized around the chromocenter. Scale bar, 12.5 μm.
Dense Brdt expression separates the chromocenter from sirtuin 1 protein
Sirtuin 1 (Sirt1) is the mammalian homologue of the yeast histone deacetylase Sir2 and like Sir2, is part of the machinery that keeps constitutive heterochromatin transcriptionally silenced (Palacios et al., 2010). Acetylated H3 lysine 9 is one of the targets of Sir2 deacetylase activity (Imai et al., 2000). Deacetylation of H3 by Sir2 can be a required step before tri-methylation of lysine 9 by histone methyltransferase Su(var)3-9 (Ekwall, 2004; Shankaranarayana et al., 2003). The chromodomain of heterochromatin protein 1 binds to tri-methylated H3K9, making this histone modification a characteristic hallmark of heterochromatin (Bannister et al., 2001; Lachner et al., 2001). The mammalian homologues of the methyltransferase, Suv39h1, and the testis-specific methyltransferase Suv39h2, are both present in round spermatids where they are restricted to the chromocenter (O'Carroll et al., 2000). Immunostaining to determine the localization of Sirt1 revealed that it was indeed expressed in round spermatids, but was completely excluded from the chromocenter (Fig. 2C, and insets in 2D,E). Double-staining for Sirt1 and Brdt showed that the dense region of Brdt staining around the chromocenter separated the region of Sirt1 localization from the chromocenter proper (Fig. 2D-E, green fluorescence only, insets in 2D,E). This created essentially a ‘buffer’ region between the chromocenter and Sirt1 protein. In mutant round spermatids, where there is no region of denser, peri-chomocentric Brdt staining, this buffer was lost and Sirt1 now also localized adjacent to the chromocenter (Fig. 2H-J, no exclusive green fluorescence).
Localization of the histone variant H2A.Z is unchanged in mutant spermatids with fragmented chromocenters
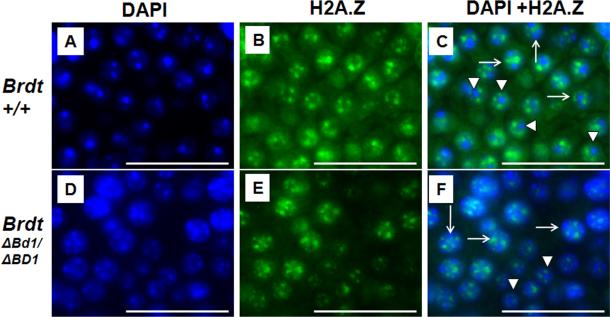
BET proteins have been implicated in delimiting heterochromatin – euchromatin boundaries in the yeast model wherein Bdf1 is involved in the deposition of the histone variant Htz1 (Krogan et al., 2003b; Mizuguchi et al., 2004; Raisner et al., 2005; Zhang et al., 2005). Loss of the mammalian homologue of Htz1, H2A.Z, results in the spread of heterochromatin into areas of euchromatin (Meneghini et al., 2003). We therefore asked if the sub-cellular localization of H2A.Z was altered in the mutant spermatids. In control round spermatids, H2A.Z surrounded the chromocenters but was excluded from the dense central region, similar to Brdt (Fig. 3C, arrowheads). In those round spermatids that had fragmented chromocenters, H2A.Z still surrounded the multiple regions of heterochromatin (Fig. 3C, arrows). This same pattern of H2A.Z localization was seen in mutant round spermatids: the intact chromocenters were still surrounded by H2A.Z (Fig. 3F, arrowheads), and severely fragmented chromocenters still had H2A.Z that was localized around the heterochromatin (Fig. 3B, arrows).
Figure 3. Localization of H2A.Z around the chromocenter is unchanged in BrdtΔBD1/ΔBD1 mutant round spermatids.
Fluorescent staining of DAPI (A,D), H2A.Z (B,E) and merge (C,F) in control (A-C) and mutant (D-F) day 24 histological sections. (A-C) In control spermatids H2A.Z is localized around the intact chromocenter (arrowheads). Fragmented chromocenters still have H2A.Z surrounding the foci of heterochromatin (arrows). (D-F) In mutant round spermatids H2A.Z is localized normally, both around intact chromocenters (arrowheads) and severely fragmented chromocenters (arrows). Scale bar, 50 μm.
