SUMMARY
Kindlin-2 belongs to an emerging class of regulators for heterodimeric (α/β) integrin adhesion receptors. By binding to integrin β cytoplasmic tail via its C-terminal FERM-like domain, kindlin-2 promotes integrin activation. Intriguingly, this activation process depends on the N-terminus of kindlin-2 (K2-N) which precedes the FERM domain. The molecular function of K2-N is unclear. We present the solution structure of K2-N, which displays a ubiquitin fold similar to that observed in kindlin-1. Using chemical shift mapping and mutagenesis, we found that K2-N contains a conserved positively charged surface that binds to membrane enriched with negatively charged phosphatidylinositol-(4,5)-bisphosphate. We show that while wild-type kindlin-2 is capable of promoting integrin activation, such ability is significantly reduced for its membrane-binding defective mutant. These data suggest a membrane-binding function of the ubiquitin-like domain of kindlin-2, which is likely common for all kindlins to promote their localization to the plasma membrane and control integrin activation.
Keywords: Kindlin, NMR, membrane, integrin activation, cell adhesion
INTRODUCTION
The binding of extracellular matrix (ECM) proteins to integrin transmembrane receptors is essential for the adhesion of cells on the ECM and, as a consequence, regulates a variety of cellular responses such as spreading, migration, and differentiation. The ligand binding function of integrins is tightly regulated through an “inside-out” activation mechanism by which the cytoplasmic face of the integrin engages intracellular proteins, which ultimately triggers conformational changes in the ectodomain of the receptor such that it acquires high affinity for ECM ligands (Qin et al., 2004; Luo et al., 2007; Shattil et al., 2010). Over the last two decades, intensive investigations have converged to demonstrate that the mechanism of “inside-out” activation depends on binding of the N-terminal head domain of talin (talin-H) to the integrin β cytoplasmic tail (CT), which disrupts its association with α CT and triggers the conformational transitions in the receptor (Vinogradova et al., 2002, 2004; Kim et al., 2003; Kim et al., 2009). However, quantitative analysis indicated that talin-H alone was insufficient to induce full integrin activation (Ma et al., 2006) and suggested that co-factors may be required for efficient integrin activation (Ma et al., 2007). Studies in cells, mice and human have now demonstrated that the kindlins function as essential regulators of integrin activation (for review, see Moser et al., 2009; Plow et al., 2009). Kindlins, a three member subfamily of FERM (four-point-one, ezrin, radixin, moesin) containing proteins (for review, see Moser et al., 2009; Plow et al., 2009) can dramatically enhance talin-mediated integrin αIIbβ3 activation, supporting their role as co-activators of integrins (Ma et al., 2008; Montanez et al., 2008; Harburger et al., 2009; Malinin et al., 2009; Svensson et al., 2009). Among the three kindlin family members, kindlin-2 is widely expressed and highly concentrated at integrin-rich cell-ECM adhesions (Tu et al., 2003), and its deficiency leads to embryonic lethality in mice and zebrafish (Dowling et al., 2008; Montanez et al., 2008). Although both kindlin-2 and talin-H contain FERM domains, which are homologous, they bind to distinct sites in integrin β3 CT (Shi et al., 2007; Ma et al., 2008; Montanez et al., 2008). Such distinct binding may promote the cooperation of the two molecules to induce a full activation of integrins. Interestingly, in addition to their FERM-like domains, kindlins have distinct N-terminal F0 domains (Shi et al., 2007; Ma et al., 2008; Montanez et al., 2008; Goult et al., 2009). Deletion of this domain dramatically impairs the kindlin-mediated integrin activation, suggesting that it is mechanistically important for the kindlin function (Ma et al., 2008; Harburger et al., 2009; Goult et al., 2009).
In this study, we have determined the solution structure of the kindlin-2 F0 domain using multidimensional NMR spectroscopy. As expected, it adopts the ubiquitin fold similar to that found in kindlin-1 (Goult et al., 2009). However, further detailed structural comparison and NMR analysis revealed surprisingly a highly conserved and positively charged surface in K2-N, which mediates association with membrane enriched with highly negatively charged lipids such as phosphatidylinositol (4,5) bisphosphate (PIP2), a known regulator of talin activation and integrin signaling (Martel et al., 2001; Goksoy et al., 2008; Saltel et al., 2009; Garcia-Bernal et al., 2009). Structure-based mutagenesis experiments demonstrate that disruption of this F0-mediated membrane binding substantially reduced the ability of kindlin-2 to cooperate with talin in inducing activation of integrin αIIbβ3. These data unravel a novel membrane-targeting function of the N-terminal ubiquitin domain of kindlin-2, which is likely common for promoting localization of kindlins in cells and for their regulation of integrin activation.
