Abstract
N-Browse is a graphical network browser for the visualization and navigation of heterogeneous molecular interaction data. N-Browse runs as a Java applet in a Web browser, providing highly dynamic and interactive on-demand access to network data available from a remote server. The N-Browse interface is easy to use and accommodates multiple types of functional linkages with associated information, allowing the exploration of many layers of functional information simultaneously. Although created for applications in biology, N-Browse uses a generic database schema that can be adapted to network representations in any knowledge domain. The N-Browse client-server package is freely available for distribution, providing a convenient way for data producers and providers to distribute and offer interactive visualization of network-based data.
Keywords: network, molecular, interaction, graph, browser, Web-based, client-server system, JAVA, functional genomics, GUI, visualization, database, MySQL
INTRODUCTION
New views of biological networks are emerging from the combination of large-scale experimental and computational approaches directed at understanding gene/protein function and functional relationships on many different levels. To help make sense of the wealth of data being generated, effective tools for visualizing and exploring these data are necessary. A natural paradigm for visualizing molecular interaction data is a network graph. However, extracting useful information about a local gene neighborhood from the entire network—which often can be very large and highly interconnected, thus colloquially termed a “giant hairball” (or “ridiculogram”)—can be challenging. The goal of N-Browse is to provide a freely available software package that allows the biology community to share and explore functional interaction networks in an efficient, interactive, and user-friendly way.
Inspired by interactive graphical interfaces for coordinate-based genome annotations such as the Generic Genome Browser (GBrowse; UNIT 9.9), we have developed a similarly intuitive, easy to use, interactive tool for navigating gene network neighborhoods based on different kinds of functional links. This “Generic Network Browser,” N-Browse, is available at http://www.gnetbrowse.org. N-Browse operates within a Web browser as a Java applet and uses a client-server system composed of a server-side MySQL database and a client-side graphical user interface (GUI). The N-Browse Web-based client allows users to quickly access and explore a variety of publicly available interaction data. In addition, the freely distributed N-Browse client-server package allows producers and providers of network-based data to employ N-Browse as a visual interface and distribution mechanism for serving their own combination of data from one or more species of interest. N-Browse seeks to provide both a user-friendly client-side interface and a straightforward procedure for server-side installation and configuration. This unit has two basic protocols that describe usage and features of the client-side GUI. Basic Protocol 1 shows how to access useful information from any N-Browse Web site using the main N-Browse site at http://gnetbrowse.org as an illustrative example. Basic Protocol 2 shows how to use advanced functions to select and configure different combinations of data for network browsing. In addition, Basic Protocols 3 and 4 describe how to set up an independent N-Browse server site from the N-Browse client-server distribution package. A fully functional N-Browse Web site will require both installing and configuring the Web server host software (Basic Protocol 3), as well as setting up and populating an N-Browse database (Basic Protocol 4). A troubleshooting section describes how to detect and fix potential problems that might be encountered during installation.
NAVIGATING THE N-Browse GUI
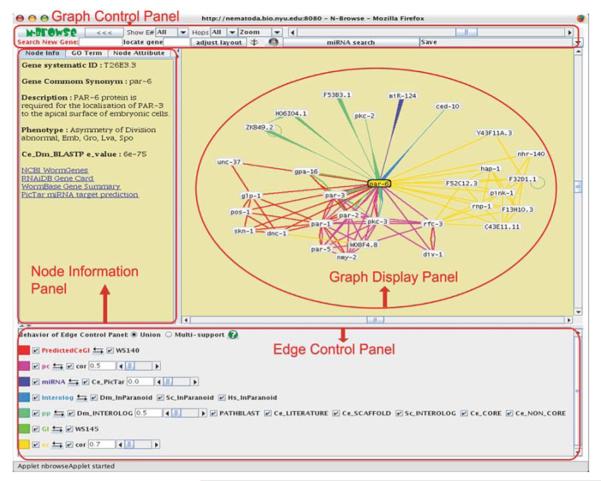
A quick tour describes the main features of the N-Browse GUI, which consists of four panels (Fig. 9.11.1). The online tutorial at http://gnetbrowse.org/ includes a more detailed description, as well as demonstration videos illustrating different aspects of the GUI; an overview of each panel's functions is described here.
Figure 9.11.1.
The N-Browse GUI. For color version of this figure see http://www.currentprotocols.com.
The Graph display panel is the central component of the GUI and provides a network representation of available interaction data. It offers a number of interactive features for manipulating the graph (described in more depth in sections below) and communicates with other panels in the N-Browse GUI.
The Edge control panel provides a menu of the different functional edge types available. Essential features of this menu include the following:
The menu is constructed automatically from stored data types. Mousing over each item in the menu will display a brief description of it.
Different edge data types are distinguished by different colors, which can be changed by clicking on the color swatches for each edge type.
For each edge data type, different datasets (e.g., from multiple independent large-scale studies) can be defined and appear as individual sub-items in the menu.
The display of each data type or dataset can be manipulated independently using a toggle switch, by checking (to show) or unchecking (to hide) the adjacent box.
Navigation within the Graph display panel is restricted to the datasets listed in this menu. Thus, at each expansion step, the number and types of edges (and nodes) drawn into the graph are limited to those datasets available in the menu. Basic Protocol 2 explains how to configure which data sets are included in this panel, which can be done by either (i) preselecting specific subsets of interest (using the Advanced tab) and/or (ii) uploading your own data. Showing/hiding edges for display and preselecting datasets for navigation produce different graphs; this is because hidden edges are still included in expansion steps, so neighbors of hidden nodes will be drawn into the graph in the former but not the latter case. See the online tutorial for more detail on this topic.