H1fnt is ectopically localized in mutant round spermatids
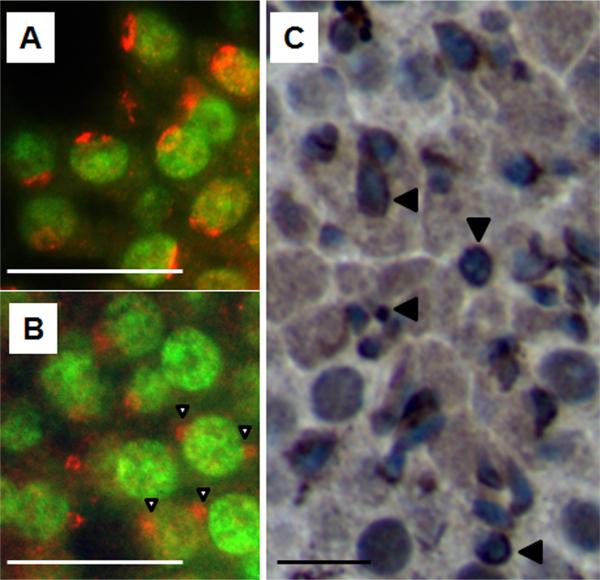
The histone 1 variant H1fnt is uniquely expressed in round spermatids, where it localizes at the apical pole in the subacrosomal region and remains there throughout spermatid elongation. An aberrant, ectopic deposition of H1fnt (described as ‘bipolar’) which is detectable in a small but distinct subset of late elongating spermatids (Stage VII-VIII) was reported in mutant strains of mice characterized as displaying fragmented chromocenters (Catena et al., 2006; Martianov et al., 2005). Fluorescent staining of histological sections of testes from young (24-day) 129 mice, which are enriched for stage VII-VIII round spermatids, was used to determine the localization of H1nft. The expected apical polar distribution of H1fnt was observed in round spermatids of the control testes (Fig.4A). In the mutants, however, the striking bipolar localization described in the other models was now also observed in late round spermatids (Fig. 4B, open arrowheads). Additionally, we observed that in later stage mutant tubules (from adult testes), many of the still round or partially elongated spermatids showed H1fnt expression surrounding the entire nucleus (Fig. 4C, arrowheads). This suggests that while H1fnt was localized in a bipolar manner in some mutant stage VII-VIII round spermatids, its localization has expanded in later stage improperly elongating spermatids.
Figure 4. H1fnt is ectopically localized in some 129 mutant round spermatids.
(A-B) Fluorescent staining to localize H1fnt in day-24 histological sections reveals a single region of apical expression in control 129 round spermatids (A) and ectopic localization in mutant 129 round spermatids (B, open arrowheads). (C) Immunostaining of H1fnt in a mutant 129 adult histological section shows non-elongated mutant spermatids with H1fnt surrounding the nucleus (arrowheads). Scale bar, 25 μm.
Levels of heterochromatin are elevated in mutant testes
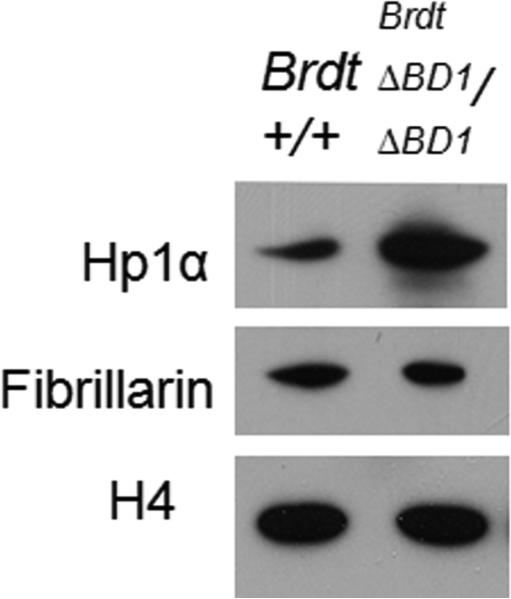
To understand the mechanisms by which loss of the first bromodomain of Brdt results in multiple chromocenters, we asked whether the fragmented chromocenters might actually reflect an increase in the overall amount of heterochromatin in mutant spermatids. Alternatively, if the aberrant structures were the result of a failure of the centromeres to coalesce properly, the amount of heterochromatin in the spermatids would be unchanged. In the Tbpl -/- mutant model, multiple chromocenters were visualized by staining for Hp1α, a known component of heterochromatin but the total levels of Hp1α in control and mutant chromatin were never measured (Martianov et al., 2002). We therefore extracted chromatin from both control and mutant testes of 129 mice and measured the levels of Hp1α protein. We observed a greater than three-fold increase in the levels of Hp1α protein in chromatin from the mutant testes (Fig. 5). In nuclei of round spermatids, HP1α is found both at centromeres and also in the nucleolus (Horakova et al., 2010). To determine whether increased levels of Hp1α were deposited at both sites, we measured levels of fibrillarin, a nucleolar structural protein (Fig. 5). Levels of fibrillarin were unchanged in the mutant testes, suggesting that the number of nucleoli is the same and thus that the increased levels of Hp1α reflected increased heterochromatin at the centromeric regions.
Figure 5. There is an increase in the amount of heterochromatin in the mutant testes.
