SUMMARY
Recent studies have shown that the Hippo-Salvador-Warts (HSW) pathway restrains tissue growth by phosphorylating and inactivating the oncoprotein Yorkie. How growth suppressive signals are transduced upstream of Hippo remains unclear. We show that the Sterile 20 family kinase, Tao-1, directly phosphorylates T195 in the Hippo activation loop and that, like other HSW pathway genes, Tao-1 functions to restrict cell proliferation in developing imaginal epithelia. This relationship appears to be evolutionarily conserved, because mammalian Tao-1 similarly affects MST kinases. In S2 cells Tao-1 mediates the effects of the upstream HSW components Merlin and Expanded, consistent with the idea that Tao-1 functions in tissues to regulate Hippo phosphorylation. These results demonstrate that one family of Ste20 kinases can activate another and identify Tao-1 as a component of the regulatory network controlling HSW pathway signaling, and therefore tissue growth, during development.
INTRODUCTION
During development, organisms must determine the overall size and shape of their individual organs through mechanisms not fully understood. The recent discovery of the evolutionarily conserved Hippo-Salvador-Warts (HSW) signaling pathway has revealed a unique mechanism to regulate proliferation independent of developmental patterning. The core members of the HSW pathway, Hippo (Hpo) and Warts (Wts), together with their scaffolding partners Salvador (Sav) and Mats, phosphorylate and inactivate the transcriptional co-activator Yorkie (Yki; reviewed in Edgar, 2006; Pan, 2007). Phosphorylation prevents Yorkie from translocating to the nucleus where it binds to TEAD-family transcription factors and drives the transcription of genes that promote growth and inhibit apoptosis (Huang et al., 2005). Loss of HSW pathway function in Drosophila leads to increased cellular proliferation resulting in tumor-like overgrowths in epithelial tissues. Similarly, knockout mouse models of HSW homologs grow tumors, and human HSW homologs have been implicated in cancers (Pan, 2010; Zhao et al., 2010). These studies suggest that HSW signaling is a crucial part of an organism’s ability to regulate cell proliferation and overall tissue size.
A central, unanswered question regarding HSW function is how the activity of Hpo, the most upstream kinase in the pathway is regulated. The atypical cadherin Fat and its ligand Dachsous can function at the plasma membrane to initiate HSW signaling, however the extracellular cues that trigger signaling and the mechanism by which Fat activates Hpo remain elusive (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006). In addition, genetic evidence strongly suggests that another source of Hpo activation functioning in parallel to Dachsous-Fat activation must exist (Cho et al., 2006). At least three different cytoplasmic proteins are believed to act upstream of Hpo to initiate signaling through the pathway, Expanded (Ex), Merlin (Mer), and Kibra (Hamaratoglu et al., 2006; Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010). Ex and Mer are members of the Four-point-one, Ezrin, Radixin, Moesin (FERM) family and Kibra is a WW-domain containing protein. Though these three proteins are thought to physically interact with each other in varying complexes (McCartney et al., 2000; Genevet et al., 2010; Yu et al., 2010), only Ex can form a complex with Hpo (Yu et al., 2010) and it is unclear how this interaction leads to activation of Hpo. Moreover, there is strong genetic evidence that Ex, Mer, and Kibra act in parallel to each other (Hamaratoglu et al., 2006; Maitra et al., 2006; Genevet et al., 2010), implying that other mechanisms for activating Hpo independently of Ex must exist.
We sought to identify genes that might function upstream of Hpo to activate the pathway using a candidate gene approach and discovered that the Sterile 20 kinase Tao-1 is a member of this signaling pathway. Tao-1 previously has been shown to destabilize microtubules and has been implicated in apoptosis in the Drosophila germline (Sato et al., 2007; Liu et al., 2010). Here, we show that loss of Tao-1 function results in increased cellular proliferation and upregulation of Yki target gene expression. We further demonstrate that Tao-1 regulates HSW pathway activity by phosphorylating Hpo at a critical activating residue. Thus, these results identify Tao-1 as a member of the HSW pathway and provide a molecular mechanism for Hpo activation.
RESULTS
Tao-1 has Tumor Suppressor Functions
Recent work has demonstrated that there are multiple, parallel inputs into the HSW pathway, but the molecular mechanisms that regulate Hpo activation remain unclear. Given that two Sterile 20 family kinases, Hpo and Slik (Harvey et al., 2003; Hughes and Fehon, 2006), have been implicated in HSW signaling, we wondered if other family members might be involved in upstream regulation of this growth control pathway. Using RNA interference in the wing imaginal disc, we tested the remaining nine Sterile 20 kinases in the Drosophila genome (Figure S1A) for effects on growth and HSW target gene expression. Of these, only Tao-1 emerged as a strong regulator of growth and HSW pathway output.