The Node information panel contains three main sections, each of which can be opened by choosing the corresponding tab, as described below. (Additional tabs may be included in future releases of the N-Browse client-server core package or in association with plug-ins.)
Node Info: Provides a brief description and some useful links about any gene in the graph. This information appears dynamically when mousing over a node with the cursor.
Node Attribute: Allows different types of node attributes to be highlighted on the graph. A menu of available node attributes is dynamically generated when this tab is opened. Both categorical (e.g. phenotypes) and ordinal data (e.g. expression levels) can be highlighted.
GO Term: Displays the Gene Ontology DAG (directed acyclic graph) and highlights terms for selected gene(s) in the current graph. At present, this tab is only available on the main N-Browse Web site (http://gnetbrowse.org), as the current implementation is experimental.
The Graph control panel provides options to manipulate the Graph display panel, including:
Locate a specific node in the current network.
Back up one navigation step in the current network (retract last expansion step).
Zoom or rotate the network.
Display a new network by entering a new query.
Auto-launch Cytoscape using Java Webstart.
Save the current network in a variety of formats (text or image).
BASICS OF THE N-Browse GUI
The N-Browse user interface provides a simple, easily accessible way to interactively browse many different kinds of functional linkages at once. The GUI was designed with several features in mind: (1) dynamic graphical interface for network browsing and expansion, (2) dynamic edge and node attribute detection, (3) easily accessible information on nodes and edges, with links to useful external resources, and (4) highly configurable selection of edge data sets and score cutoffs.
This protocol will take the user through the main features of the N-Browse client-side user interface. Below we present the general idea of each function and some examples of how to view and explore network data, navigate local neighborhoods, and visualize properties of interest. The N-Browse online tutorial also includes demonstration videos illustrating each of these functions.
Necessary Resources
Hardware
Any computer with internet access
Software
Java-compatible internet browser
Java Runtime Environment (JRE) 1.4 or above
Files
N-Browse homepage: http://gnetbrowse.org
N-Browse tutorial: http://gnetbrowse.org/N-Browse tutorial.html
N-Browse system requirements: http://gnetbrowse.org/info.html
Browsing the network neighborhood around a single query gene
- 1. Start a Java-compatible Web browser and open the N-Browse homepage at http://gnetbrowse.org (Fig. 9.11.2).Make sure Javascript and Java are enabled in your browser preferences, since N-Browse requires both (Javascript is needed to render the homepage and Java for the GUI).Follow steps 2 to 4 below to generate a network display centered around a single gene/protein query.Basic Protocol 2 provides instructions on selecting specific datasets for display and integrating user-defined data uploads in network navigation.
- 2. Type in the name of a gene.Sample queries for different species are provided for reference.
3. Select the species from the drop-down menu.
- 4. Click the GO button.This will open a new window containing the N-Browse GUI described above. Since the Java applet requires access to your computer's hard drive, you will also need to click Trust when prompted about the Java applet's certificate.In Windows operating systems running Explorer, the GUI window will occupy the full screen (pressing Ctrl+Esc will exit full-screen mode).If you do not see the new window pop up, check that your browser preferences are set to allow pop-up windows for the N-Browse Web site. If a new window is launched but you do not see the N-Browse GUI (Fig. 9.11.2), double-check your browser preferences to make sure Java is enabled.All subsequent operations described below refer to the main N-Browse GUI window.
5. Inspect a node: Mousing over a node causes relevant information about that node (gene) to be displayed in the Node information panel, including links to external databases (Fig. 9.11.1). Right-clicking (or Control-clicking with a one-button mouse) on a node causes a drop-down menu to appear with several control options to manipulate the selected node(s). To select multiple nodes, press and hold the Shift button on the keyboard while clicking nodes in the current network.
- 6. Inspect an edge: Mousing over an edge will display the edge type, edge dataset, and its numerical value, if any. Right-clicking (or Control-clicking with a one-button mouse) on an edge will open a drop-down menu with options to hide the edge or to show any external links associated with that edge (e.g., in other databases), which will appear in a new browser window. In order to see information resulting from mousing over, the size of the network can be adjusted by selecting Zoom in the Graph control panel and moving the adjacent slider bar as described in step 16.If you do not see a new window pop up when following external links, please make sure that your pop-up blockers are disabled for the N-Browse Web site.
- 7. Select a new query: In the Graph control panel, look for Search New Gene. Type a gene ID or name in the box and press Enter/Return to build a new network graph around this gene as the starting point.Ambiguous or unidentifiable IDs will trigger a warning prompt with a new window offering a list of available choices. Select one of these and click the Submit button to generate a new query.
Figure 9.11.2.
The N-Browse homepage.
Expanding the network around a node in the graph
- 8. Expand the graph: Double-click any node to retrieve any additional functional links to that gene/protein that are not currently displayed (Fig. 9.11.3). With the default view setting (“matrix” view), this will display all edges between any newly retrieved first-degree neighbors of this node and all other nodes in the current graph. Browse the local network neighborhood by sequentially expanding around successive nodes as illustrated in Figure 9.11.3.Expanding the network in the default matrix view setting may generate a tremendous number of new edges. To avoid this, users can switch to a “spoke” view, which displays only links between the selected node and its direct first-degree neighbors. See Basic Protocol 2 for information on invoking the alternate “spoke” display view.
9. Step backward: Recent steps in the network expansion sequence can be sequentially reversed by clicking the “<<<” button in the Graph control panel, causing all nodes and edges gathered in each step to disappear from view.
- 10. Select “miRNA search:” This will cause all microRNAs predicted to target genes in the current graph to be displayed.miRNA-target relationships at gnetbrowse.org are currently based on predictions by the PicTar algorithm, available at http://pictar.org.This function is hidden in the N-Browse distribution package, as it depends on the presence of a particular data type in the corresponding species database. It can be restored by uncommenting out this part of the code and recompiling.