Chromatin was extracted from control and mutant spermatogenic cells and immunoblotted with anti-Hp1α antibody, anti-fibrillarin antibody and anti-H4 antibody. Over 3-fold more Hp1α is present in mutant chromatin. Fibrillarin levels were unchanged in the mutant testes. H4 was used as a loading control.
All aspects of the BrdtΔBD1/ΔBD1 mutant phenotype detected are background dependent
Fragmented chromocenters
Fragmented chromocenters were also observed in the round spermatids of homozygous mutants on the B6 background, however at a lower frequency (Fig 1C, arrowheads). The number of spermatids with fragmented chromocenters was quantified for the various genotypes on both genetic backgrounds (Table 1). The basal level of fragmented chromocenters was significantly higher in the control 129 background than in the control B6 background (17% vs 7%; pValue ≤ 5.0×10-3). The number of cells exhibiting fragmented chromocenters in the BrdtΔBD1/ΔBD1 mutant on the 129 background was significantly increased compared to the controls (67% vs 17%; pValue ≤5.0 × 10-8), and also compared to the BrdtΔBD1/ΔBD1 mutant on the B6 background (67% vs 23%; pValue ≤5.0 × 10-7). Severely fragmented chromocenters were rarely seen in control spermatids of either the 129 (2%) or the B6 (<1%) backgrounds but were more frequent in the BrdtΔBD1/ΔBD1 mutants, significantly so in the129 background (55% vs 6% pValue ≤5.0×10-17) (Table 1).
Spermatid elongation
Starting in stage IX, none of the spermatids in the 129 mutant testes elongate properly. Even in later stages of spermiogenesis, many spermatids remain round with no obvious change to the morphology of the nucleus (Fig. 6C arrow, and inset). Those cells that do begin to elongate remain aberrantly short and do not develop the characteristic hook that should be seen in later stages (Fig.6E, and G). Compaction of the chromatin appears to be occurring as the size of the nucleus is greatly reduced when compared to round spermatids; however, without proper elongation this results in the presence of abnormal, very small round cells (Fig. 6C, E, G-arrowheads, inset in E and G). These cells are unlikely to be anything other than spermatids as the progression from the presence of round spermatids to the appearance of small, non-elongate spermatids can be followed directly in testes from mutant mice in the first wave of spermatogenesis that occurs during ensuing days of post-natal development. All of the various abnormalities described in the mixed background mutant testes (Shang et al., 2007) are now consistently seen in all tubules of strain 129 mice homozygous for the BrdtΔBD1 mutation (Table 2). Homozygous male mutants on the B6 background were still infertile but spermiogenesis proceeded more normally. Although aberrant elongation can still be seen in mutant tubules (Fig. 6D, F and H arrowheads, and inset in D and F), approximately 70% of the tubules contain some spermatids that appear to be elongating and compacting fairly normally and some later stage tubules look grossly morphologically normal (Fig. 6H). Some aberrantly short spermatids are present and 85% of all tubules still contain non-elongated small compacted spermatids (Fig. 6D, F, H arrowheads, and inset in D and F), but these cells are not prevalent (Table 2).
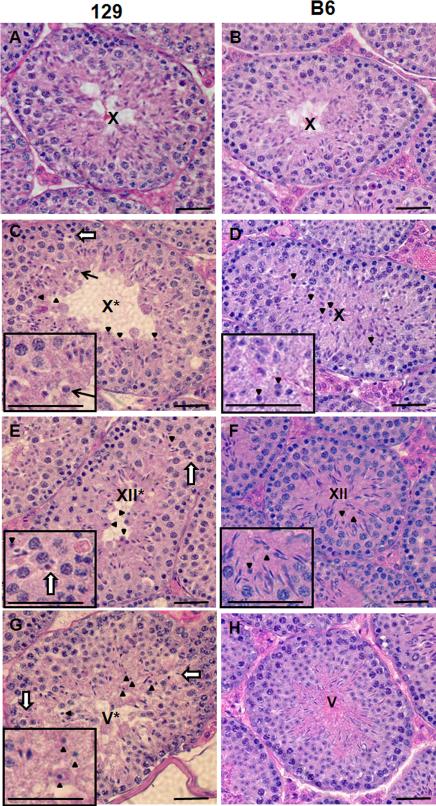
Figure 6. The abnormalities in BrdtΔBD1/ΔBD1 mice are more severe and uniform on the 129 background.
(A-H) H&E staining of histological sections of (A) control129 stage X tubule, (B) control B6 stage X tubule, (C) mutant 129 stage X* tubule, (D) mutant B6 stage X tubule, (E) mutant 129 stage XII* tubule, (F) mutant B6 stage XII tubule, (G) mutant 129 stage V* tubule, and (H) mutant B6 stage V tubule. Grossly normally elongating spermatids are absent in the mutant 129 testes, but predominate the B6 mutant testis. Conversely, compacted but not elongated spermatids are present (C-H, arrowheads and D-G inset) in B6 testes, but they predominate the 129 testes. (see Table 2 for quantification). Spermatids which have neither compacted nor elongating (C, arrow and inset) can be found only in the 129. In 129 testes some spermatids can be found in the spermatocyte layer of the tubules (C,E, and G, open arrows and E inset). Asterisk: although the severe disruption of the order of the seminiferous epithelium in the 129 mutant made precise staging difficult, we refer to approximately staged tubules with a roman numeral followed by an asterisk. Scale bar, 25 μm.