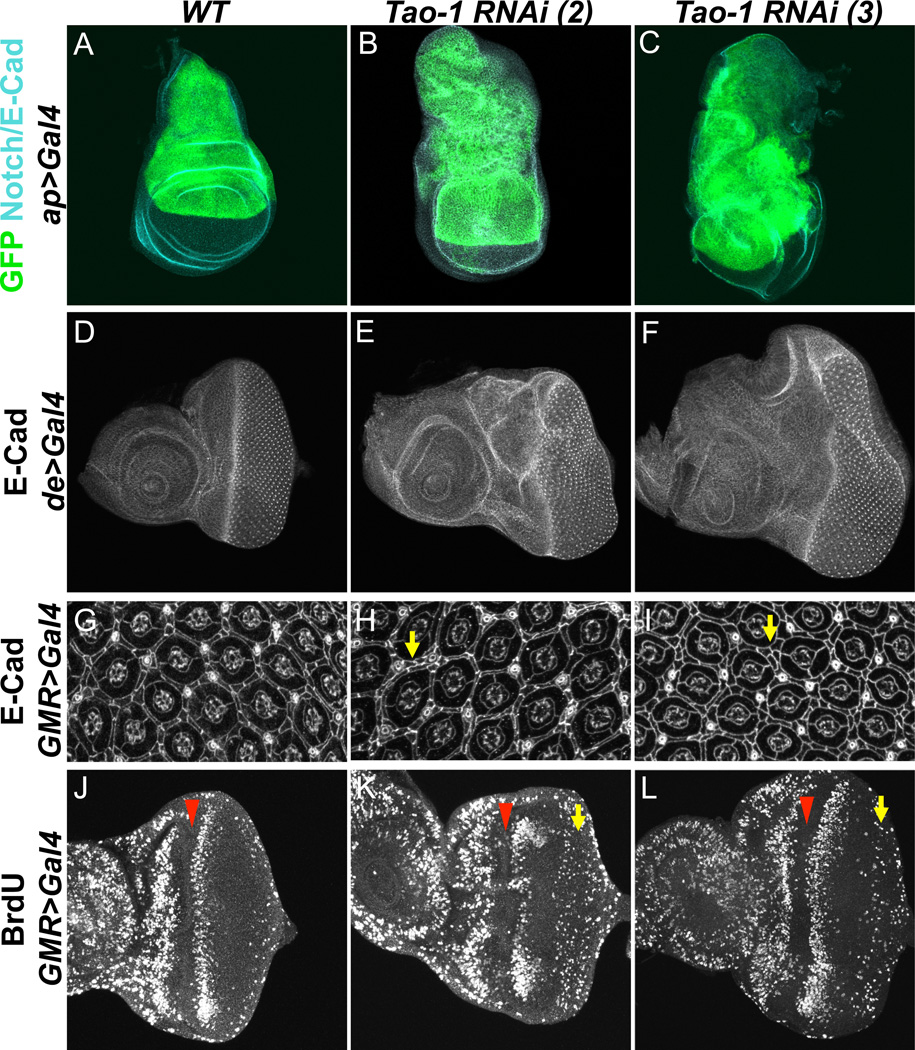
Compared to wild-type controls, the dorsal compartment of wing imaginal discs from animals expressing Tao-1 RNAi under the control of the apterous-Gal4 driver (ap>Tao-1 RNAi) was larger than normal and misshapen (Figure 1A–C). In adult wings from animals expressing Tao-1 RNAi under the decapentaplegic-Gal4 driver (dpp>Tao-1 RNAi), extra folds and vein material were present (Figure S1B–C). Quantifying the area between veins three and four of these wings revealed a statistically significant increase in size relative to wild-type wings (**p<0.001, Figure S1D). Using a driver specific for the dorsal compartment of the eye (de-Gal4), we also noted an increase in the size of eye imaginal disc tissue lacking Tao-1 function. de>Tao-1 RNAi eyes had grossly overgrown dorsal compartments relative to both the ventral compartment and to wild-type controls (Figure 1D–F). The increased size in eye imaginal discs can be attributed to increased cell number, as demonstrated by the higher number of interommatidial cells in pupal eyes from GMR >Tao-1 RNAi flies (Figure 1G–I). These cells are normally eliminated through apoptosis during early pupal development (Wolff and Ready, 1991), but persist in animals mutant for components of the HSW pathway, suggesting that Tao-1 may function in the HSW pathway.
Figure 1. Loss of Tao-1 function causes overproliferation.
(A–C) ap>Tao-1 RNAi causes massive overgrowth of 3rd instar wing imaginal discs compared to wild-type discs. GFP marks the apterous domain, and Notch or E-Cadherin is in blue. Ventral is down. (D–F) Tao-1 knockdown causes overproliferation in the eye imaginal disc. Larval eye imaginal discs expressing Tao-1 RNAi under the control of the dorsal enhancer driver (de>Tao-1 RNAi) are overgrown dorsally compared to wild-type eye discs. Anterior is to the left and ventral down in all eye disc images. (G–I) E-cadherin staining of pupal eyes 43 hr APF showing increased numbers of interommatidial cells in GMR>Tao-1 RNAi eyes compared to GMR-Gal4/+ eyes. Yellow arrows point out regions where interommatidial cell layer is doubled. (J–L) Representative images of GMR-Gal4/+ and GMR>Tao-1 RNAi third instar eye discs labeled with BrdU to indicate mitotically active cells. Note increased incorporation of BrdU posterior to the second mitotic wave (yellow arrow) in GMR>Tao-1 RNAi discs. The morphogenetic furrow is marked by the red arrowhead. Tao-1 RNAi transgenes on the second chromosome (images in B, E, H, K) and third chromosome (images in C, F, I, L) give similar phenotypes. Unless otherwise noted, all subsequent experiments using Tao-1 RNAi were conducted with the transgene on the third chromosome, which was used primarily for convenience in building strains.
HSW pathway mutants are characterized by inappropriate persistence of cell division in the developing third instar eye imaginal disc. In a wild-type eye disc, cells become quiescent after a final synchronous round of cell division (termed the second mitotic wave, SMW) following the passing of the morphogenetic furrow, which moves anteriorly across the eye disc to initiate differentiation and patterning. In animals lacking hpo or wts function, cells fail to exit the cell cycle and undergo division after the SMW, shown by BrdU incorporation during S phase (Harvey et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003). In GMR>Tao-1 RNAi eyes, but not in controls, BrdU incorporation occurs posterior of the SMW (Figure 1J–L). This suggests that the defect in animals lacking Tao-1 function may be related to the defect observed in HSW signaling mutants.