RESULTS
Structure of the kindlin-2 F0 subdomain
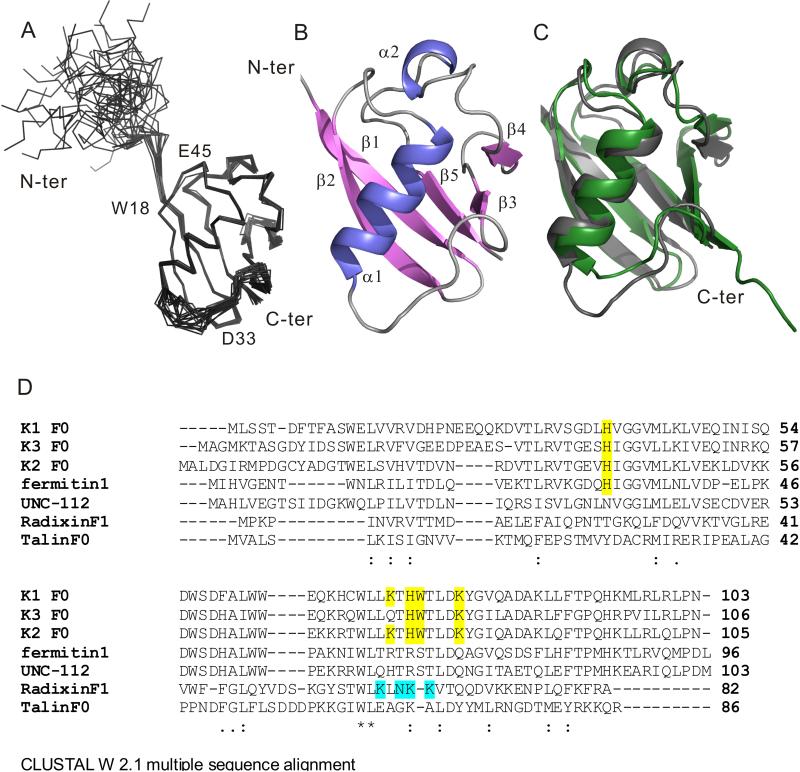
Multiple constructs were explored for the optimal expression and solubility of kindlin-2 N-terminus containing the F0 subdomain. These constructs included kindlin-2 1-105, 1-134, 1-154, 11-143, and 1-218; among these, 1-105 (termed K2-N hereafter) yielded a highly soluble and stable protein suitable for detailed NMR structural analysis. The superposition of 20 lowest energy structures is shown in Fig 1A (see also Table 1 for structural statistics and Fig S1A for the backbone assignment of excellent HSQC spectrum of K2-N). The structures are well converged except the N-terminal M1-D15 that appears to be highly flexible with few long range NOEs. Fig 1B provides the ribbon representation, which reveals an ubiquitin fold with a long α1 helix packing against a five-stranded β-sheet. A short 310 α2 helix seen in ubiquitin is also present in K2-N sitting on the top of the α/β fold (Fig 1B). The overall backbone r.m.s.d between K2-N and ubiquitin (PDB code 1UBQ) is only 1.7Å despite little sequence homology (Fig 1C). K2-N is also very similar to the homologous kindlin-1 F0 (Goult et al., 2009) (sequence identity 66%, see Fig 1D) with the backbone r.m.s.d of 1.8Å (Fig S1B). However, the conformations of two loop regions T25-N28 and D53-H61 in K2-N differ from those in kindlin-1 D19-Q26 and N51-Y58 respectively (Fig S1B). K2-N T25-N28 is much shorter than the corresponding kindlin-1 D19-D26 loop. The overall structure of Kindlin-3 F0 is also expected to be similar to kindlin-1 (61% sequence identity) and kindlin-2 (59% sequence identity) (Fig 1D).
Fig. 1.
Structure of K2-N and its comparison with other ubiquitin-like domains. (A). Superposition of 20 lowest energy structures of K2-N. See also supplementary Fig S1A and Table 1. (B). A cartoon model diagram of residues 16-93 of the lowest energy structure. (C) K2-N has a similar beta grasp fold (grey) to ubiquitin (green). (D) Clustal W 2.1-based multiple sequence alignment of K2-N with homologous ubiquitin domains including kindlin-1,3 F0 domains, Ferminitin1 from Drosophila, UNC-112 from C. elegance, radixin F1 domain, and talin F0 domain. Residues highlighted in yellow are potentially involved in binding to membrane (see text). Residues highlighted in cyan are involved in membrane binding for radixin F1 (Hamada et al., 2000). See also supplementary Fig S1B.
Table 1.
Structural Statistics of K2-N
| Complex | |
|---|---|
| Number of NOEs | 1412 |
| intra-residual, | 447 |
| sequential, |i-j|=1 | 436 |
| medium range, 1<|i-j|<5 | 222 |
| long range, |i-j|≥5 | 307 |
| Number of dihedral restraints | 108 |
| Rmsd deviationa | |
| NOEs (no violations > 0.5 Å) | 0.0786±0.0032 |
| dihedrals (°) (no violations > 5°) | 1.5291±0.1257 |
| bonds (Å) | 0.0057±0.0001 |
| angles (°) | 0.6908±0.0154 |
| impropers (°) | 0.5335±0.0191 |
| EL-J (kcal/mol, based on CHARMM19 parameters) | -310.5±9.8 |
| Ramachandran plot | |
| allow regions(%) | 99.3 |
| disallowed regions (%) | 0.7 |
| Coordinate precision (Å)b | |
| Backbone atoms | 0.52±0.14 |
| All heavy atoms of TMCD region to the mean | 1.06±0.14 |
Mean ± standard deviation over 20 structures with lowest energies.