Figure 9.11.3.
Network navigation: sequential expansion around selected network nodes. In this example, the user has entered an initial query par-6 (A) and subsequently expands the network around par-5 (B) and par-3 (C). For demonstration video see the N-Browse online tutorial. For color version of this figure see http://www.currentprotocols.com.
Customizing the display
- 11. Select specific data types, datasets, and/or numerical thresholds for display.By default, all available data types and data sets (listed in the Edge control menu) are included for display in the network graph. The Edge control panel provides the ability to selectively hide or show edges, either by toggling check boxes for different data types and datasets or by changing numerical cutoffs (where present).See Basic Protocol 2 for more information on preselecting basis datasets for network navigation.
- 12. Display Gene Ontology terms: Switching to the GO Term tab in the Node information panel displays a hierarchical term list from the Gene Ontology DAG (directed acyclic graph). There are two ways to highlight genes annotated with particular GO terms on the current graph (see UNIT 7.2 for information on GO):
- Clicking on a specific GO term in the term list displayed in this panel will highlight any nodes in the graph that are annotated with that term.
- Selecting one or more nodes in the graph and choosing Show GO Term (*) from the drop-down menu that appears in response to a right-click (or control-click with a one-button mouse) on the selected node(s) will also activate the GO Term panel. In this case, all GO terms associated with the selected set of genes will then be highlighted in the DAG term list. Right-clicking selected nodes in the graph will also open a second window that displays a graphical hierarchy of the GO terms associated with those genes.This function is not included in the N-Browse distribution package as the current implementation is experimental, but future releases will contain a corresponding feature.
- 13. Highlight node attributes on the network graph: Switching to the Node Attribute tab in the Node information panel allows the visualization of various properties associated with genes/proteins in the current graph. When the Node Attribute tab is opened, a list of available attributes will be displayed. Select a checkbox to load the corresponding attribute class for highlighting on the graph. Node attributes may be highlighted with either a graded scale of color intensity (for ordinal data) or a solid color (for categorical data). A description of this control menu is also available when clicking the “?” icon on the top-right corner of the Node Attribute tab.Node attributes can be any type of data associated with nodes (genes/proteins), such as phenotypes, expression levels, protein domains, etc.
14. Anchor or weigh nodes in the graph: All nodes in the graph can be frozen in place or unfrozen by clicking the Anchor icon in the Graph control panel. Alternatively, selected node(s) can be pasted to the background using the right-click drop-down menu that becomes available when mousing over nodes in the graph and selecting Anchor node or Weigh anchor, with the former anchoring the node and the latter removing the anchor, respectively.
- 15. Adjust the current graph layout: Clicking the “adjust layout” button in the Graph control panel resets the rendering of edges in the graph, resulting in the straightening or bowing of different edges.Normally, multiple edges between node pairs are splayed to avoid superimposition, and single edges appear straight. Because the rendering method is not automatically adjusted after each operation, hiding or unhiding selected edge types can sometimes leave, for example, single edges that appear bowed. Adjusting the layout may take a long time for a large graph with numerous edges, so be patient.
16. Zoom, rotate, or move the graph: The size and orientation of the network view can be adjusted by selecting Zoom or Rotate in the Graph control panel and moving the adjacent slider bar. The entire graph can be moved by positioning the cursor anywhere in the background field and dragging it while holding down the mouse button.
Saving and exporting network information
- 17. Save network data: Information about the current graph can be saved in a variety of formats:
- A list of nodes (as a tab-delimited text file).
- A list of interactions (as a tab-delimited text file).
- A Cytoscape .sif network file (see UNIT 8.13).
- A screenshot image (Save PNG image).
- An Encapsulated PostScript (EPS) file (Save EPS image).The EPS format provides a vector representation of the image, producing very high quality views that are especially useful for publications such as posters.
- 18. Auto-launch Cytoscape: Further analysis of the currently displayed network can be performed in the stand-alone graph layout application Cytoscape (also see UNIT 8.13 and http://cytoscape.org). Clicking on the Cytoscape icon in the Graph control panel will cause all of the data in the current graph to be packaged for export to and displayed using Cytoscape, which will be launched automatically on the user's computer using Java Web Start.The Cytoscape-compatible files created by N-Browse do not automatically specify the geometry of the graph layout, so a new layout will need to be generated from within Cytoscape after import.
WORKING WITH DATASETS AND USER-DEFINED UPLOADS
By default, network navigation in N-Browse includes all data in an N-Browse database and operates using a “matrix,” or complete, view of all defined edges. N-Browse provides the ability to configure the range of data used for network navigation, either by: (1) preselecting specific subsets of data and thresholds from an N-Browse database, or (2) uploading user-defined data for integrated visualization with publicly available data.
In addition, N-Browse allows users to configure the display method for visualizing links between selected nodes and their first-degree neighbors by toggling between the default “matrix” view and an alternative “spoke” view. This protocol describes each of these features. The N-Browse online tutorial also includes demonstration videos illustrating each of these functions.
Necessary Resources
Hardware
Any computer with internet access
Software
Java-compatible internet browser
Java Runtime Environment (JRE) 1.4 or above
Files
N-Browse homepage: http://gnetbrowse.org
N-Browse Advanced page: http://gnetbrowse.org (select “Advanced” tab)
N-Browse tutorial for user-defined data upload: http://gnetbrowse.org/upload_tutorial.html
Start a Java-compatible Web browser (such as Firefox, Safari, or Internet Explorer) and open the N-Browse homepage at http://gnetbrowse.org.
Select the Advanced tab (Fig. 9.11.5).
Type in the name of a gene.
Select the species from the drop-down menu.
Figure 9.11.5.