Two lines of evidence verify the identity and specificity of the RNAi transgenes used in these experiments. First, two non-overlapping Tao-1 RNAi transgenes (Dietzl et al., 2007) were tested and gave similar phenotypes (Figure 1). Second, these transgenes reduced antibody staining of endogenous Tao-1 (Figure S1E–E’). Taken together, these results indicate that these transgenes specifically knockdown Tao-1 expression.
Previous studies have identified three alleles of Tao-1, which is located very proximally (18D) on the X chromosome, but it has not been possible to test their effects on growth. Sato et al. (2007) described a null allele that was lethal and sterile in germline clones, but this allele has been lost (Kobayashi, pers. comm.). King et al. (2011) recently described two alleles. One is associated with a 2 kb deletion at the 5’ end that appears to affect an adjacent gene (Tao-150), and the other is viable and only slightly affects Tao-1 expression (Tao-1EP1455). Unfortunately, Tao-150 mutants die at the second larval instar, precluding any analysis of overgrowth in imaginal discs (data not shown).
Loss of Tao-1 Causes Upregulation of HSW Pathway Targets
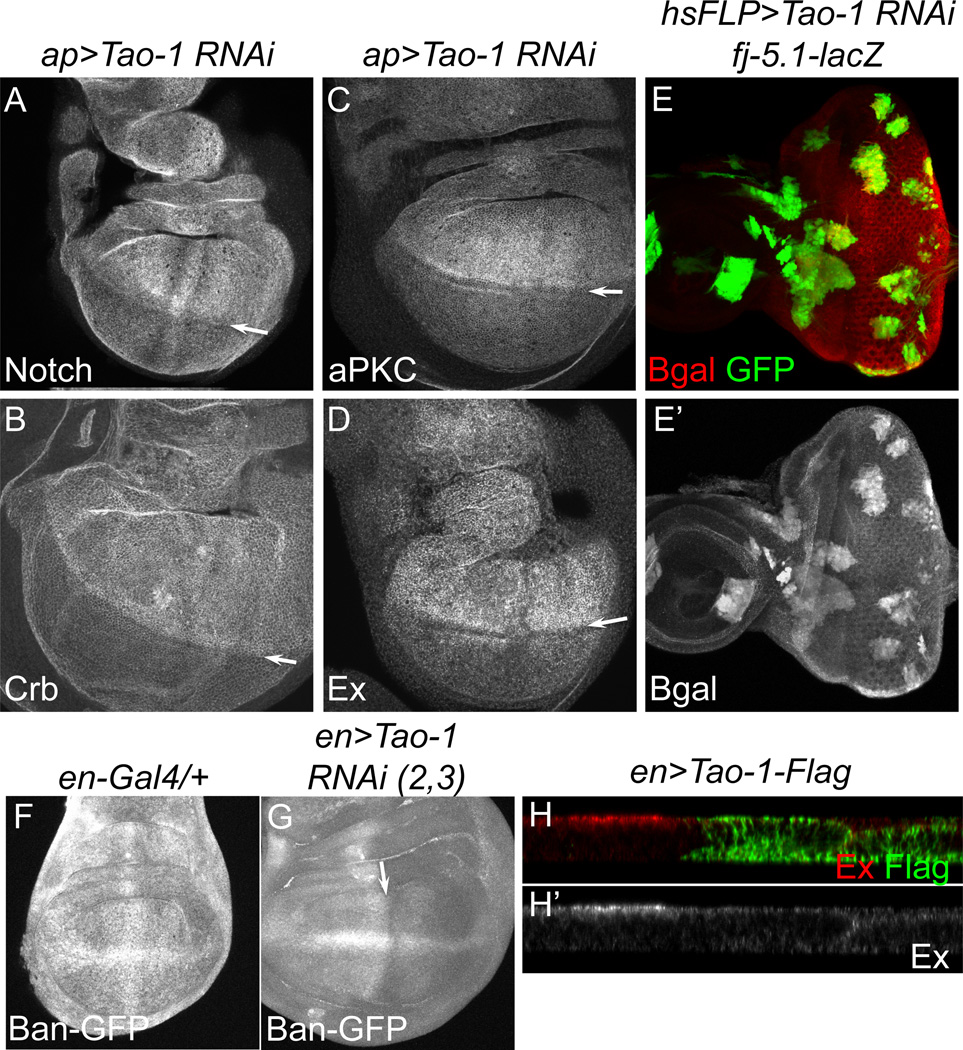
Another hallmark of defects in HSW signaling is an increase in levels of cell surface proteins including receptors and adhesion proteins, as well as juxtamembrane proteins involved in cell polarization and intracellular signaling. We found elevated levels of Notch, Crumbs, and aPKC (Figure 2A–C), as well as phospho-Moesin, E-Cadherin, and PatJ (Figure S2D–F) in ap>Tao-1 RNAi wing discs. Proteins that act as upstream activators of the HSW pathway are also transcriptional targets of the pathway and are upregulated apically in hpo or wts mutant cells (Hamaratoglu et al., 2006; Genevet et al., 2010). Consistent with this, we found increased apical levels of Ex, Mer, Fat, and Kibra (Figure 2D and Figure S2A–C) in ap>Tao-1 RNAi wing discs.
Figure 2. Tao-1 depletion causes upregulation of many known targets of the HSW pathway.