Structured region: G16 – T93
K2-N has a conserved positively charged surface that binds to PIP2-enriched negatively charged membrane
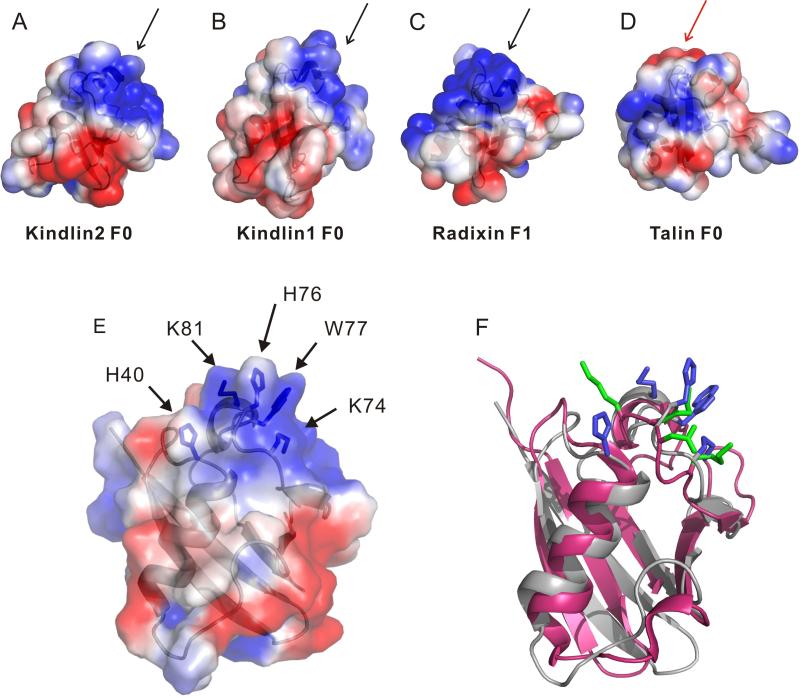
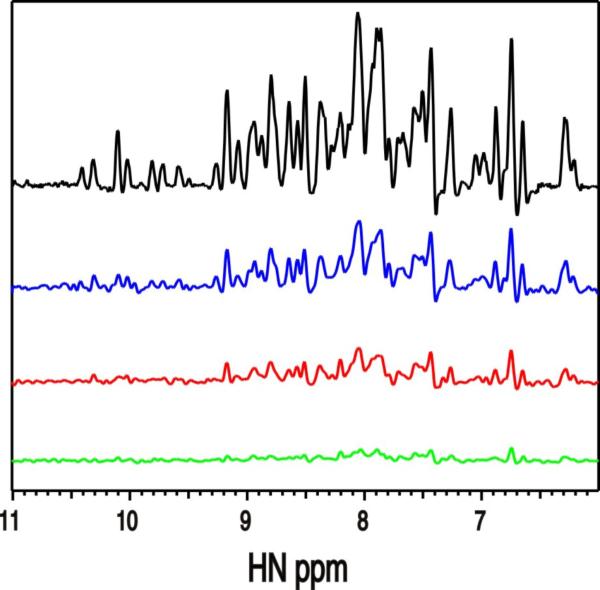
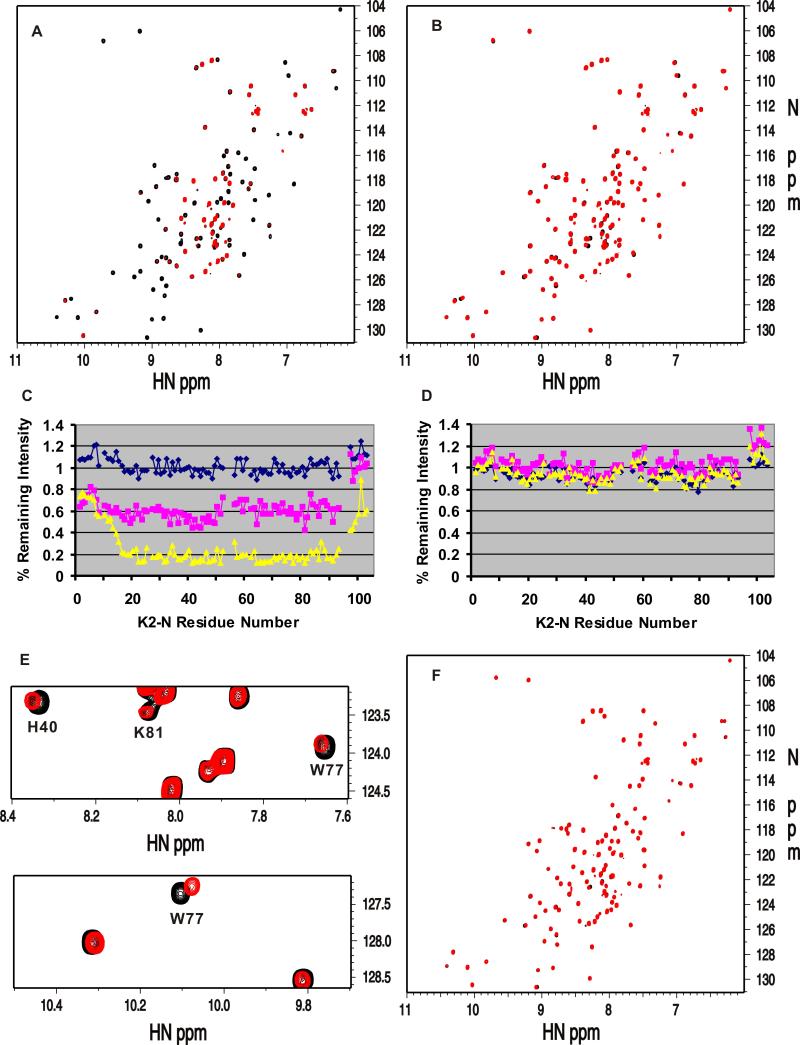
While kindlins all contain the ubiquitin-like N-terminus, a key question is how it controls the function of the FERM domain that binds integrin via the C-terminal F3 or PTB subdomain (Shi et al., 2007; Ma et al., 2008; Harburger et al., 2009). To address this issue, we first examined the possibility of K2-N binding to known positive regulators of integrin activation: talin-H and migfilin (Shattil et al., 2010). The former directly binds to integrin β CT and co-activates integrin with kindlin-2 (Ma et al., 2008; Montanez et al., 2008; Goult et al., 2009) whereas the latter has been suggested to promote integrin activation by dissociating filamin – a negative regulator of talin-integrin interaction, from integrin β CT (Lad et al., 2008; Ithychanda et al., 2009). However, detailed NMR analysis revealed that K2-N exhibits little interaction with either talin-H or migfilin (Fig S2). Next, we performed an in silico comparative analysis of K2-N, kindlin-1 F0, and structurally homologous talin F0 and radixin F1 (Goult et al., 2009). Interestingly, we found that K2-N, kindlin-1 F0, and radixin F1 but not talin F0 share a positively charged surface (Fig 2A-D) despite that F0 of both kindlin-1 and kindlin-2 have little sequence homology with the corresponding F1 subdomain of radixin (Fig 1D). Fig 2E displays a cluster of residues H40, K74, H76, W77, and K81, which are contained in this positively charged surface. This cluster corresponds to a similar cluster in radixin F1 involving K60, N62, K63, and K64 (Fig 2F). Because radixin F1 is well-known as part of a FERM domain that utilizes these residues to bind phosphatidylinositol (4,5) bisphosphate (PIP2) and possibly surrounding membrane surface (Hamada et al., 2000), we wondered if K2-N also has the capacity to bind PIP2-containing membrane via its corresponding positively charged surface. The involvement of PIP2 is plausible at functional level because: (i) PIP2 is highly enriched in the inner membrane of the integrin adhesion site (Ling et al., 2002; di Paolo et al., 2002; Cluzel et al., 2005; Saltel et al., 2009) where kindlin-2 is a key player; and (ii) PIP2 is involved in activating talin (Martel et al., 2001; Goksoy et al., 2008; Saltel et al., 2009; Garcia-Bernal et al., 2009) that cooperates with kindlin-2 for inducing integrin activation (Ma et al., 2008; Montanez et al., 2008; Harburger et al., 2009). To examine this possibility, we first performed NMR-based binding analyses in which we added K2-N into a large unilamilar vesicle (LUV) that consisted of 80% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 20% PIP2. The 1D trace of the HSQC of 15N-labeled K2-N shows that K2-N signals experienced increasing line-broadening effect with increasing concentration of the vesicle at the protein/vesicle molar ratio of 1:5, 1:20, and 1:50 respectively, indicating that K2-N indeed interacts with the negatively-charged membrane (Fig 3). To investigate the lipid specificity, we compared the effect of PIP2 and POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-L-serine]) with the latter having different head group and being much less negatively charged. We chose the vesicle condition where protein/lipid molar ratio is 1:20 that caused substantial line-broadening of K2-N in Fig 3 (colored in red) and replaced the PIP2 in the vesicle with the same amount of POPS. While PIP2 caused the substantial line-broadening of K2-N (Fig 4A), the same amount of POPS caused little line-broadening of K2-N (Fig 4B). We further found that the line-broadening effect of K2-N was PIP2 concentration-dependent (Fig 4C). In contrast, K2-N had little line-broadening effect regardless of the POPS concentration (Fig 4B). These data demonstrate that K2-N is highly specific to the negatively charged PIP2. At lower lipid/protein ratio where line-broadening is less, we were able to map a cluster of residues whose chemical shifts are perturbed by PIP2 including H40, K74, H76, W77, and K81 (Fig 4E). Remarkably, these perturbed residues are mapped to the positively charged surface we have identified in