Table 2. Summary of morphological abnormalities in BrdtΔBD1/ ΔBD1 mutant mice.
BrdtΔBD1/ ΔBD1 mutant mice on the 129 background never have normally elongating spermatids and all tubules contain very small condensed but not elongated spermatids. BrdtΔBD1/ ΔBD1 mutant tubules on the B6 background often contain normally elongating spermatids, but only 15% of all tubules look grossly morphologically normal. Defects in the order of the seminiferous epithelium occur at a greater frequency in 129 mutant tubules than in control tubules.
| B6 +/+ | B6 ΔBD1/ΔBD1 | 129 +/+ | 129 ΔBD1/ΔBD1 | |
|---|---|---|---|---|
| Percent of stage IX-XII tubules containing any normally elongating spermatids | 100% | 73% | 100% | 0% |
| Percent of all tubules containing very small compacted but not elongated spermatids | 0% | 85% | 0% | 100% |
| Percent of stage IX-XII tubules containing round spermatids | 0% | 0% | 0% | 12% |
| Percent of all tubules containing condensed spermatids in the spermatocyte layer | 2% | 9% | 4% | 23% |
| Percent of all tubules containing round spermatids in the tubule lumen | 2% | 3% | 1% | 23% |
Disruption of cellular associations
We also observed that there was a loss of order of the cellular layers in the 129 mutant seminiferous epithelium which had not been reported in the mutant on a mixed background (Shang et al., 2007). For example, round and aberrantly elongating spermatids can be seen in the spermatocyte layer of the tubule (Fig. 6C, E, G- open arrows, inset in E) and round spermatids are sometimes found in the very center of tubules (Table 2). In contrast, on the B6 background, the loss of order of the seminiferous epithelium was observed at a low frequency, comparable to control testes, and these phenotypes were consistent with all tubules containing overtly properly elongating spermatids (Table 2).
Epididymal sperm
We had previously reported that there was heterogeneity in the presence of epididymal sperm on a mixed background: some animals had considerable numbers of epididymal sperm while others had very nearly empty epididymides (Shang et al., 2007). Histological examination of the 129 mutant animals consistently revealed that all three major parts of the epididymis were virtually devoid of sperm, with only occasional sloughed off spermatocytes and round spermatids and cytoplasmic bodies and very small and round spermatids (Fig. 7A). In contrast, on the B6 background sperm are consistently present in the three regions of the epididymis (Fig. 7A). Morphologically, there was still a great deal of heterogeneity. Some sperm looked fairly ‘normal’, with close to the proper hook shape to the head and containing midpieces and full tails (Fig. 7C arrow). Others however, were highly aberrant and resembled the 129 mutant sperm (Fig. 7C).
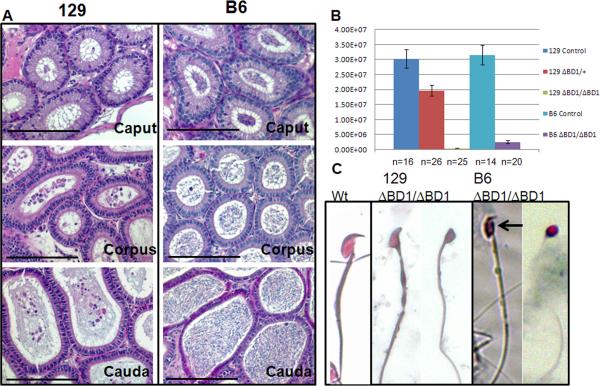
Figure 7. Differences in epididymal sperm content of BrdtΔBD1/ΔBD1 mutants on 129 and B6 backgrounds.
(A) H&E staining of histological sections of mutant 129 and mutant B6 caput, corpus and caudal epididymides, showing the nearly complete absence of sperm in the 129 animal and the abundance of sperm in the B6 animal. (B) Caudal sperm counts from control, heterozygous and homozygous mutant 129 mice and control and homozygous mutant B6 mice. There is a statistically significant loss of sperm in the 129 heterozygous cauda compared to control and a statistically significant difference between the 129 and B6 mutant counts. (C) Light microscope photomicrographs of 129 control and BrdtΔBD1/ΔBD1 and B6 BrdtΔBD1/ΔBD1 sperm. Although all sperm are abnormal, there is greater heterogeneity in the B6 sperm, with some sperm that appear close to normal (arrow). Scale bar, 50 μm.