(A–D) ap>Tao-1 RNAi results in upregulation of known HSW targets within the dorsal compartment of the wing. (A) Notch, (B) Crumbs, (C) aPKC, and (D) Expanded are apically enriched in Tao-1 RNAi wing discs. Arrows mark the D–V boundary. (E–E’) In the eye imaginal disc, four-jointed-LacZ, a transcription reporter for HSW signaling, is upregulated in heat-shock flip-out cell clones expressing Tao-1 RNAi (marked by GFP). (F–G) In the wing, Tao-1 knockdown under the en-Gal4 driver results in increased activity of the bantam miRNA (as assayed by the bantam-GFP sensor, Brennecke et al., 2003), a yki target gene, suggesting increased bantam transcription. Arrow indicates A–P boundary. (H–H’) Expression of Flag-tagged Tao-1 under the engrailed promoter (en>Tao-1-Flag) results in lower Ex levels in the posterior compartment of wing discs. In all images ventral is down and posterior to the right.
To confirm that loss of Tao-1 results in transcriptional upregulation of known HSW target genes, we examined the expression of lacZ transcriptional reporters for four-jointed and ex (Cho et al., 2006; Hamaratoglu et al., 2006). In the absence of Tao-1 function, transcription of four-jointed and ex was increased in eye and wing imaginal discs, respectively (Figure 2E-E’ and Figure S2G–G’). Activity of another HSW target gene, the miRNA bantam, was also higher (as assayed by a GFP bantam reporter; Brennecke et al., 2003) in tissues lacking Tao-1, suggesting elevated levels of bantam (Figure 2F–G). Overexpression of Tao-1 lowered levels of Ex at the apical surface, consistent with overexpression of other HSW pathway genes (Figure 2H–H’). These results are similar to those of known HSW pathway genes, suggesting that Tao-1 is also a member of this signaling pathway.
Tao-1 Functionally Interacts with Members of the HSW Pathway and Promotes Wts Phosphorylation
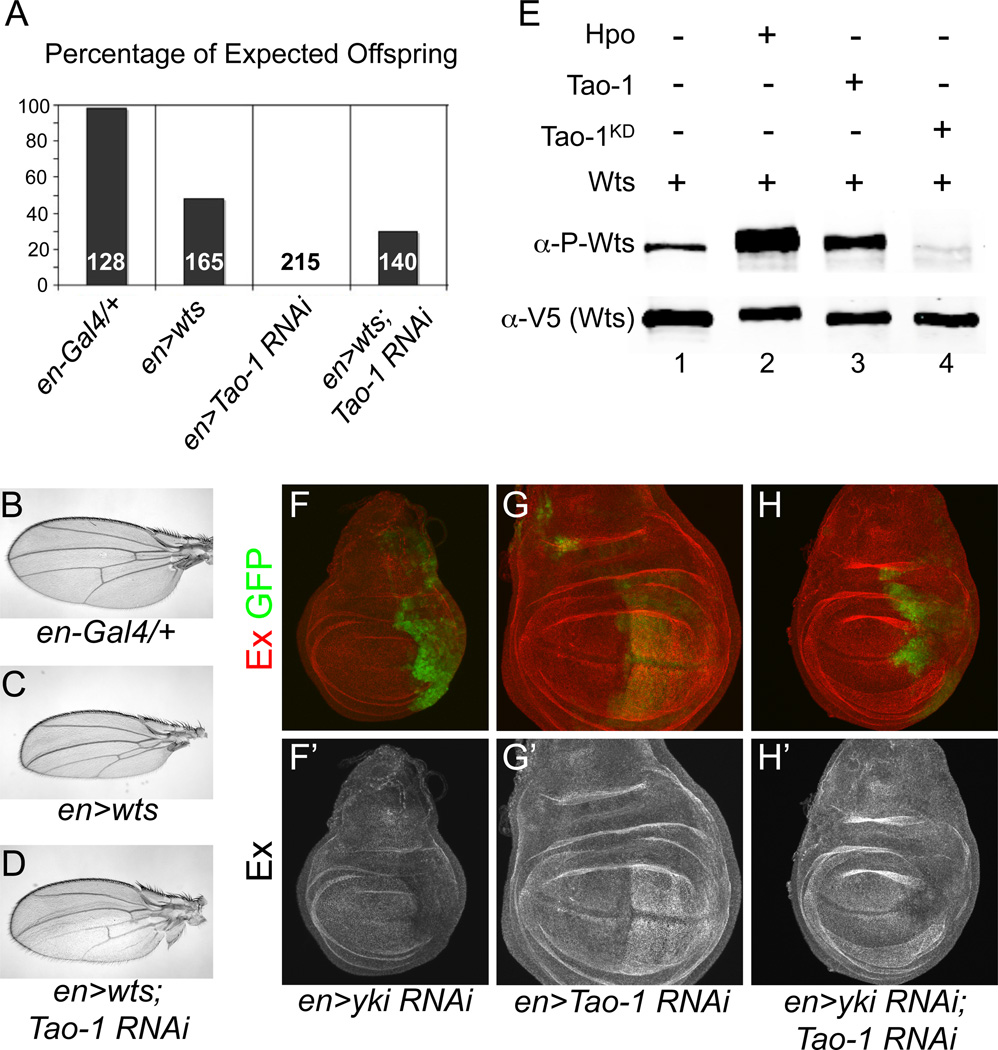
Given that Tao-1 loss-of-function phenotypes are similar to HSW pathway mutants, we asked whether Tao-1 genetically interacts with any of the previously identified HSW pathway genes. Previous studies in Drosophila have shown that wts overexpression can rescue the lethality of fat and ex null mutations (Feng and Irvine, 2007). Similarly, we found that co-expression of wts in the wing strongly suppressed the lethality of Tao-1 RNAi driven by engrailed-Gal4 (en>Tao-1 RNAi), suggesting that Tao-1 functions through HSW signaling (Figure 3A). Additionally, while en>wts wings appeared under proliferated, having smaller posterior compartments than en-Gal4 controls, this under proliferation was partially suppressed in en>wts; Tao-1 RNAi wings (Figure 3B–D). This result suggests that complete activation of Wts in the wing requires Tao-1 activity. Taken together, these results suggest that Tao-1 is required for complete HSW pathway activation in developing epithelial tissues.