Fig 2E and 2F. To be more definitive, we made a mutant where the clustered residues are all mutated into Ala, i.e., K40A/K74A/H76A/W77A/K81A. Contrasting to the wild type K2-N that caused substantial line-broadening in the PIP2 vesicle (Fig 4A), the mutant spectra had little change with and without the same vesicle (Fig 4F), confirming the specificity of this positively charged surface to the PIP2-enriched membrane. Importantly, the structural integrity of the mutant was not affected by these surface mutations as judged by its HSQC spectrum that is very similar to that of wild type K2-N (Fig 4F), demonstrating that the mutation effect was solely due to the disruption of the K2-N binding to membrane. Since PIP3 is the product of PIP2, which also regulates the kindlin-2 function in a temporal manner (Qu et al., 2011), we wondered if PIP3 with one additional phosphate group as compared to PIP2 would also bind K2-N. Fig S3 shows that the head group of PIP3 induced larger chemical shift changes of K2-N than that of PIP2 suggesting that the more negatively charged PIP3 binds better to K2-N than PIP2, a scenario that also occurs to the kindlin-2 PH domain (Qu et al., 2011).
Fig. 2.
Electrostatic surface property of K2-N as compared to other ubiquitin domains. (A) Representation of the electrostatic surface of Kindlin-2 F0 (K2-N, 13-93); (B) kindlin-1 F0 (10-96, PDB code: 2KMC); (C) radixin F1 (PDB code: 1GC6); and (D) talin F0 (PDB code: 3IVF). Black arrows indicate the similar positively charged regions (blue) in kindlin 1,2 and radixin F1, contrasting to the red arrow in talin F0. All proteins were oriented in the same way as radixin F1 that is positioned to bind to PIP2. (E) The electrostatic surface of K2-N highlighting the cluster of residues included in the positively charged surface H40, K74, H76, W77 and K81. (F) Comparison of the clustered residues (blue) in the positively charged surface of K2-N (grey) with those (blue) in radixin F1 (pink). The latter green residues are involved in binding to membrane (Hamada et al., 2000).
Fig. 3. Interaction between K2-N and membrane surface (LUV/PIP2).
One dimensional projections of 2D HSQC spectra of 0.1mM 15N-labeled K2-N in the absence (black, top) and presence of the LUV consisting of 80% POPC and 20% PIP2 at the protein:total lipid rations of 1:5 (blue), 1:20 (red), and 1:50 (green), respectively. Each sample contains 0.1mM K2-N and 0.5mM (1:5 ratio) or 2mM (1:20 ratio) or 5mM (1:50 ratio) total lipid, respectively. The spectra reveal increasing line broadening effect with increasing vesicle concentrations. See supplementary Fig S2.
Fig. 4. K2-N is highly specific to negatively charged PIP2 containing membrane surface.
(A) Superposition of 2D HSQC spectra of 0.1mM 15N-labeled K2-N in the absence (black) and presence (red) of 2mM LUV composing of POPC and PIP2 at 4:1 molar ratio. Substantial line-broadening occurs by the PIP2 containing LUV. (B) Superposition of 2D HSQC spectra of 0.1mM 15N-labeled K2-N in the absence (black) and presence (red) of 2mM LUV composing of POPC and POPS at 4:1 molar ratio. As compared to (A), no line-broadening occurs by the POPS-containing LUV. (C)-(D) Line-broadening analysis of K2-N as a function of PIP2 (C) or POPS (D) concentration in LUV. Plotted is the percentage of remaining intensity of individual residues of K2-N in LUV as compared to that of free form K2-N. Membranes used in (C) are 2mM total POPC/PIP2 mixture containing PIP2 at 5% (blue line, top), 10% (pink, middle), and 20% (yellow, bottom). Membranes used in (D) are 2mM total POPC/POPS mixture containing POPS at 5% (blue line), 10% (pink), and 20% (yellow). PIP2 causes line-broadening in (C) but not POPS (D) indicating that more negatively charged PIP2 is more specific to K2-N than POPS. (E) Representative zoomed regions of the HSQC spectra of 0.1mM wild type K2-N in the absence (black) and presence of 0.5mM POPC:PIP2 LUV containing 20% PIP2 showing that some residues are selectively shifted and broadened. (F) Superposition of 2D HSQC spectra of 0.1mM K2-N mutant H40A/K74A/H76A/W77A/K81A with (red) and without (black) 2mM POPC/PIP2 LUV containing 20% PIP2, showing that the mutations completely abolished the membrane binding to K2-N as compared to wild-type K2-N in (A) at the same experimental condition. See supplementary Fig S3.
