Abstract
The incidence of serogroup D Salmonella has been increasing in Taiwan. Most of these isolates belonged to Salmonella enterica serovar Enteritidis and showed a relatively higher rate of resistance to sulfamethoxazole-trimethoprim than to other antimicrobial agents. The results of molecular experiments indicated that genes responsible for the resistance were located on plasmids. The resistance may occur via horizontal gene transfer. Furthermore, the first identification of ciprofloxacin and ceftriaxone resistance in serogroup D Salmonella in our hospital is also than they did to other antimicrobial agents cause for concern.
Nontyphoidal Salmonella is recognized as one of the principal causes of foodborne infections worldwide. Among the more than 2,000 Salmonella serovars, serovar Enteritidis was one of the top two serovars reported in the United States (6). Most cases of gastroenteritis caused by serovar Enteritidis occur sporadically or as limited outbreaks, but recent reports of large, hospital- and nursing home-associated outbreaks emphasize the importance of serovar Enteritidis infections as a major public health problem (4, 7).
Antimicrobial resistance among nontyphoidal Salmonella has been a serious problem worldwide. In the United States, the number of Salmonella organisms that were resistant to one or more antimicrobials rose significantly from 16% in the 1980s to 31% in the 1990s (3). A similar situation has been found in the United Kingdom (10). Serovar Enteritidis is more susceptible to available antimicrobial agents than other common Salmonella serotypes (3, 10).
In a previous study, Su et al. found that for nontyphoidal serogroup D Salmonella isolates (serovar Enteritidis in particular), the level of resistance to ampicillin and chloramphenicol was approximately 10% and to sulfamethoxazole-trimethoprim was approximately 20% (8). To further study the associated mechanism, records of clinical Salmonella isolates in the Department of Clinical Pathology of Chang Gung Memorial Hospital (CGMH) (a 3,500-bed university-affiliated teaching hospital) in Taoyuan, Taiwan, obtained between 1997 and 2002 were retrospectively reviewed. Throughout the study period, standard methods were used for the isolation and identification of bacteria and no major changes were made. Antimicrobial susceptibility levels were investigated, and results were defined according to those suggested by the National Committee for Clinical Laboratory Standards (5). The chi-square test was used to determine the significance of differences.
Furthermore, a total of 20 isolates of serogroup D Salmonella, including 18 serovar Enteritidis and 2 serovar Dublin isolates, were randomly collected between 2001 and 2002 for molecular analysis. Of the isolates, 15 were from blood, 3 were from feces, 1 was from pus, and the remaining 1 was from sputum. Plasmid profiles were determined by a method described earlier (2). The oligonucleotide primers synthesized according to the published DNA sequences of spvC, a conserved gene located on the virulence plasmid of Salmonella, were used in PCR to detect the presence of the plasmid (1). The following resistance genes were detected by PCR using methods described earlier (1, 2): sulI and sulII for sulfamethoxazole resistance and dfr for trimethoprim resistance. Moreover, class I integron gene cassettes were detected using PCR with primers derived in previous work (11). PCR products were sequenced using an ABI 377 automatic sequencer (Perkin-Elmer, Applied Biosystems). The search for homologous sequences was done using FASTA software available at the GenBank database website on the Internet. DNA-DNA hybridization was performed by a method described earlier (2) to examine whether the resistance was plasmid mediated.
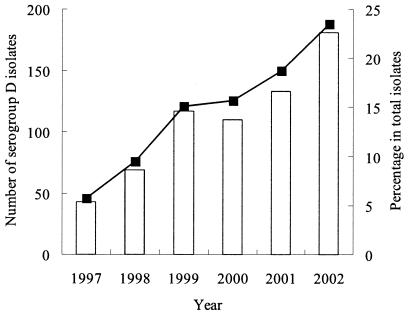
According to the laboratory records available in CGMH, serogroup D formed the third largest group (after serogroups B and C) among all Salmonella isolates. In contrast to serogroups B and C, however, infections caused by serogroup D Salmonella have been increasing in recent years; the number of isolates obtained was 43 in 1997, while after 1999 a significant (P < 0.01) increase to more than 100 in each year was noted (Fig. 1). Compared to total Salmonella isolates, the percentage of serogroup D isolates increased approximately fourfold (from 6 to 24%). Su et al. have confirmed that most serogroup D isolates belonged to serovar Enteritidis (9).
FIG. 1.
Prevalence and antimicrobial susceptibility of serogroup D Salmonella isolates in CGMH between 1997 and 2002. Annual numbers of isolates obtained (bars) and the annual incidence (per total Salmonella isolates) (line) of serogroup D Salmonella infections are indicated.