Correspondingly, caudal epididymal sperm counts were extremely low. Mutant 129 epididymides had an average of 4.02×105 total sperm (roughly 4 sperm per slide), which was 10-fold below the bottom threshold for fertility (4×106) (Meistrich, 1982; Reid et al., 1981) and 75-fold below the 129 control average (3.03×107) (Fig. 7B). The few sperm that were present in the 129 epididymis appear very abnormal with excess cytoplasm, misshapen heads, and deformed tails and midpieces (Fig. 7C). Interestingly, the average sperm count for the heterozygous BrdtΔBD1/+ animals on the 129 background was only 1.96×107 or about two-thirds that of the wild-type. Using a standard two-tailed t-test this was a significant loss of sperm at a pValue of 0.005. This is the first observed heterozygous phenotype; however, this number of sperm is still well above the threshold of fertility. In BrdtΔBD1/ΔBD1 mutants on the B6 background, caudal sperm counts yielded an average of 2.41×106 sperm, also below the threshold for fertility but almost six-fold more than the 129 mutant (Fig. 7B). This difference in the number of sperm between the two mutants was statistically significant by a two-tailed t-test at a pValue of 0.5×10-6.
Hmgb2 is expressed at higher levels in B6 round spermatids
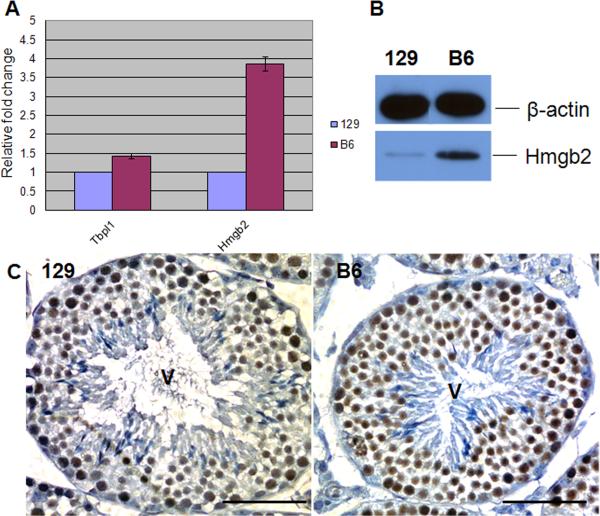
Two other genes, Tbpl1 and Hmgb2, have been shown by targeted mutagenesis to be required for proper chromocenter formation and/or maintenance (Catena et al., 2006). To ask whether either protein may be involved in the difference in the levels of fragmented chromocenters detected between the two backgrounds in both control and BrdtΔBD1/ΔBD1 mutant spermatids, we investigated their expression in testes from the two backgrounds. Quantitative real-time PCR revealed that expression of Tbpl1 was comparable in the two backgrounds, but that Hmgb2 expression was nearly 4-times higher in the B6 testes (Fig. 8A). Immunoblotting with anti-Hmgb2 antibody showed that this mRNA difference was also reflected in the protein levels (Fig. 8B). Immunostaining of histological sections of testes from both strains placed side by side on one slide, confirmed that anti-Hmgb2 antibodies localized more intensely in both round spermatids and pachytene spermatocytes in the B6 testis as compared to 129 testes (Fig. 8C).
Figure 8. Hmgb2 is expressed at higher levels in the B6 testis.
(A) Quantitative real-time PCR showing that Hmgb2 but not Tbpl1 transcription is higher in the B6 testis than in the 129 testis. The expression levels are shown as relative levels of the 129 expression which is set as 1. (B) Western blot of Hmgb2 confirming 4 times greater protein in the B6 testis. β-actin is used as loading control. (C) Hmgb2 expression in stage V tubules. Protein levels are higher in the B6 cells, specifically in round spermatids. Scale bar, 50 μm.
Discussion
This study reports heretofore unknown chromatin defects in BrdtΔBD1/ΔBD1 mutant spermatids and also illustrates the strongly genetic background-dependent nature of the phenotype. Our earlier analysis of the BrdtΔBD1/ΔBD1 mutant phenotype on a mixed 129/B6 background focused on defects in spermatid elongation and chromatin condensation (Shang et al., 2007). Upon breeding the mutation onto a genetically pure 129 background, a striking and highly penetrant phenotype was revealed—that of a high proportion of round spermatids with multiple foci of dense heterochromatin, indicative of fragmented chromocenters. There were still no apparent defects during meiosis, raising the possibility that ΔBD1 may represent a hypomorphic allele. It is interesting to recall that in the yeast model, mutations in Bdf1 that were only in the first bromodomain did not have an effect on sporulation (Chua and Roeder, 1995). Rather it was BD2 and the ET domain that were required. It is possible that we have yet to detect a subtle defect in pachytene spermatocytes, and we are continuing to investigate meiosis in the mutant testis. However it is equally possible that the truncated Brdt protein is sufficient for proper meiosis and progression to round spermatids.