The membrane/kindlin-2 F0 interaction promotes the kindlin-2 mediated co-activation of integrin αIIbβ3 with talin
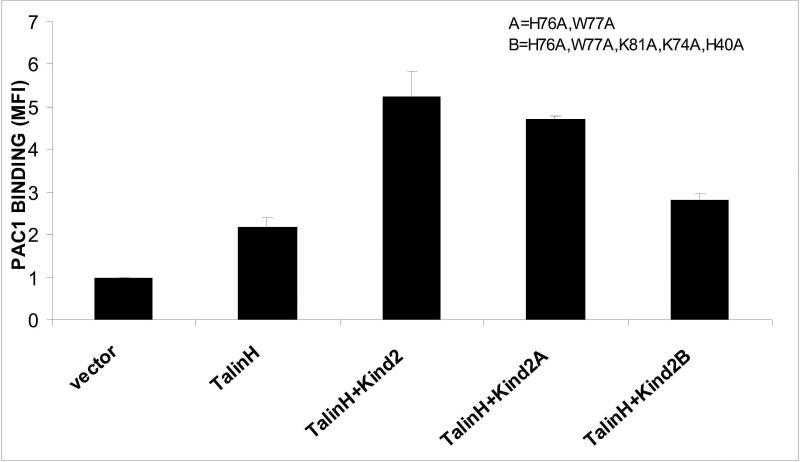
What is then the functional significance of the K2-N binding to the highly negatively charged membrane? To address this critical issue, we compared the effects of wild type and kindlin-2 mutants in integrin activation assays since the major function of kindlin-2 is to bind to and activate integrins (Moser et al., 2009; Plow et al., 2009). We chose two types of mutants as derived from the positively charged surface in Fig 2E: (A) H76A/W77A that does not significantly alter the surface charge; (B) H76A/W77A/H40A/K74A/K81A that dramatically reduces the surface charge and the binding of K2-N to membrane (Fig 3D). Fig 4 shows that wild type kindlin-2 potently enhances talin-mediated integrin αIIbβ3 activation, consistent with prior studies (Ma et al., 2008; Montanez et al., 2008; Harburger et al., 2009). By contrast, the membrane binding defective H76A/W77A/H40A/K74A/K81A mutant exhibited significantly reduced co-activator effect (P<0.003 compared to wild-type kindlin-2 and talin-H). In comparison, H76A/W77A had no significant effect on the co-activator activity of kindlin-2 (Fig 4). These data provide strong mechanistic evidence that the positively charges surface of K2-N is crucial for targeting kindlin-2 to highly negatively charged membrane for regulating integrin activation. It is conceivable that such binding would promote the kindlin-2 localization to the plasma membrane for effective integrin binding and cooperation with talin which is also regulated by negatively charged membrane (Martel et al., 2001; Goksoy et al., 2008; Saltel et al., 2009), thereby leading to the potent integrin activation.
DISCUSSION
Kindlins belong to a distinct class of FERM containing proteins that are composed of F0, F1, F2 with an inserted PH, and F3 subdomains. Here, we showed that kindlin-2 F0, which resides N-terminal to the F1 subdomain, has the capacity to bind to highly negatively-charged PIP2-enriched vesicle. Despite little sequence homology between kindlin-2 F0 and radixin F1 which both have ubiquitin fold, the two proteins remarkably share a positively charged surface that recognizes PIP2. Compared to many known phospholipid binding domains such as PH and FYVE (Lemmon, 2008), such ubiquitin fold in binding to phospholipid is interesting since ubiquitin is typically involved in protein-protein interaction and enzyme reaction. Functionally, we demonstrated that the membrane binding via this ubiquitin domain is crucial for the kindlin-2 mediated integrin activation. K2-N seems to be highly specific to the negatively charged PIP2 and PIP3 with the latter being more preferential. This feature is reminiscent of the PH domain inserted into the F2 domain of kindlin-2, which was also found to bind PIP2 and preferentially PIP3 (Qu et al., 2011). How the kindlin-2 binding events to PIP2 and PIP3 are temporally regulated remains to be determined. Since both PIP2-producing enzyme (PIPKIγ) (Bolomini-Vittori et al., 2009; Garcia-Bernal et al., 2009) and PIP3-producing enzyme PI3K (Shimizu et al., 1995; Zhang et al., 1996; Byzova et al., 2000; Qu et al., 2011) are involved in promoting integrin activation, it is possible that kindlin-2 is initially anchored to membrane via PIP2 and the anchoring is further strengthened by the PI3K-mediated conversion of PIP2 to the higher affinity PIP3.