Figure 3. Tao-1 functionally interacts with HSW pathway components.
(A) Lethality caused by loss of Tao-1 is suppressed by expressing a wts transgene. Surviving adults were scored 1–2 days after eclosion and the percentage of expected offspring was plotted for each genotype. The number within each bar represents the expected number of adult progeny for each genotype. (B–D) Tao-1 RNAi suppresses the effect of Wts overexpression in the wing. (B–C) Male en-Gal4/+ control wings appear normal, whereas en>wts wings have a smaller posterior compartment. (D) en>wts; Tao-1 RNAi animals survive to adults and have an intermediate wing phenotype. (E) Tao-1 promotes Wts phosphorylation in Drosophila S2 cells. Tao-1 and Wts were transiently expressed in S2 cells and Wts phosphorylation was measured using a phospho-specific antibody (Yu et al., 2010). Levels of Wts phosphorylation (lane 1) were increased in the presence of Hpo and Tao-1 (lanes 2 and 3). In contrast, Wts phosphorylation was severely diminished in the presence of a kinase-dead form of Tao-1 (lane 4). (F–H’) HSW pathway target gene upregulation in Tao-1 RNAi knockdown requires yki. (F–G’) Reduction of yki using the en-Gal4 driver leads to a reduction in Ex staining, while Tao-1 RNAi results in increased Ex expression. (H–H’) Double knockdown of Tao-1 and yki causes decreased levels of Ex, similar to yki RNAi alone.
Wts activity is regulated by the upstream kinase Hpo, and Wts phosphorylation is a commonly used readout of HSW pathway activation (Huang et al., 2005; Dong et al., 2007). We next asked whether Tao-1 promotes Wts phosphorylation in Drosophila S2 cells using a phospho-specific Wts antibody that recognizes a critical regulatory residue (T1077; Yu et al., 2010). We found that Wts phosphorylation was substantially increased in the presence of Tao-1 and that expression of a kinase-dead form of Tao-1 strongly reduced Wts phosphorylation (Figure 3E). Thus, like several other upstream activators of HSW signaling Tao-1 regulates pathway activation by modulating Wts phosphorylation.
Most, if not all, HSW pathway function is dependent on the transcriptional co-activator Yki, which is thought to promote growth through target genes such as Cyclin E, DIAP1 and the miRNA bantam (Huang et al., 2005; Nolo et al., 2006; Thompson and Cohen, 2006). In addition to these growth-promoting genes, Yki also regulates expression of the tumor suppressor genes ex, Mer, and kibra as part of a negative feedback loop (Hamaratoglu et al., 2006; Genevet et al., 2010). To ask if overgrowth and Ex upregulation in cells lacking Tao-1 function is Yki-dependent, we generated flies expressing both yki and Tao-1 RNAi and found that both overproliferation and Ex staining were suppressed (Figure 3F–H’). In fact, en>yki RNAi and en>yki RNAi; Tao-1 RNAi wing discs were nearly indistinguishable from one other, indicating that Tao-1 functions through Yki to regulate organ size.
Tao-1 Phosphorylates Hpo to Activate the HSW Pathway
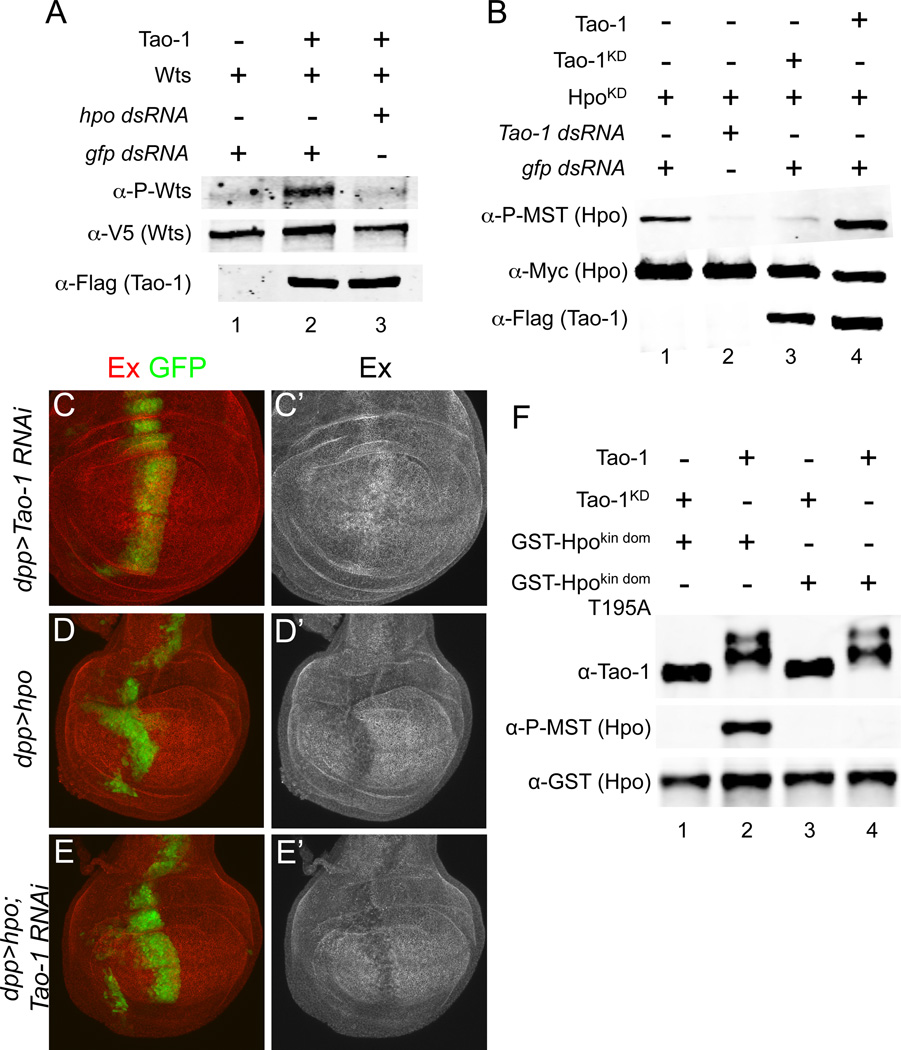