The N-Browse Advanced Web page. For color version of this figure see http://www.currentprotocols.com.
Selecting “matrix” versus “spoke” views
Two display options are available for visualizing functional links between selected nodes of interest and their first-degree neighbors (Fig. 9.11.4): (i) Matrix view: This is the default for N-Browse and displays all links between all nodes in the graph (Fig. 9.11.4, left-hand panel). The matrix view guarantees that if any known edge exists between any two nodes in the current graph, it will appear in the network diagram (unless any of these have been manually hidden by the user). (ii) Spoke view: In contrast, the “spoke” view (Fig. 9.11.4, right-hand panel) shows only links directly attached to query nodes (either the initial query or a node selected for expansion). Since any edges between two neighbors of a query node will not be shown, many existing edges are typically not displayed in this view. The spoke view reduces visual “clutter,” but makes a trade-off with information content (since many potentially interesting functional links will not be revealed). In some cases the spoke view may be preferred, particularly where relationships between neighbors are not clear (e.g., co-immunoprecipitation can recover many proteins pulled down by a single query protein, but no information is available on whether any of these directly interact with each other).
Figure 9.11.4.
Matrix (A) and Spoke (B) views for the query par-6 in C. elegans at http://gnetbrowse.org. For color version of this figure see http://www.currentprotocols.com.
5. In the Configure Datasets section on the Advanced page, select either Matrix View (Gather all interactions between neighbors) or Spoke View (Only show interactions from the requested gene/protein to its 1-hop neighbors).
Selecting specific datasets and thresholds for network navigation
If some data types available in the database are not of interest to the user for some reason, it is possible to exclude them from the base data sets used for network navigation. For example, a user may be interested only in physical or genetic interactions, or may consider some datasets unreliable, or may wish to impose a more stringent cutoff than the default threshold for a correlation coefficient or other score.
- 6. Configure datasets for network navigation: The Configure Datasets section on the Advanced page will dynamically list the available datasets in the N-Browse database for the selected species (Fig. 9.11.5). Selecting a different species from the menu in the Search section will automatically refresh the contents of the list. Configure datasets as follows:
- To restrict the use of specific data types or datasets, deselect them by unchecking the corresponding checkbox.
- To change the threshold cutoff used for data with numerical ranges, enter a new number in the text box for a specific dataset (the ranges present in the database are shown for reference).
- For more information about each data type and dataset available in the database, click on the Types and Datasets link.
- 7. Press Go in the Search section.If no prior query has been entered, a new N-Browse GUI window will open containing only the selected data types, datasets, or data subsets. If a previous query was entered in the Advanced page, the open N-Browse GUI will be refreshed and the edge menu in the Edge control panel will now contain only the selected sets. Eliminated datasets will no longer be considered during network navigation.Different network information is retrieved from the database when navigating using the full database contents versus selected subsets of the data. Using preselected subsets limits the edges and nodes gathered during network expansion steps to those subsets matching the preselection criteria. Thus, data that are eliminated from network navigation by preselection will never be displayed in the network graph and are not considered for expansion steps. Typically, this results in gathering fewer new edges and nodes at each step. Users should be aware that preconfiguring datasets is very different from toggling datasets for display in the Edge control panel menu, in which case all data listed in the edge menu (including hidden data) are still gathered in expansion steps. This is necessary so that data hidden from view can still be retrieved for display at any point in the session.
Uploading data for integrated viewing
Users are often interested in visualizing data that are not available in an N-Browse database, either from their own laboratory's work or other data sources. N-Browse allows users to upload their own data for integrated viewing with the publicly available data in an online N-Browse database. Currently, the file upload function accepts a simple tab-delimited file format; descriptions and a sample file for C. elegans are provided in the N-Browse online tutorial at http://gnetbrowse.org/upload_tutorial.html.
- 8. Open either the N-Browse homepage or Advanced page on the N-Browse Web site.Both provide the ability to integrate with available data in the N-Browse database. If data are uploaded using the Advanced page, you can simultaneously configure the network view (matrix or spoke) and the datasets in the N-Browse database to be included for network navigation.
- 9. Specify a file containing network data: In the section User-Defined Network, click Choose File and select a file located on your computer that contains network data in one of the accepted file formats described on the N-Browse Web site.User-defined data are not stored by the application, but are temporarily cached during an active session. Thus these data are no longer accessible once the current session is closed or expires.
10. Upload the data file: Select the species network with which you wish your data to be integrated, and press Upload. If a previous query was entered in the Advanced page and the corresponding N-Browse GUI is still open, the GUI will be refreshed and the edge menu in the Edge control panel will now contain one or more new menu items listing the data you provided. If your file explicitly specified one or more data types or datasets, each of these will be listed by name with the prefix UD_(for user-defined), for example UD_Y2H; otherwise, your data will appear under the moniker UD_unknown.
INSTALLING AND CONFIGURING THE N-BROWSE CLIENT-SERVER PACKAGE
As described in the Introduction, a fully functional N-Browse site will require completing Basic Protocols 3 and 4. This protocol describes how to install the N-Browse client-server package in the Unix/Linux environment. It is assumed that the user has proper knowledge and privileges to install software in the Unix/Linux environment.
After installation, the N-Browse Web pages should appear through an HTTP connection. To test the N-Browse GUI, you will need to populate an N-Browse database with either the test data provided with the distribution or your own data, as described in Basic Protocol 4.