Although genes involved in chromatin organization, such as Tbpl1 and Hmgb2, are required for proper formation and/or maintenance of the chromocenter (Catena et al., 2006), the mechanisms involved are as yet unknown. The question of whether fragmented chromocenters result of a spreading of centromeric heterochromatin or alternatively, a failure of the centromeres to coalesce into one area is also not known. What also emerges from the studies on our BrdtΔBD1/ΔBD1 mutant is that the term “fragmented chromocenter” is somewhat misleading. That is, multiple heterochromatin foci are seen as early as Stage I in our mutant, and the frequency of their appearance does not change throughout round spermatid development. It therefore does not seem that a single chromocenter is forming and then fragmenting apart, but rather that an intact singular chromocenter does not form. We propose that going forward, this phenomenon should be termed ‘multiple chromocenters’ because although the centromeric heterochromatin has partially coalesced, it did not form one unique chromocenter.
In our BrdtΔBD1/ΔBD1 mutant, the increased levels of HP1α in chromatin suggest that there is an expansion of heterochromatin. No such increase in the levels of HP1α was reported in either Hmgb2-/- or Tbpl1-/- mutant testes (Catena et al., 2006) although this may not have been investigated. The strong effect that genetic background has on the chromocenter defect, in combination with the differing levels of Hmgb2 expression between the two strains, suggests that in the 129 mutant animals, the lower levels of Hmgb2 may exacerbate the presence of multiple chromocenters. Indeed there is a slightly higher basal number of multiple chromocenters in control 129 round spermatids compared to control B6 spermatids. The loss of function of the first bromodomain of Brdt not only increases the frequency with which multiple chromocenters occur (total number of cells with multiple chromocenters), but also increases the number of partial chromocenters per cell (number of cells with 4 or more foci of heterochromatin). We do not yet know whether Brdt and Hmgb2 work in the same pathway to create and/or maintain the chromocenter or if there are multiple independent processes at work. For example, it is possible that the abnormal chromatin architecture in the Tbpl1 and Hmgb2 mutants may cause a failure of the centromeres to position together, but that the aberrant chromocenters in Brdt mutant spermatids result from expansion of centromeric heterochromatin. In Tbpl1 and Hmgb2 mutants, the defects in round spermatids lead to wide-spread apoptosis in elongating spermatids with almost no sperm being produced. In our BrdtΔBD1/ΔBD1 mutant, a wave of apoptosis is not observed, and although 129 animals produce almost no sperm, sperm (albeit highly abnormal) can be found in both mixed and B6 animals. While the properties of the multiple chromocenters are grossly similar among the three mutant models (BrdtΔBD1, Tbpl1, and Hmgb2), the mechanisms involved may be quite distinct and may contribute to the higher level of spermatid survival in the BrdtΔBD1/ΔBD1 mutants.
It has been previously reported in Saccharomyces cerevisiae that the BET family homologue Bdf1 binds acetylated H4 and H3 at the heterochromatin-euchromatin boundary outside of telomeres and the mating locus and competes with the Sir2 deacetylase (Ladurner et al., 2003). It is speculated that the physical presence of Bdf1 on chromatin halts the spreading of Sir2 and as a result the spreading of constitutive heterochromatin. It is important to note that in Saccharomyces cerevisiae centromeres do not have large heterochromatic repeats and thus telomeres and the mating locus are the only constitutive heterochromatin in that organism (Buhler and Gasser, 2009). In Schizosaccharomyces pombe, which has silenced constitutive heterochromatin at the centromere, the Sir2 homologue spSir2 also functions to silence the centromeric heterochromatin (Shankaranarayana et al., 2003). The first bromodomain of Brdt has been shown to strongly bind acetylated H4 and to a lesser extent acetylated H3 (Moriniere et al., 2009) in much the same way as the first bromodomain of Bdf1. We have now shown that Brdt is localized around the centromeric regions that form the chromocenter, which is also a constitutive heterochromatin-euchromatin boundary, and that this localization is dependent on the presence of the first bromodomain. This supports our hypothesis that Brdt is binding to acetylated H4 or H3 in the same way as Bdf1. We also showed that a mammalian Sir2 homologue, Sirt1, localized within euchromatin around the chromocenter but was excluded from the region of stronger Brdt localization adjacent to the chromocenter. In the absence of perichromocentric Brdt localization resulting from the loss of the first bromodomain, Sirt1 protein was present adjacent to the chromocenter, suggesting that this localization of Brdt might normally impede Sirt1's access to peri-centromeric heterochromatin. We further speculate that the first bromodomain-dependent localization of Brdt creates a buffer region between the Sirt1 deacetylase and the Suv39h1/2 methyltransferases, and loss of this localization allows the deacetylase and methyltransferases access to the same chromatin. This could then contribute to creating ectopic heterochromatin beyond the centromeric region. This expansion of heterochromatin might block the centromeres from coming together properly to a single structure, resulting in the observed multiple chromocenters. Alternatively, a single chromocenter might be formed transiently but due to the expanded heterochromatin, be very unstable.