We next sought to determine where Tao-1 functions within the HSW pathway using a combination of biochemical assays in S2 cells and genetic epistasis experiments in vivo. Because the Sterile 20 kinase Hpo is known to phosphorylate Wts (Wu et al., 2003), we looked at whether Hpo is required for Tao-1 to promote Wts phosphorylation. We used double stranded RNA (dsRNA) to deplete S2 cells of endogenous Hpo and found that Tao-1 required Hpo to promote Wts phosphorylation (Figure 4A). This strongly suggested that Tao-1 functions upstream of Hpo. To test this hypothesis, we next asked whether Tao-1 promotes Hpo phosphorylation in S2 cells. For these experiments we used a kinase-dead form of Hpo (HpoKD) to prevent auto-phosphorylation, which could obscure Tao-1 effects on Hpo phosphorylation. We found that levels of HpoKD phosphorylation were increased in the presence of wild-type, but not kinase-dead Tao-1. Furthermore, knockdown of Tao-1 using dsRNA decreased levels of HpoKD phosphorylation (Figure 4B). These results suggest that Tao-1 functions through Hpo to modulate pathway activity and that endogenous Tao-1 is required for normal levels of Hpo phosphorylation found in unstimulated S2 cells.
Figure 4. Tao-1 phosphorylates Hpo to activate the HSW pathway.
(A) Hpo is required for Tao-1 to promote Wts phosphorylation in S2 cells. Normally, Wts is phopshorylated in the presence of Tao-1 (lane 2); however, transfecting hpo dsRNA prevents Wts phosphorylation (lane 3). (B) Tao-1 promotes Hpo phosphorylation at a critical regulatory residue (T195). Cells were transfected with a kinase-dead form of Hpo (HpoKD) to prevent auto-phosphorylation. A low level of Hpo phosphorylation was detected in these cells using a commercially available phospho-MST1/2 antibody (lane 1), which specifically recognizes phosphorylation at T195. This level of phosphorylation can be diminished by depleting endogenous Tao-1 (lane 2), or by expressing a kinase-dead form of Tao-1 (Tao-1KD; lane 3). Hpo phosphorylation is greatly increased in the presence of wild-type Tao-1 (lane 4). (C–E’) Tao-1 functions through hpo to regulate HSW pathway target genes. (C–C’) As shown previously, loss of Tao-1 results in increased Ex levels. (D–D’) UAS-hpo expressed using a dpp-Gal4 driver (dpp>hpo) results in decreased Ex expression. (E–E’) dpp>hpo; Tao-1 RNAi also results in decreased Ex staining, suggesting that hpo functions downstream of Tao-1. All flies raised at 18°C. (F) Tao-1 directly phosphorylates T195 within the activation loop of Hpo. Flag-tagged Tao-1 was immunoprecipitated from S2 cells and incubated with recombinant Hpo kinase domain (aa 1–367; GST-Hpokin dom). Immunoprecipitated Tao-1 displayed a mobility shift (lanes 2 and 4) relative to Tao-1KD (lanes 1 and 3), indicating that Tao-1 was catalytically active. Staining with phospho-MST1/2 antibody revealed that Tao-1 phosphorylates the kinase dead mutant of Hpo at T195 (lane 2). The T195A mutation confirmed that the phospho-MST1/2 antibody specifically recognizes phosphorylation at T195 (lane 4).
Previous studies have shown that overexpression of Hpo results in decreased levels of Ex in the imaginal epithelia (Hamaratoglu et al., 2006). If Tao-1 functions upstream of Hpo, then overexpression of wild-type Hpo should suppress the upregulation of Ex in Tao-1 RNAi wing discs. To test this hypothesis, we simultaneously expressed UAS-hpo and Tao-1 RNAi in the wing using the dpp>Gal4 driver. These crosses were done at 18ºC to prevent excessive tissue loss resulting from Hpo induced apoptosis. We found that dpp>hpo wing discs were indistinguishable from dpp>hpo; Tao-1 RNAi discs and that both had decreased levels of Ex staining, whereas dpp>Tao- 1 RNAi discs had elevated levels of Ex (Figure 4C–E). Additionally, in the reciprocal experiment we found that overexpression of Tao-1 could not suppress the upregulation of Ex resulting from reduction in hpo function (data not shown). We conclude from these results that Tao-1 functions upstream of Hpo to regulate cell proliferation and target gene expression.