Necessary Resources
Hardware
Any Unix (Linux, Solaris or other) workstation or Macintosh OS X
A minimum of 500 Mb RAM
Internet connection
Software
Most of the required software can be installed using a package manager for your OS platform or downloaded directly from the providers at the Universal Resource Locators (URLs) listed below
Standard software
N-Browse client-server package [the package can be downloaded either from the main N-Browse Web site (http://gnetbrowse.org) or from SourceForge (http://sourceforge.net/projects/nbrowse/)]
Tomcat4 or higher (Tomcat5 is preferred; the package is available at http://tomcat.apache.org/)
Java Software Development Kit (Java SDK) 1.4 or higher (the package is available at http://java.sun.com)
Perl v. 5.0 or higher (Perl 5.8 or higher is preferred; Perl will generally be installed on most machines with a Unix-like operating system, but it is also available at http://www.perl.org)
MySQL Server 4.1 or higher. MySQL Server 5 is preferred (MySQL is available at http://www.mysql.com/)
JDBC Driver for MySQL (Connector/J) (the library is available at http://dev.mysql.com/downloads/connector/j/5.0.html)
Nonstandard software
These packages will be needed if you want to set up Cytoscape auto-launch using Java Web Start:
Apache Web server (available at http://httpd.apache.org/)
Perl CGI package (available at http://search.cpan.org/dist/CGI.pm/)
Perl Driver for MySQL (DBD::mysql) (the library is available at http://search.cpan.org/dist/DBD-mysql/)
Files
The install.pl and README files are located in the nbrowse_server_client/ directory after unpacking the N-Browse tarball
Download and install N-Browse
- 1. Download and install the required software: Download N-Browse from either of the locations listed above to the prospective N-Browse server machine.
- The file is in .tar.gz format and will need to be uncompressed and unpacked before installation.
- 2. Configure the install_conf file in the nbrowse_server_client/ directory. Edit the file so that the required parameters suit your machine's configuration.This configuration file MUST be modified before installing the N-Browse package. Table 9.11.1 provides an explanation of each parameter with example values.
- 3. Install the N-Browse package from source: Run the install.pl script located in the nbrowse_server_client/ directory:
- $ perl install.plThe “$” symbol represents a command line prompt. The prompt may be represented by other symbols on different systems.This will install the required software components in the locations and using the parameter setting specified in the install_conf file.
- 4. Test the installation: Check if the installation went correctly by opening the following pages in your favorite browser (replacing the text string TOMCAT_APP_FOLDER in the URL below with the parameter value you specified in the install_conf file):
- http://localhost:8180/TOMCAT_APP_FOLDER/NBrowse.htmlIf the installation was successful, you should see your customized N-Browse homepage at this URL (similar to Fig. 9.11.2). If you do not see the NBrowse.html page, it is likely that your Tomcat is not configured properly. In this case, see the Troubleshooting section below.
Table 9.11.1.
Required Parameters in the install conf Filea
| Parameter | Description |
|---|---|
| TOMCAT_SERVER [nematoda.bio.nyu.edu] | Domain name of the Tomcat Web server. This will serve as the base URL for HTTP connections. |
| TOMCAT_PORT [8180] | Port number for Tomcat HTTP connections. The default port for Tomcat5 is 8180. |
|
TOMCAT_WEBAPPS_PATH [/var/lib/tomcat5/webapps/] |
Physical (directory) location of the Tomcat Web application in the file system. The default location for Tomcat5 is /var/lib/tomcat5/webapps/. |
| TOMCAT_APP_FOLDER [NBrowse] | Directory name to be used for the N-Browse Web application. If you plan to run multiple N-Browse servers on the same machine, this name can be customized to distinguish different instances. |
| MYSQL_SERVER [localhost] | MySQL server location for database connections. If the server resides on the same machine as the N-Browse server package, you can use “localhost.” If the database resides on a different machine, a domain name is required to make remote database connections. |
| MYSQL_PORT [3306] | MySQL server port number. You can leave it empty if you connect to “localhost” (MySQL default setting). |
| MYSQL_DATABASE_NAME [nbrowse] | Name of the MySQL database containing the N-Browse database schema (see Basic Protocol 4). |
| MYSQL_USERNAME [handler] | MySQL username that the N-Browse package will use as a handler for database connections. |
| MYSQL_PASSWORD [] | MySQL password for the above MySQL user. It can be empty if the MySQL user has no password. |
| INSTALL_CYTOSCAPE_AUTOLAUNCH [Y/N] | Choose Y (yes) or N (no) to set up the Cytoscape auto-launch function. If Y, the next 3 parameters must be specified (JNLP_CODEBASE, JAVA_LOCATION, and CYJNLP_LOCATION). |
|
JNLP_CODEBASE [nematoda.bio.nyu.edu/cgi-bin/nbTest/] |
Web address (URL) for a directory on the N-Browse server machine in which Perl CGI scripts have permission to run (specified in Tomcat or Apache config files). This is essential to create the files for Cytoscape auto-launch. |
| JAVA_LOCATION[/usr/bin/] | Physical (directory) location of the JAVA binary. |
|
CYJNLP_LOCATION [/usr/lib/cgi-bin/nbTest/] |
Physical (directory) location of the Perl CGI scripts required for Cytoscape auto-launch. |
|
WEBSITEHOSTBY [NYU Center for Genomics & Systems Biology] |
Text for customizing the N-Browse Web site at your institution, to be displayed in an iFrame container at the top of the N-Browse Web pages at your site. If desired, you can further customize the Web site (with graphics etc.) by directly editing the HTML code in the containerInfo.html file included in the distribution. |
These parameters must be customized for your server machine prior to N-Browse installation. Example values for each parameter are provided in square brackets.