We also observed ectopic expression of H1fnt in a small but distinct subset of step VII-VIII BrdtΔBD1/ΔBD1 mutant spermatids, very similar to what was seen in the Tbpl1 and Hmgb2 knockout models which also exhibit multiple chromocenters. We further noted that BrdtΔBD1/ΔBD1 spermatids that failed to elongate often had nuclei entirely surrounded by H1fnt. Since the apical localization of H1fnt blocks the formation of foci of heterochromatin at the nuclear envelope under the acrosome (Martianov et al., 2005), this ectopic distribution throughout the periphery of the nucleus could explain, at least in part, the previously observed loss of such foci in the spermatids of our BrdtΔBD1/ΔBD1 mutant (Shang et al., 2007). It has been speculated that such foci may connect chromatin to the cytoskeleton and could couple chromatin condensation with morphological changes in the cytoplasm and the spermatid head. Indeed, excess cytoplasm, and defective head morphology are two the most striking defects of BrdtΔBD1/ΔBD1 sperm.
The connection between the presence of multiple chromocenters, aberrant localization of H1fnt and abnormal sperm morphology is further supported by our studies of the BrdtΔBD1/ΔBD1 mutation on pure backgrounds. Multiple chromocenters are more prevalent in 129 mutant spermatids than B6 or mixed mutant spermatids, which explains why we did not observe this defect in our initial analysis. The defect in spermatid elongation is also much more severe and penetrant on the 129 background. The loss of polarity of H1fnt localization might reflect an overall loss of polarity in the nucleus that impedes proper elongation. Further, ectopic H1fnt localization is probably the cause of the loss of perinuclear heterochromatin foci. It has been hypothesized that the foci connect chromatin to the actin network in the sperm head and may couple chromatin condensation to overall morphological change (Martianov et al., 2005). Thus their loss may disconnect condensation from proper elongation. Both these aspects of H1fnt mis-localization tie chromocenter formation to elongation. It is important to note that completely morphologically normal sperm are never produced. This suggests that Brdt has additional functions beyond chromocenter formation, such as transcriptional or post-transcriptional regulation of genes that are required for elongation and sperm head and body formation. We are currently investigating such a possible role for Brdt with microarrays and Chip-Seq approaches.
The dynamic chromatin changes that occur during spermiogenesis are still poorly understood, but we have clearly shown the Brdt, and specifically its first bromodomain is essential for this process to proceed normally. The chromocenter is a central organizing structure for the chromatin of the spermatid and mature spermatozoa and it is now apparent that without complete Brdt function the proper architecture of the sperm nucleus cannot be achieved. The BrdtΔBD1/ΔBD1 mutant is unique relative to other mutations affecting sperm chromatin organization in that many spermatids survive, and as a result it may be ideal for further studies of the interconnections of chromatin architecture and sperm head formation. Further study of the mechanism of chromocenter formation and Brdt's role therein will help to understand the highly complex and ordered restructuring of the chromatin of the sperm nucleus.
Research highlights.
- Brdt protein is localized to nuclei in round spermatids, but is excluded from the chromocenter.
- An increased localization of Brdt surrounds the chromocenter, separating it from Sirt1 protein.
- This unique Brdt localization is lost in BrdtΔBD1/ΔBD1 mutant spermatids.
- The BrdtΔBD1/ΔBD1 mutant testis has increased levels of heterochromatin.
- Multiple chromocenters are a prominent feature of the BrdtΔBD1/ΔBD1 mutant phenotype.
Research highlights.
-Brdt protein is localized to nuclei in round spermatids but is excluded from the chromocenter.
-There is an increased localization of Brdt surrounding the chromocenter, separating it from the Sirt1 protein. This localization is lost in BrdtΔBD1/ΔBD1 mutant spermatids.
-Multiple chromocenters are a prominent feature of the BrdtΔBD1/ΔBD1 mutant phenotype.
-The BrdtΔBD1/ΔBD1 mutant testis has increased levels of heterochromatin.
-All aspects of the BrdtΔBD1/ΔBD1 mutant phenotype are more severe on the 129Sv/Ev as compared to the C57BL/6J background.
Acknowledgements
We thank Claire Egan for her assistance in epididymal sperm counting, and both Claire and Justin DeGrazio for their work in maintaining the BrdtΔBD1/ΔBD1 129 mouse colony. We also are grateful to Xiangyuan Wang for preparing histological sections. Additionally, we are grateful for the input and discussion concerning the chromocenter abnormalities provided by Dr. Margarita Vigodner during her sabbatical stay in our lab and Drs. Marcia Manterola and Li Wang for comments and suggestions on the manuscript.