Our observation that Hpo phosphorylation in S2 cells is Tao-1 dependent suggests that Hpo could be a direct substrate for Tao-1 kinase activity. To test this hypothesis we examined the ability of purified Tao-1 to phosphorylate recombinant Hpo protein in vitro. In control experiments using P32 and autoradiography, we found that full-length Tao-1 immunoprecipitated from S2 cells was able to autophosphorylate and therefore was catalytically active. In this experiment we failed to detect any other co-purifying autocatalytic kinases, suggesting that Tao-1 does not co-purify with an intermediary kinase (Supplemental Figure 3A). We then tested the ability of purified Tao-1 to phosphorylate the isolated kinase domain of Hpo (GST-Hpokin dom), which contains the activation loop with a critical regulatory residue (T195) that controls Hpo activity (Colombani et al., 2006). Using an antibody specific for phosphorylation at T195, we found that purified Tao-1 clearly phosphorylated Hpo at this site (Figure 4F, lane 2). This phosphorylation was specific because it was not observed with a kinase dead form of Tao-1 (Figure 4, lane 1), and a mutation in the activation loop that blocks Hpo activation, T195A, completely blocked this signal (Figure 4F, lanes 3–4). Additional in vitro kinase experiments using P32 confirmed that Tao-1 directly phosphorylates Hpo (Supplemental Figure 3A). These results indicate that Tao-1 directly activates Hpo by phosphorylating at a critical regulatory site.
To further confirm the specificity of Hpo as a substrate for Tao-1 we next asked whether a mammalian ortholog of Tao-1, TAOK3, was able to directly phosphorylate MST1 and 2, the mammalian orthologs of Hpo. Interestingly we found that human TAOK3, which shares 70% identity with the kinase domain of Tao-1, was able to directly phosphorylate the activation of loop of human MST1 and 2 in vitro (Supplemental Figure 3B). This result strongly suggests that Tao-1’s role in HSW signaling is functionally conserved in mammals.
Merlin and Expanded function with Tao-1 to Activate HSW Signaling
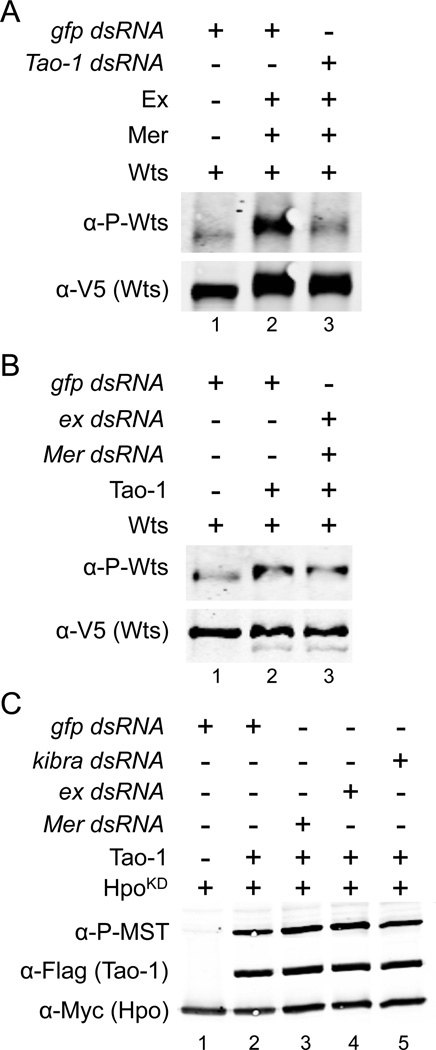
The cytoplasmic proteins Mer and Ex have been reported to promote HSW pathway activation (Hamaratoglu et al., 2006), so we next asked if Tao-1 mediates their effect on HSW activation in cultured cells. As a readout of HSW activation, we examined phosphorylation of Wts, which is increased by expression of Mer and Ex (Yu et al., 2010; Figure 5A, lane 2). This effect is substantially blocked by co-transfection with dsRNA against Tao-1 (Figure 5A, lane 3; controls for Tao-1 dsRNA knockdown efficiency are shown in Figure S4B), suggesting that Tao-1 mediates the effects of Mer and Ex on HSW signaling. Conversely, we found that activation of Wts by Tao-1 was not affected by depletion of Mer and Ex (Figure 5B), suggesting that Tao-1 functions downstream of these proteins. We also found that depletion of Mer or Ex had no effect on Tao-1’s ability to promote Hpo phosphorylation (Figure 5C), further arguing that Tao-1 functions independently of Mer and Ex (the converse experiment was not possible because we were unable to detect an effect of Mer and Ex expression on Hpo phosphorylation state, data not shown). A similar absence of effect was seen with depletion of Kibra (Figure 5C), but we were unable to unambiguously test whether Wts phosphorylation induced by Kibra is Tao-1 dependent because in our experiments expression of Kibra resulted in greatly decreased Wts expression. Attempts to co-immunoprecipitate Flag-tagged Tao-1 from S2 cells with Mer, Ex or Kibra proved unsuccessful (data not shown), suggesting that these proteins do not form a complex.
Figure 5. Mer and Ex require Tao-1 to activate the HSW pathway.
(A) Mer and Ex require Tao-1 to induce increased Wts phosphorylation. Normally Wts phosphorylation increases in the presence of Mer and Ex (lane 2). However, depleting Tao-1 with dsRNA eliminates this effect (lane 3). (B) Tao-1 induced Wts phosphorylation is not dependent on Mer and Ex. Depletion of Mer and Ex by dsRNA (lane 3) does not affect Tao-1 induced p-Wts staining (lane 2). (C) Tao-1 promotes Hpo phosphorylation independently of Mer, Ex and Kibra. S2 cells were transfected with dsRNA targeting Mer (lane 3), ex (lane 4) or kibra (lane 5). Knockdown of these genes had no effect on Tao-1’s ability to promote Hpo phosphorylation (compare lane 2 to lanes 3–5).