INSTALLING AND POPULATING THE N-Browse GENERIC DATABASE
The design considerations for the N-Browse database schema included the need to accommodate a diversity of data types without prior knowledge of their content or structure, and the ability of the system to automatically discover the types of data and ranges of values present across the entire database. To facilitate populating an N-Browse database, the N-Browse distribution package provides a set of Perl scripts that will automatically populate the generic N-Browse database schema using user-supplied data in simple tab-delimited (.cvs) files. This protocol provides a step-by-step guide to setting up and populating an N-Browse database by running these scripts. With default parameters, these scripts will load a set of sample data included in the distribution package. The resulting sample database can be used as a test version for the installation process. For convenience, this protocol also includes an optional shortcut for generating the sample database directly from a MySQL data dump to facilitate testing other aspects of the installation.
Necessary Resources
Hardware
Any Unix (Linux, Solaris or other) workstation or Macintosh OS X
A minimum of 500 Mb RAM
Internet connection
Software
All necessary software should be installed if Basic Protocol 3 has been completed
Files
After unpacking the N-Browse tarball, the Perl dataloader scripts and README file are located in the nbrowse_dataloader/ directory
- Dataloader Perl scripts:
- dataloader_node_syn.pl
- dataloader_edge_meta.pl
- dataloader_url.pl
- dataloader_node_attr.pl
- dataloader_truncate_tbs.pl
- Data file format specification:
- dataloader_csv_format.txt
- README file containing short descriptions of the Perl dataloader scripts:
- README.txt
Create an N-Browse generic database
- 1. Create an empty MySQL database called nbrowse and make it accessible to the N-Browse MySQL database handler. This can be accomplished using the following commands:
- $ mysql -u root -p
- Enter password: ********
- The “$” symbol represents a command line prompt. The prompt may be represented as other symbols on different systems. The MySQL user does not need to be “root,” but must have the privilege to create databases.
- mysql> create database nbrowse;
- Query OK, 1 row affected (0.01 sec)Here we use “nbrowse” as a database name for demonstration purposes. This name can be anything but should be the same as the one indicated in the install_conf file described in Basic Protocol 3.
- mysql> grant all privileges on nbrowse.*
- to handler@localhost;
- Query OK, 0 rows affected (0.01 sec)
- When granting privileges to the user, replace handler with the username of the nbrowse database handler and nbrowse with the name of your database. These should be the same as specified in the install_conf file described in Basic Protocol 3.
- mysql> quit
- Bye
- 2. Load the N-Browse database schema into the newly created database. Start MySQL as the N-Browse database handler:
- $ mysql -u handler -p
- Enter password: ********
- Switch to the nbrowse database and load the schema file nbrowse_schema.sql located in the nbrowse_dataloader/ directory:
- mysql> use nbrowse;
- mysql> source /home/bob/nbrowse_install_package/nbrowse_dataloader/nbrowse_schema.sql;
- This example uses the Unix username “bob” as the N-Browse package administrator. When loading the schema into the database, replace the above path with the absolute path of the nbrowse_schema.sql file on your system.
- mysql> quit
- Bye
Load a sample N-Browse database from a MySQL data dump
This section is optional. It will allow you to immediately test the N-Browse GUI for your server setup using a prebuilt database. You can generate the same database by skipping this section and following the instructions in the next section.
NOTE: If you chose to carry out steps 3 and 4, make sure to either truncate all the tables in the nbrowse database (e.g., using the provided script dataloader_truncate_tbs.pl), or drop the database completely and repeat steps 1 and 2 prior to proceeding with the next section.
- 3. Load the sample N-Browse database dump into the newly created database. Start MySQL as the N-Browse database handler:
- $ mysql -u handler -p
- Enter password: ********
- Switch to the nbrowse database and load the SQL dump for the sample database (nbrowse_sample_data.sql located in the nbrowse_dataloader/ directory):
- mysql> use nbrowse;
- mysql> SOURCE /home/bob/nbrowse_install_package/nbrowse_dataloader/ nbrowse_sample_data.sql;
As in step 2, replace the Unix path above with the absolute path of the nbrowse_schema.sql file on your system.
4. Test the GUI for your N-Browse installation: Go to the N-Browse URL on your system, enter a query (e.g., “par-6”), and click GO. The N-Browse GUI should appear with a small sample network.
Populate an N-Browse database from flat files
This section provides instructions for populating an N-Browse database using a set of Perl scripts provided with the distribution package for the user's convenience. However there are many other ways to populate the database, and users should feel free to use whatever method works best for them. Other methods will typically involve generating the appropriate SQL commands with customized scripts. To load the database with sample data included in the N-Browse distribution, follow the steps outlined in this section using default configuration parameters. Upon completion of these steps (assuming you have previously completed Basic Protocol 3) you should be able to navigate network data on your site using the N-Browse GUI.
In addition to the table definition file, a diagram of the N-Browse database schema is included in the N-Browse distribution package. To help users learn to understand the schema, examples of SQL queries to retrieve different kinds of data from a populated N-Browse database are also provided. The N-Browse database schema uses autoincrement IDs as primary keys in many of the data tables. Users who prefer to load data using scripts that automatically generate and cross-reference these IDs should remove the autoincrement flags from the corresponding table definitions.
- 5. Prepare tab-delimited .csv files containing the data you wish to load to the database: The required fields for each .csv file are described in the dataloader_csv_format.txt file in the nbrowse_dataloader/ directory. The directory example_data/ contains the sample data files.The .csv format can be generated programmatically with a script, manually using a text editor, or automatically by Microsoft Excel or OpenOffice Calc. To export data in this format, place the data in different columns and then save as a “tab-delimited” file. (Data in each column can include spaces, but should not include the “tab” character.)
- 6. Configure the dataloader_conf file in the nbrowse_dataloader/ directory: The Perl dataloader scripts consult this configuration file for the names of the various data files to be loaded to the database. Specify the names of the data files you have prepared as values for the corresponding parameters in the configuration file (or, to load the sample database, use the default parameters).Table 9.11.2 presents parameters in the dataloader_conf file.