The involvement of membrane binding via both K2-N and PH domains of kindlin-2 suggests that kindlin-2 is likely anchored onto membrane via a multivalent mechanism. Such membrane anchoring topology seems to be different from radixin FERM domain which utilizes F1 and F3 to bind to one PIP2 (Hamada et al., 2000). It is also distinct from talin-H that also has the F0-FERM architecture for the following reasons: (i) Although talin-F0 has a ubiquitin fold and precedes the FERM domain, it does not have the corresponding membrane binding surface found in K2-N (Fig 2). Consistently, talin F0 displays little binding to the same PIP2 vesicles that bind to K2-N (data not shown). (ii) Unlike kindlins, talin-H does not have PH domain inserted into the F2 domain. Instead, talin-H directly utilizes its F2 subdomain to bind PIP2 (Saltel et al., 2009). Despite these differences, our data about kindlin-2 and previous studies on talin (Martel et al., 2001; Cluzel et al., 2005; Goksoy et al., 2008; Saltel et al., 2009; Garcia-Bernal et al., 2009) suggest that both talin and kindlins are co-regulated by locally enriched PIP2 and the regulation is further strengthened by the PIP2 to PIP3 conversion, thereby leading to the synergistic integrin activation. The distinct membrane anchoring topology of kindlin-2 may be spatially required to insure effective binding to integrin β CT and cooperation with talin-H. How does the cooperation occur? As mentioned above, in integrin αIIbβ3, kindlin-2 binds to the C-terminal membrane-distal region of the β3 CT and this site is distinct from the talin-H binding site (Shi et al., 2007; Ma et al., 2008). Because the C-terminal membrane distal regions of the β3 CT associate with membrane (Vinogradova et al., 2004; Metcalf et al., 2010) and dissociate upon binding to talin-H (Anthis et al., 2009), one can envisage that the membrane associated kindlin-2 binding to integrin would further facilitate dissociation of the C-terminal β3 CT from membrane thus leading to effective αIIb/β3 CT unclasping along the membrane surface and integrin αIIbβ3 activation (Ma et al., 2008; Montanez et al., 2008; Harburger et al., 2009).
In conclusion, we have identified a novel mechanism by which the N-terminal ubiquitin-like domain helps to target kindlin-2 to membrane enriched with highly negatively-charged lipids. Using structure-based mutational assays, we showed that such targeting is crucial for the kindlin-2 mediated integrin αIIbβ3 co-activation with talin. In view of the high conservation of this membrane binding site among the kindlins (Fig 1D), we propose that the N-terminal ubiquitin domain-mediated membrane binding is applicable to all three mammalian kindlins and serves as a general mechanism to facilitate targeting and regulation of integrin activation and signaling.
EXPERIMENTAL PROCEDURES
Protein subcloning, expression, purification, and sample preparation
The cDNA of human Kindlin-2 1-105 (K2-N) and other longer fragments were amplified by PCR from a full length human Kindlin-2 cDNA and cloned into the expression vector parallel pGST-1 (Sheffield et al., 1999) using EcoRI and XhoI restriction enzyme sites. GST-K2-N was expressed in BL21 (DE3) pLys E. coli strain using 0.5mM IPTG at room temperature overnight. For isotopic 15N or 15N/13C labeling of the protein, a minimal media culture containing 15NH4Cl (1.1g/L) or 15NH4Cl(1.1g/L)/13C glucose (3g/L of culture) was used, respectively. Bacterial cells were harvested by centrifugation at 3500rpm for 15min, and the cell pellet was re-suspended in a chilled phosphate buffered saline solution (PBS) containing 1mM DTT. The re-suspended cells were frozen in liquid nitrogen and lyzed overnight at 4°C, containing lysozyme 10mg/L, DNase 5mg/L, a cocktail of complete protease inhibitors, and 1mM MgCl2. The fusion protein in the clarified lysate was purified using glutathione sepharose 4B resin (GE Healthcare, Inc) according to manufacturer's protocols. GST fusion tag was cleaved using AcTEV protease (Invitrogen) and the cleaved protein was isolated and purified using size exclusion chromatography, superdex 75 (GE Healthcare, Inc). PBS with 1mM DTT was used throughout the purification protocol. Protein concentrations were measured by a UV-visible spectrophotometrically by absorbance at 280nm. The unlabelled K2-N protein yield more than 15 mg per liter of LB culture and was concentrated to 2mM without any precipitation. Migfilin LIM1 (172-242) and LIM2-3 (241-367) were cloned into pGST-1 vector and the proteins were expressed overnight using 0.5mM IPTG at 15oC. PBS buffer was used for the purification of the migfilin LIM domains using the same procedure as K2-N. Talin-H was expressed and purified as previously described (Vinogradova et al., 2002).
LUV (large unilamellar vesicle) preparation
POPC, POPS, and PIP2 were purchased from Avanti Polar Lipids, Inc (Alabaster, AL). Large unilamellar vesicles of mixed lipid were prepared by extrusion. Briefly, lipids were first dissolved together in chloroform. The chloroform was then removed under liquid nitrogen followed by overnight vacuum. The lipid film was hydrated in the desired buffer followed by homogenization with several freeze-thaw cycles. The LUV was then formed by extruding the lipid suspension ~20 times through two stacked 0.1 μm polycarbonate filters (Avanti Polar Lipids).
Sample preparation, NMR spectroscopy and structure calculation
1mM 15N/13C-labeled K2-N was prepared for NMR data collection, resonance assignment, and NOE analysis. The protein was dialyzed into a buffer solution containing 50mM sodium phosphate, 100mM NaCl, pH 6.8, and 0.05% of NaN3, and 10% (v/v) D2O was added to the sample. An unlabeled 1mM K2-N sample was prepared for the collection of NOESY and TOCSY spectra to analyze aromatic residues. A 0.1mM 15N protein solution was prepared in 100% D2O for deuterium exchange experiments. All data were collected at 298K on 600, 800, and 900 MHz BRUKER AVANCE Spectrophotometers equipped with cryoprobes. HNCA, HNCO, HNCACB, CBCACONH, H(CCO)NH, and C(CCO)NH three dimensional NMR spectra were acquired for assignment of all 1H, 13C and 15N side-chain and backbone resonances. The PASA protocol was used for the initial semi-automatic sequential backbone assignment.(Xu et al., 2006). The ambiguous assignments were further analyzed manually using HNCO - HNCACO and HNCA - HNCACB for the sequential assignment of backbone amides. 15N and 13C 3D NOESY spectra with mixing times 150ms were acquired for NOE distance restraints for structure calculation. NMR spectra were processed using NMR-PIPE (Delaglio et al., 1995) and analyzed by PIPP (Garrett et al., 1991) and NMR View (Johnson, 2004).