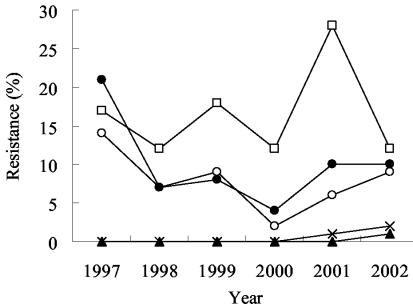
Figure 2 shows the trend of antimicrobial resistance to ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole among nontyphoidal serogroup D Salmonella isolates over the study period. The average rate of resistance to sulfamethoxazole-trimethoprim was 16.5%, which was higher than that to ampicillin (7.8%) or chloramphenicol (10%) (P < 0.05). None of the serogroup D isolates was resistant to ciprofloxacin or ceftriaxone until 2001 to 2002, when one ciprofloxacin-resistant isolate and three ceftriaxone-resistant isolates were first identified.
FIG. 2.
Secular trends in antimicrobial resistance to ampicillin (○), chloramphenicol (•), sulfamethoxazole-trimethoprim (□), ciprofloxacin (▴), and ceftriaxone (×) in serogroup D Salmonella isolates over time.
Analysis of plasmid profiles showed that all 18 of the serovar Enteritidis isolates examined harbored a 60-kb serotype-specific virulence plasmid. Plasmids of other sizes were found in some isolates (Table 1). Of the 18 serovar Enteritidis isolates, 4 were resistant to sulfamethoxazole-trimethoprim (Table 1) and 3 (isolates 336, 338, and 340) carried dfr and sulI genes, which were responsible for trimethoprim and sulfamethoxazole resistance, respectively. DNA-DNA hybridization showed that both dfr and sulI were located on a 125-kb plasmid. PCR targeted at the class I integron produced a 1.5-kb amplicon in the three strains that carried the dfr and sulI genes, and the amplicon was found (by sequencing) to contain the dfr and aadA2 genes that confer resistance to trimethoprim and streptomycin, respectively. DNA-DNA hybridization showed that the integron was located on the 125-kb plasmid as well. One serovar Dublin isolate (isolate 189) and three serovar Enteritidis isolates (isolates 329, 334, and 343) also contained the dfr gene. Among these four isolates, three (isolates 189, 334, and 343) remained susceptible to sulfamethoxazole-trimethoprim because a sul gene was lacking. Serovar Enteritidis strain 329 was resistant to sulfamethoxazole-trimethoprim, as this strain contained a sulII gene along with the dfr gene. Another serovar Enteritidis strain (isolate 332) that was susceptible to sulfamethoxazole-trimethoprim also harbored a sulII gene but not a dfr gene. DNA-DNA hybridization showed that the sulII gene was located on a smaller, 40-kb plasmid in the two isolates.
TABLE 1.
Characteristics of the 20 isolates of serogroup D Salmonella studied
| Strain | Serotype | No. of plasmids | Size(s) (kb) | Resistance gene
|
Class I integron | Resistance to SXTa | ||
|---|---|---|---|---|---|---|---|---|
| sulI | sulII | dfr | ||||||
| 189 | Dublin | 1 | 80 | − | − | + | − | − |
| 295 | Dublin | 1 | 80 | − | − | − | − | − |
| 294 | Enteritidis | 1 | 60 | − | − | − | − | − |
| 328 | Enteritidis | 1 | 60 | − | − | − | − | − |
| 329 | Enteritidis | 4 | 60, 40, 25, 20 | − | + | + | − | + |
| 330 | Enteritidis | 1 | 60 | − | − | − | − | − |
| 332 | Enteritidis | 4 | 60, 40, 25, 20 | − | + | − | − | − |
| 333 | Enteritidis | 2 | 60, 45 | − | − | − | − | − |
| 334 | Enteritidis | 1 | 60 | − | − | + | − | − |
| 335 | Enteritidis | 1 | 60 | − | − | − | − | − |
| 336 | Enteritidis | 2 | 125, 60 | + | − | + | + | + |
| 338 | Enteritidis | 3 | 125, 60, 45 | + | − | + | + | + |
| 339 | Enteritidis | 1 | 60 | − | − | − | − | − |
| 340 | Enteritidis | 2 | 125, 60 | + | − | + | + | + |
| 342 | Enteritidis | 1 | 60 | − | − | − | − | − |
| 343 | Enteritidis | 3 | 125, 60, 45 | − | − | + | − | − |
| 346 | Enteritidis | 2 | 60, 10 | − | − | − | − | − |
| 347 | Enteritidis | 2 | 60, 10 | − | − | − | − | − |
| 348 | Enteritidis | 1 | 60 | − | − | − | − | − |
| 350 | Enteritidis | 1 | 60 | − | − | − | − | − |
SXT, sulfamethoxazole-trimethoprim.