- 7. Populate tables in the nbrowse database: Each Perl dataloader script populates the database with a different type of information, as described below. Only the first two are essential for network display: (i) information about the identity (and descriptions) of nodes and (ii) information about edges (the types of functional links and any associated numerical values). Node data should be loaded first.
- Load information about nodes (genes and/or proteins) and their synonyms:
- $ cd nbrowse_dataloader
- $ perl dataloader_node_syn.plThis script populates the tables node, synonym, and attribute in the nbrowse database. The .csv files TABLE_NODES and TABLE_SYN are required to run this script.
- Load information about edges:
- $ perl dataloader_edge_meta.plThe script populates the tables (gnb_interactions, edge_group, edge_attribute, attribute, and metadata) in the nbrowse generic database. The .csv files TABLE_EDGEDEF and TABLE_GNBI are required to run this script. Currently, this script only populates binary interactions in the generic schema (vs. interactions with multiple partners, such as obtained from co-IP data).
- Load information about external links:
- $ perl dataloader_url.plThis is optional. The script populates the tables (external_url, url_attribute, attribute, and metadata) in the nbrowse database. The .csv file TABLE_URL is required to run this script. This is useful for you to show your client-users more detailed information on nodes or edges.
- Load information about node attributes:
- $ perl dataloader_node_attr.plThis is optional. The script populates the tables (node_attribute, attribute, and metadata) in the nbrowse generic database. The .csv file TABLE_NODE_ATTR is required to run this script. This is for auto-constructing menu options for the Node Attribute menu (see Basic Protocol 1: Highlight node attributes on the network graph.).If you completed steps 4-7 above using the default parameters and thus have loaded the sample database, make sure to truncate all the tables in the nbrowse database or drop the database completely and repeat steps 1 and 2 above prior to loading the actual data you wish to use.
Table 9.11.2.
Parameters in the dataloader conf Filea
| Parameter | Description |
|---|---|
| TAXON_ID [6239] | NCBI Taxonomy ID. N-Browse uses this ID to distinguish network data from different species within the same database and to retrieve species names from NCBI for display. A current list of species IDs and names can be found at ftp://ftp.ncbi.nih.gov/pub/taxonomy/ in the names.dmp file contained in the taxdump archive (distributed in various formats: .zip,.tar.Z,.tar.gz). |
|
TABLE_EDGEDEF [./example data/table edgedef.csv] |
Edge type definition file. |
|
TABLE_GNBI [./example data/table gnbi.csv] |
Binary interaction data file. |
|
TABLE_NODES [./example data/table nodes.csv] |
File containing node primary names and descriptions. |
|
TABLE_SYN [./example data/table syn.csv] |
Node synonyms for search and display functions. A priority score specifies the preferred names for display. |
|
TABLE_URL [./example data/table url.csv] |
Optional data file specifying the construction of call strings for external URLs that can be attached to nodes or edges (e.g. links to other database resources). |
|
TABLE_NODE_ATTR [./example data/table node attr.csv] |
Optional data file of node attributes (e.g. BlastP E-values, phenotypes, domains, expression levels, etc.) |
| MYSQL_SERVER [localhost] | MySQL server location. |
| MYSQL_PORT [3306] | MySQL server port number. You can leave it empty if you connect to “localhost” (MySQL default setting). |
| MYSQL_DATABASE_NAME [nbrowse] | MySQL N-Browse database name. |
| MYSQL_USERNAME [handler] | MySQL username with database write privilege. |
| MYSQL_PASSWORD [] | MySQL password for the above MySQL user. It can be empty if the MySQL user has no password. |
These parameters must be customized for your server machine and data filenames. Using the default parameter values shown in square brackets to run the Perl dataloader scripts will populate an empty N-Browse database with the sample data provided with the N-Browse distribution package.
COMMENTARY
Background Information
The central idea behind N-Browse is to develop an easily accessible, simple yet powerful tool that enables biomedical researchers to quickly extract data and generate hypotheses from the results of large-scale analyses in diverse organisms. Inspired by GBrowse (UNIT 9.9), an open-source software package that provides a Web-based GUI for coordinate-based genome annotations supported by a light-weight database, N-Browse aims to provide an analogous intuitive portal for network exploration and an easily configurable client-server package for distribution. The data content available from an N-Browse server, in terms of both functional linkage types and species-specific data, will vary at different providers' sites, but any data that can be described as nodes and can be displayed.
Several applications now provide similar network visualization tools, including Cytoscape (UNIT 8.13), Osprey, VisANT (UNIT 8.8), and STRING (http://string.embl.de/). Each was designed with differing goals and implemented independently. Different tools share certain features with the vision of N-Browse, such as navigating functional relationships based on data available from a remote server (e.g., Osprey applet version, VisANT, and STRING) or providing an open-source package for distribution (Cytoscape). N-Browse occupies a unique niche as a simple yet powerful on-demand navigation tool that allows researchers access to heterogeneous data through a Web browser in a highly interactive way and in a rich contextual environment. N-Browse can be easily integrated with other Web resources via URL links and its functionality is extensible through the integration of new data types and software plug-ins. Among the tools mentioned above, N-Browse is unique in offering an open-source client-server system supported by a generic database schema that is freely available for distribution. The N-Browse client-server package is suitable as a data distribution and visualization mechanism for any research group that wishes to serve network-related data to the public. N-Browse is affiliated with the Generic Model Organism (GMOD) project, which provides open-source software components for distribution of genomic and functional genomic data for any organism. A description of N-Browse and links to other N-Browse resources can be found on the GMOD Wiki site at http://www.gmod.org/wiki/index.php/nbrowse.
Critical Parameters and Troubleshooting
Below are addressed some common issues and questions encountered during use or installation of the N-Browse package:
Expansion steps are taking a long time to appear after double-clicking on a selected node. Is there a way to improve the querying process?