The structure of K2-N 1-94 was calculated according to standard protocols (95-105 were disordered and therefore excluded from the calculations). All distance restraints were manually selected from 3D NOESY spectra. Dihedral angle restraints φ/ψ were obtained with TALOS program (Cornilescu et al, 1999). H-bond restraints were identified from the initial structure and confirmed by deuterium exchange experiments. Protein structure was calculated by NIH-XPLOR. (Schwieters et al., 2003). A total of 20 lowest energy structures were selected for detailed analysis from 100 calculated structures. The stereo-chemical quality of the calculated ensemble of structures was assessed with the PROCHECK program. (Laskowski et al., 1993). All structural models were generated by PYMOL (www.pymol.org).
Membrane binding studies and site directed mutagenesis
The interaction between wild type K2-N and lipid was investigated by acquiring HSQC spectra of 0.1mM 15N-labeled K2-N in the presence and absence of PIP2 (20%) integrated LUV at 1:5, 1:20 and 1:50 molar ratios. To examine the lipid specificity, HSQC data were also acquired for 0.1mM 15N-labeled K2-N in the absence and presence of PIP2 or POPS at different concentrations in a fixed 2mM total lipid POPC/PIP2 or POPC/POPS mixture. To investigate the effect of negative charge on K2-N/lipid interaction, HSQC spectra of K2-N protein were recorded in the presence of head groups of PIP2 and PIP3, i.e., IP3 and IP4 respectively (Echelon, Inc). According to NMR data, the most perturbed residues on Kindlin 2 1-105 due to the membrane interaction were chosen for mutagenesis. The cluster of residues H40, K74, H76, W77, and K81 on K2-N fused to pGST-1 vector and full length Kindlin-2 fused to EGFP-C2 vector were sequentially mutated to alanine residues using Quick-Change-MultiSite-Directed mutagenesis kit (Stratagene). All mutations were verified by sequencing the entire constructs. HSQC experiments for the mutated K2-N protein with and without LUV were performed under the same experimental conditions as for the wild type K2-N.
Integrin αIIbβ3 activation assay
Activation of integrin αIIbβ3 was assessed as previously described (Ma et al., 2008) using PAC1, an antibody specific for the active conformation of the integrin. EGFP-fused Kindlin-2 WT or its mutants were co-transfected with Talin-H into αIIbβ3-CHO (A5) cells using Lipofectamine 2000 (Invitrogen). Cells were collected 24 h after transfection, treated with 10μg/ml PAC1 IgM and Alexa Fluor 633 goat anti-mouse IgM conjugated antibody, and fixed with 4% paraformaldehyde. Integrin activation was analyzed by flow cytometry (FACS) measuring the median fluorescence intensity (MFI) of PAC1 binding. Background staining due to secondary antibody alone was subtracted from all values. To obtain relative MFI values, median fluorescence intensities of PAC1 binding were normalized based to PAC1 binding to cells transfected with EGFP vectors alone.
Supplementary Material
HIGHLIGHTS.
Kindlin-2 adaptor has a FERM domain that binds and activates integrin
A ubiquitin-like N-terminus preceding the FERM domain controls the kindlin-2 function
The kindlin-2 ubiquitin domain binds to negatively charged membrane surface
The membrane binding to the kindlin-2 ubiquitin domain promotes integrin activation
Fig. 5.
K2-N/PIP2 binding promotes the kindlin-2 mediated integrin αIIbβ3 activation. While the WT kindlin-2 dramatically enhances talin-H-mediated αIIbβ3 activation, the kindlin-2 mutant H76A/W77A/K81A/K74A/H40A, which displayed little binding to the PIP2-containing vesicle (Fig 3F), had significantly reduced effect. In comparison, the double mutant H76A/W77A did not have significant effect, suggesting that K74/K81/H40 are crucial for binding to the negatively charged PIP2. The experiments were repeated three times with the P value <0.003 compared to WT kindlin-2 and talin-H.
ACKNOWLEDGMENTS
This work was supported by NIH grants GM62823 to J.Q. and P01 HL073311 to EFP. We thank Saurav Misra, Koichi Fukuda, Zhen Xu, Richard Page, and Xian Mao for technical assistance and useful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Atomic coordinates and NMR constraints have been deposited with the World-wide Protein Data Bank, accession code 2lgx.
REFERENCES
- Adamian L, Nanda V, DeGrado WF, Liang J. Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins. 2005;59:496–509. doi: 10.1002/prot.20456. [DOI] [PubMed] [Google Scholar]
- Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, Campbell ID. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–32. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolomini-Vittori M, Montresor A, Giagulli C, Staunton D, Rossi B, Martinello M, Constantin G, Laudanna C. Regulation of conformer-specific activation of the integrin LFA-1 by a chemokine-triggered Rho signaling module. Nat Immunol. 2009;10:185–94. doi: 10.1038/ni.1691. [DOI] [PubMed] [Google Scholar]
- Byzova TV, Goldman CK, Pampori N, Thomas KA, Bett A, Shattil SJ, Plow EF. A mechanism for modulation of cellular responses to VEGF: activation of the integrins. Mol Cell. 2000;6:851–60. [PubMed] [Google Scholar]
- Cluzel C, Saltel F, Lussi J, Paulhe F, Imhof BA, Wehrle-Haller B. The mechanisms and dynamics of {alpha}v{beta}3 integrin clustering in living cells. J Cell Biol. 2005;171(2):383–92. doi: 10.1083/jcb.200503017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, De Camilli P. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- Dowling JJ, Gibbs E, Russell M, Goldman D, Minarcik J, Golden JA, Feldman EL. Kindlin-2 is an essential component of intercalated discs and is required for vertebrate cardiac structure and function. Circ Res. 2008;102:423–31. doi: 10.1161/CIRCRESAHA.107.161489. [DOI] [PubMed] [Google Scholar]