This work was supported by a grant from the NIH to DJW (GM081767).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Buhler M, Gasser SM. Silent chromatin at the middle and ends: lessons from yeasts. Embo J. 2009;28:2149–61. doi: 10.1038/emboj.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catena R, Ronfani L, Sassone-Corsi P, Davidson I. Changes in intranuclear chromatin architecture induce bipolar nuclear localization of histone variant H1T2 in male haploid spermatids. Dev Biol. 2006;296:231–238. doi: 10.1016/j.ydbio.2006.04.458. [DOI] [PubMed] [Google Scholar]
- Chua P, Roeder GS. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol. 1995;15:3685–96. doi: 10.1128/mcb.15.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SS, Wang X, Roberts SS, Griffey SM, Reczek PR, Wolgemuth DJ. Oral administration of a retinoic Acid receptor antagonist reversibly inhibits spermatogenesis in mice. Endocrinology. 2011;152:2492–502. doi: 10.1210/en.2010-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SS, Wolgemuth DJ. Role of retinoid signaling in the regulation of spermatogenesis. Cytogenet Genome Res. 2004;105:189–202. doi: 10.1159/000078189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J, Miyaike M, Kikuchi A, Handel MA. Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase II alpha localization and chromosome condensation. Chromosoma. 1999;108:412–25. doi: 10.1007/s004120050393. [DOI] [PubMed] [Google Scholar]
- Ekwall K. The roles of histone modifications and small RNA in centromere function. Chromosome Res. 2004;12:535–42. doi: 10.1023/B:CHRO.0000036584.40567.e5. [DOI] [PubMed] [Google Scholar]
- Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front Biosci. 2001;6:D1008–18. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–69. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. The X and Y chromosomes assemble into H2A.Z-containing [corrected] facultative heterochromatin [corrected] following meiosis. Mol Cell Biol. 2006;26:5394–405. doi: 10.1128/MCB.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res. 1995;219:604–11. doi: 10.1006/excr.1995.1270. [DOI] [PubMed] [Google Scholar]
- Horakova AH, Bartova E, Galiova G, Uhlirova R, Matula P, Kozubek S. SUV39h-independent association of HP1 beta with fibrillarin-positive nucleolar regions. Chromosoma. 2010;119:227–41. doi: 10.1007/s00412-009-0252-2. [DOI] [PubMed] [Google Scholar]
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer-Fender S, Singh PB, Motzkus D. The murine heterochromatin protein M31 is associated with the chromocenter in round spermatids and Is a component of mature spermatozoa. Exp Cell Res. 2000;254:72–9. doi: 10.1006/excr.1999.4729. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003a;12:1565–76. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003b;23:4207–18. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Ladurner AG, Inouye C, Jain R, Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell. 2003;11:365–76. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Conesa C, Lesage P, Swanson RN, Ruet A, Carlson M, Sentenac A, Seraphin B. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 1994;22:5332–40. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A. 2005;102:2808–13. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Gansmuller A, Parvinen M, Davidson I, Sassone-Corsi P. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development. 2002;129:945–55. doi: 10.1242/dev.129.4.945. [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–63. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Meistrich ML. Quantitative correlation between testicular stem cell survival, sperm production, and fertility in the mouse after treatment with different cytotoxic agents. J Androl. 1982;3:58–68. [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–36. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca M, Muller-Navia J, Scherthan H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci. 1998;111(Pt 10):1363–70. doi: 10.1242/jcs.111.10.1363. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–8. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Muller CW, Petosa C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–8. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei MG, Denny P, Brown SD, Schweizer D, Jenuwein T. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423–33. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol. 2010;191:1299–313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–48. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BO, Mason KA, Withers HR, West J. Effects of hyperthermia and radiation on mouse testis stem cells. Cancer Res. 1981;41:4453–7. [PubMed] [Google Scholar]
- Shang E, Nickerson HD, Wen D, Wang X, Wolgemuth DJ. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development. 2007;134:3507–15. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- Shang E, Salazar G, Crowley T, Wang X, Lopez RA, Wang X, Wolgemuth DJ. Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expression Patterns. 2004;4:513–519. doi: 10.1016/j.modgep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238:908–17. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–6. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- Travis AJ, Jorgez CJ, Merdiushev T, Jones BH, Dess DM, Diaz-Cueto L, Storey BT, Kopf GS, Moss SB. Functional relationships between capacitation-dependent cell signaling and compartmentalized metabolic pathways in murine spermatozoa. J Biol Chem. 2001;276:7630–6. doi: 10.1074/jbc.M006217200. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Allen MJ, Kobayashi A, Zalenskaya IA, Balhorn R, Bradbury EM. Well-defined genome architecture in the human sperm nucleus. Chromosoma. 1995;103:577–90. doi: 10.1007/BF00357684. [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–31. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]