Everything is calculated on-the-fly when a client-end user queries a node. If the subnetwork of each node you have in your database is very large, you may want to activate a function that caches the edge number around each node. This will potentially save querying time if the hairball around the querying node is humongous. To implement this option (after deploying the package and populating the database), perform the following steps.
Change directory to the servelets location (TOMCAT_WEBAPPS_PATH/TOMCAT_APP_FOLDER/WEB-INF/classes), replacing TOMCAT_WEBAPPS_PATH and TOMCAT_APP_FOLDER with the parameters specified in your install.conf file (see Basic Protocol 3 and Table 9.11.1).
- Run the following command (you must have permission to run sudo):
- $ sudo java -classpath . databaseProcess.UpdateEdgeNum
Note that this script might take a while to run, depending on how many nodes and edges are contained in the database.
No data shows in the network browser GUI. What happened?
This is most likely a problem with the Tomcat security policy. To check this, perform the following steps.
Examine whether your Tomcat opens the SocketPermission.
Using your Web browser, try linking to the following URL (substituting the uppercase text with appropriate values specified in your install_conf file): http://TOMCAT_SERVER:TOMCAT_PORT/TOMCAT_APP_FOLDER/database.jsp.
If you see the text “1 2 3” appear in your browser window, your Tomcat server is communicating well with your MySQL server. If you see “1 2” and tons of exception messages, you may need to change your Tomcat policy to allow the connection to establish. Each version of Tomcat might behave differently. One possible fix you can try is to change the tomcat5 security policy as follows: (a) Find the file policy.d/04webapps.policy on your machine. (b) Copy the following lines and paste into the 04webapps.policy file:
//allow MySQL connect
permission java.net.SocketPermission “localhost”, “connect, resolve”;
//allow getting species information from NCBI
permission java.net.SocketPermission “www.ncbi.nlm.nih.gov:80”, “connect, resolve”;
- (c) Run the following command to restart the Tomcat Web server (you must have sudo permission):
- $ sudo /etc/init.d/tomcat5 restart
Why is my user upload function not working?
Again, this may be a problem with the tomcat5 policy. Here are some suggestions you may want to try:
Modify the tomcat5 policy:
Find the file policy.d/04webapps.policy on your machine.
- Copy the following lines and paste into the 04webapps.policy file:
- permission java.io.FilePermission “/var/lib/tomcat5/temp/-”, “read,write,delete”;
- permission java.io.FilePermission “/tmp/-”, “read,write,delete”;
- permission java.io.FilePermission “./temp/-”, “read,write,delete”;
- permission java.io.FilePermission “./uploads/-”, “read,write,delete”;
- permission java.util.PropertyPermission “java.io.tmpdir”, “read”;
- Run the following command to restart Tomcat Web server (you must have sudo):
- $ sudo /etc/init.d/tomcat5 restart
- Re-compile the servelets for the data upload function: Change directory to the Tomcat Web application directory (NBrowse or whatever name you gave it in the install_conf file), and run the following commands:
- $ cd WEB-INF/classes
- $ javac -classpath . com/raditha/megaupload/*.java
- $ sudo /etc/init.d/tomcat5 restart
Test the user upload function again. You can use the example file for user uploads user_upload_example.txt located in the nbrowse_server_client/ directory to test the user upload function.
Why is my Cytoscape auto-launch function not working?
Check the permissions of the cgi-bin/ directory. The default setting for apache2 is to allow execution of all file extensions in this directory as CGI scripts. If you change these permissions, you must at least allow files with .cgi,.pl, and .jnlp extensions to run as executable CGI scripts in CYJNLP_LOCATION/ (this directory is specified in the install_conf file; see Basic Protocol 3 and Table 9.11.2).
ACKNOWLEDGEMENTS
We thank Fabio Piano and Yih-Shien Chiang for invaluable brainstorming sessions, advice, and suggestions during the development process and Leslie Greengard for his encouragement and support. We thank the following beta testers of the N-Browse software for helpful feedback on the installation process: Payan Canaran, Todd Harris, and Igor Antoshechkin (from WormBase) and Nicolas Simonis and Changyu Fan at the Center for Cancer Systems Biology (Dana Farber Cancer Center, Harvard). This work was supported by Department of the Army Award W81XWH-04-1-0307 and NYSTAR Contract #C040066.
KEY REFERENCE
- Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, MacMenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. This is the first article in the literature to describe the use of N-Browse for integrating a new genome-scale dataset with other available molecular interaction data. N-Browse was used to integrate microRNA-target predictions with multiple types of functional links in C. elegans gathered from a variety of sources (these datasets are described on the gnetbrowse.org Web site). [DOI] [PubMed] [Google Scholar]
INTERNET RESOURCES
- http://gnetbrowse.org The main N-Browse Web site, currently providing access to heterogeneous functional data in E. coli, C. elegans, D. melanogaster, and H. sapiens (see the Web site for details on available datasets). Provides a link to the downloadable N-Browse client-server distribution package.
- http://sourceforge.net/projects/nbrowse The N-Browse client-server distribution package can be downloaded from here.
- http://www.gmod.org/wiki/index.php/nbrowse Provides a description of the N-Browse project with news and links to other N-Browse resources.
- http://www.wormbase.org The first example of an independent N-Browse client-server installation. WormBase currently uses N-Browse as a graphical interface to server molecular interaction data curated there. Links to the N-Browse GUI at WormBase are available on the Gene Summary pages. Also see UNIT 1.8.
- http://interactome.dfci.harvard.edu/C elegans/host.php An N-Browse portal is provided by the CCSB Interactome Database to visualize C. elegans protein-protein interaction data in the context of other functional genomic data.