- Elliott PR, Goult BT, Kopp PM, Bate N, Grossmann JG, Roberts GC, Critchley DR, Barsukov IL. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–99. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bernal D, Parmo-Cabañas M, Dios-Esponera A, Samaniego R, Hernán-P de la Ossa D, Teixidó J. Chemokine-induced Zap70 kinase-mediated dissociation of the Vav1-talin complex activates alpha4beta1 integrin for T cell adhesion. Immunity. 2009;31:953–64. doi: 10.1016/j.immuni.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two- three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J.Magn.Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Goksoy E, Ma YQ, Wang X, Kong X, Perera D, Plow EF, Qin J. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol Cell. 2008;31:124–33. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Bouaouina M, Harburger DS, Bate N, Patel B, Anthis NJ, Campbell ID, Calderwood DA, Barsukov IL, Roberts GC, Critchley DR. The structure of the N-terminus of kindlin-1: a domain important for alphaiibbeta3 integrin activation. J. Mo. Biol. 2009;394:944–56. doi: 10.1016/j.jmb.2009.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 2000;19(17):4449–62. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harburger DS, Bouaouina M, Calderwood DA. Kindlin-1 and -2 directly bind the C-terminal region of beta integrin cytoplasmic tails and exert integrin-specific activation effects. J Biol Chem. 2009;284:11485–97. doi: 10.1074/jbc.M809233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithychanda SS, Das M, Ma YQ, Ding K, Wang X, Gupta S, Wu C, Plow EF, Qin J. Migfilin, a molecular switch in regulation of integrin activation. J. Biol. Chem. 2009;284:4713–22. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. 2004. pp. 313–52. [DOI] [PubMed]
- Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- Kim C, Lau TL, Ulmer TS, Ginsberg MH. Interactions of platelet integrin {alpha}IIb and {beta}3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. 2009;113:4747–53. doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad Y, Jiang P, Ruskamo S, Harburger DS, Ylänne J, Campbell ID, Calderwood DA. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J. Biol. Chem. 2008;283:35154–63. doi: 10.1074/jbc.M802592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9(2):99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–47. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YQ, Yang J, Pesho M, Vinogradova O, Qin J, Plow EF. Molecular insight IIb 3 activation by distinct regions of its cytoplasmic tails. Biochemistry. 2006;45(21):6656–62. doi: 10.1021/bi060279h. [DOI] [PubMed] [Google Scholar]
- Ma YQ, Qin J, Plow EF. Platelet integrin alpha(IIb)beta(3): activation mechanisms. J Thromb Haemost. 2007;5:1345–52. doi: 10.1111/j.1538-7836.2007.02537.x. [DOI] [PubMed] [Google Scholar]
- Ma YQ, Qin J, Wu C, Plow EF. Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J Cell Biol. 2008;181:439–46. doi: 10.1083/jcb.200710196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinin, et al. A point mutation in KINDLIN3 ablates activation of three integrin subfamilies in humans. Nat Med. 2009;15(3):313–8. doi: 10.1038/nm.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, Galmiche A, Block MR, Albiges-Rizo C. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 2001;276:21217–21227. doi: 10.1074/jbc.M102373200. [DOI] [PubMed] [Google Scholar]
- Metcalf DG, Moore DT, Wu Y, Kielec JM, Molnar K, Valentine KG, Wand AJ, Bennett JS, DeGrado WF. NMR analysis of the alphaIIb beta3 cytoplasmic interaction suggests a mechanism for integrin regulation. Proc. Natl. Acad. Sci. U S A. 2010;107:22481–6. doi: 10.1073/pnas.1015545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanez E, Ussar S, Schifferer M, Bosl M, Zent R, Moser M, Fassler R. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008;22:1325–30. doi: 10.1101/gad.469408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–9. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- Plow EF, Qin J, Byzova T. Kindling the flame of integrin activation and function with kindlins. Curr Opin Hematol. 2009;16:323–8. doi: 10.1097/MOH.0b013e32832ea389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Vinogradova O, Plow EF. Integrin bidirectional signaling: a molecular view. PLoS Biol. 2004;2:e169. doi: 10.1371/journal.pbio.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu H, Tu Y, Shi X, Larjava H, Saleem MA, Shattil SJ, Fukuda K, Qin J, Kretzler M, Wu C. Kindlin-2 regulates podocyte adhesion and fibronectin matrix deposition through interactions with phosphoinositides and integrins. J. Cell Sci. 2011;124:879–91. doi: 10.1242/jcs.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltel F, Mortier E, Hytönen VP, Jacquier MC, Zimmermann P, Vogel V, Liu W, Wehrle-Haller B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell. Biol. 2009;187:715–31. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15(1):34–9. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shi X, Ma YQ, Tu Y, Chen K, Wu S, Fukuda K, Qin J, Plow EF, Wu C. The MIG-2/Integrin Interaction Strengthens Cell-Matrix Adhesion and Modulates Cell Motility. J Biol Chem. 2007;282:20455–66. doi: 10.1074/jbc.M611680200. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Mobley J, Finkelstein L, Chan A. A role for phosphatidylinositol 3-kinase in the regulation of beta 1 integrin activity by the CD2 antigen. J. Cell Biol. 1995;131:1867–1880. doi: 10.1083/jcb.131.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, et al. Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat Med. 2009;15(3):306–12. doi: 10.1038/nm.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Wu S, Shi X, Chen K, Wu C. Migfilin and mig-2 link FAs to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas T, Plow E, Qin J. A structural mechanism of integrin alpha(IIb)beta(3) “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Vaynberg J, Kong X, Haas TA, Plow EF, Qin J. Membrane-mediated structural transitions at the cytoplasmic face during integrin activation. Proc. Natl. Acad. Sci. U S A. 2004;101:4094–4099. doi: 10.1073/pnas.0400742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang X, Yang J, Vaynberg J, Qin J. PASA--a program for automated protein NMR backbone signal assignment by pattern-filtering approach. J Biomol NMR. 2006;34:41–56. doi: 10.1007/s10858-005-5358-0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang J, Shattil SJ, Cunningham MC, Rittenhouse SE. Phosphoinositide 3-kinase gamma and p85/phosphoinositide 3-kinase in platelets. Relative activation by thrombin receptor or beta-phorbol myristate acetate and roles in promoting the ligand-binding function of alphaIIbbeta3 integrin. J. Biol. Chem. 1996;271:6265–72. doi: 10.1074/jbc.271.11.6265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.