The study confirmed the emergence and rapid increase in numbers of nontyphoidal serogroup D Salmonella (particularly serovar Enteritidis) infection in Taiwan in the past 6 years. Concurrently, in our hospital the number of serogroup B isolates began to decline gradually after 1995 (8). If this trend continues, the incidence of nontyphoidal serogroup D-induced salmonellosis in Taiwan will soon surpass that of serogroup B infection, as has been the situation in the United States and Europe (6, 7). Serovar Enteritidis is known to be closely associated with layer and broiler flocks, and the infection is generally believed to be derived from chicken and chicken products, including eggs (6, 7). In Taiwan, there was an epidemic of swine mouth-foot disease in 1996 to 1997. This resulted in a decreased consumption of pork and, in turn, an increased consumption of poultry in the community. The results of a recent study have shown that approximately 88% of broiler flocks and 49% of broilers in Taiwan were contaminated with Salmonella (H. J. Tsai and C. H. Chou, Abstr. Proc. 4th Int. Symp. Typhoid Fever and Other Salmonellosis, Taipei, Taiwan, abstr. P12, 1999). Popularized Western-style food such as mayonnaise may also be a factor contributing to the increase. From the public health standpoint, such a rapid increase in serovar Enteritidis infection necessitates a more detailed surveillance of the situation and better methods for precise identification of the organism.
Our findings confirm those of earlier studies indicating that most serogroup D strains (serovar Enteritidis in particular) were susceptible to a wide range of antimicrobial agents (3, 4, 10). Nevertheless, a higher rate of resistance to sulfamethoxazole-trimethoprim was observed in Taiwan (8). This study showed that all resistance genes found were located on plasmids. Furthermore, a class I integron carrying a dfr-aadA2 gene cassette was detected in the resistant isolates. The location of the integron on the plasmid might contribute to horizontal dissemination of the antibiotic resistance gene cassette. In an earlier study (9), a predominant genotype among clinical isolates of serovar Enteritidis was identified by pulsed-field gel electrophoresis, indicating that the emergence of serovar Enteritidis in Taiwan was mainly due to dissemination of clones that were endemic in Taiwan. Taken together, these results provide evidence suggesting that the resistance to sulfamethoxazole-trimethoprim developed due to the acquisition of resistance genes by the preexisting susceptible serovar Enteritidis strains via horizontal gene transfer.
Although most isolates remained susceptible, the first identification of ciprofloxacin and ceftriaxone resistance in serogroup D Salmonella isolates in 2001 and 2002 is cause for concern. Active monitoring of serogroup D Salmonella for resistance to antimicrobial agents is crucial because of the public health implications derived from the increasing prevalence of such organisms.
Acknowledgments
This study was supported by research grants CMRP1313 and CMRPG32033 from Chang Gung Memorial Hospital, Taoyuan, Taiwan.
REFERENCES
- 1.Chiu, C. H., and J. T. Ou. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 34:2619-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, C., C.-H. Chiu, W.-Y. Wu, C.-H. Chu, T.-P. Liu, and J. T. Ou. 2001. Large drug resistance virulence plasmids of clinical isolate of Salmonella enterica serovar Choleraesuis. Antimicrob. Agents Chemother. 45:2299-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee, L. A., N. D. Puhr, E. K. Maloney, N. H. Bean, and R. V. Tauxe. 1994. Increase in antimicrobial-resistant Salmonella infections in the United States, 1989-1990. J. Infect. Dis. 170:128-134. [DOI] [PubMed] [Google Scholar]
- 4.Ling, J. M., I. C. Koo, K. M. Kam, and A. F. Cheng. 1998. Antimicrobial susceptibility and molecular epidemiology of Salmonella enterica serotype Enteritidis strains isolated in Hong Kong from 1986 to 1996. J. Clin. Microbiol. 36:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 6.Olsen, S. J., R. Bishop, F. W. Brenner, T. H. Roels, N. Bean, R. V. Tauxe, and L. Slutsker. 2001. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987-1997. J. Infect. Dis. 183:753-761. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigue, D. C., R. V. Tauxe, and B. Rowe. 1990. International increase in Salmonella enteritidis: a new pandemic? Epidemiol. Infect. 105:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su, L. H., C. H. Chiu, A. J. Kuo, J. H. Chia, C. F. Sun, H. S. Leu, and T. L. Wu. 2001. Secular trends in incidence and antimicrobial resistance among clinical isolates of Salmonella at a university hospital in Taiwan, 1983-1999. Epidemiol. Infect. 127:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su, L. H., C. H. Chiu, T. L. Wu, C. Chu, J. H. Chia, A. J. Kuo, C. C. Lee, C. F. Sun, and J. T. Ou. 2002. Molecular epidemiology of Salmonella enterica serovar Enteritidis isolated in Taiwan. Microbiol. Immunol. 46:833-840. [DOI] [PubMed] [Google Scholar]
- 10.Threlfall, E. J., L. R. Ward, J. A. Skinner, and B. Rowe. 1997. Increase in multiple antibiotic resistance in nontyphoidal salmonellas from humans in England and Wales: a comparison of data for 1994 and 1996. Microb. Drug Resist. 3:263-266. [DOI] [PubMed] [Google Scholar]
- 11.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence of transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]