DISCUSSION
In an effort to identify additional regulators of HSW signaling we have examined the role of Tao-1 in growth control during development. Tao-1 depletion in either the eye or wing epithelium results in overgrowth phenotypes as well as transcriptional upregulation of HSW targets. Using a combination of genetic epistasis, experiments in cultured S2 cells, and in vitro biochemistry, we have demonstrated that Tao-1 directly phosphorylates the critical T195 regulatory residue in the activation loop of Hpo to promote HSW pathway activation. The observation that a mammalian orthologue of Tao-1, TAOK3, can phosphorylate MST kinases at the same residue further suggests that this regulatory function is conserved in mammals. Taken together, these results implicate Tao-1 as a component of HSW signaling and reveal a mechanism for regulation of Hpo activity (Figure 6).
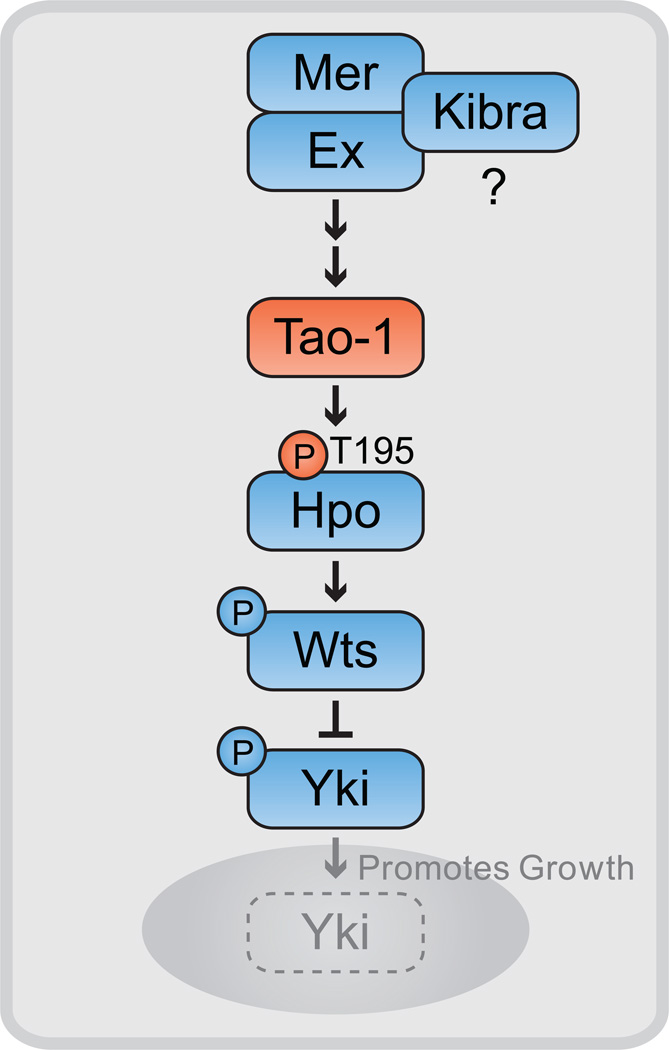
Figure 6. A model for Tao-1’s function in the HSW pathway.
Tao-1 directly phosphorylates Hpo at T195 in the kinase activation loop, leading to the activation of Wts and inhibition of Yki, which remains cytoplasmic when phosphorylated. In the absence of HSW pathway activity, Yki translocates to the nucleus to promote transcription of target genes that promote growth and inhibit cell death. Genetic and biochemical experiments position Tao-1 upstream of Hpo and suggest that Mer and Ex function through Tao-1 to activate HSW signaling. Kibra, Mer and Ex can form a protein complex and promote HSW activation, but it remains unclear if Kibra functions upstream or in parallel to Tao-1.
While Tao-1 depletion results in overgrowth phenotypes that are similar to mutations in other HSW pathway genes, these phenotypes are less severe than those of core components such as hpo and wts. One likely explanation for this is that the RNAi transgenes we have used in these studies do not completely remove Tao-1 function.. It is also possible that there are multiple mechanisms for activating HSW signaling, including, but not limited to, Tao-1 phosphorylation of Hpo. Indeed, previous studies have demonstrated that the upstream components Mer, Ex, and Kibra act, at least in part, in parallel to activate Hpo (Baumgartner et al.; Genevet et al., 2010; Yu et al., 2010). Our biochemical evidence indicates that two of these proteins, Mer and Ex, function with Tao-1 to activate HSW signaling. While it is probable that Kibra functions upstream of Tao-1, we cannot rule out the possibility that Kibra functions independently of Mer and Ex to activate HSW signaling in a Tao-1-independent manner (Figure 6). Further genetic analysis using a Tao-1 null allele would be helpful in defining Tao-1’s role relative to other HSW components, but unfortunately the deletions associated with the sole existing Tao-1 null allele, Tao-150, also appear to affect an adjacent gene (CG14218; King et al., 2011). In addition, Tao-1 maps very close to the most proximal FRT element on the X chromosome, making it difficult to generate recombinant chromosomes for somatic mosaic analysis.
How do Mer, Ex and Tao-1 cooperate to regulate Hpo phosphorylation? Given that Ex has been shown to interact with Hpo (Yu et al., 2010), one possibility is that Mer and Ex function to scaffold Tao-1 together with Hpo, thereby promoting the ability of Tao-1 to phosphorylate and activate Hpo. However, despite repeated attempts we have been unable to detect Tao-1 in a complex with either Mer or Ex, and knockdown of Mer, ex or kibra does not diminish the ability of Tao-1 to promote Hpo phosphorylation in S2 cells. For these reasons, we favor the possibility that Mer and Ex indirectly affect Tao-1 function, perhaps by interacting with other proteins that in turn directly regulate Tao-1. For example, Tao-1 activity could be directly regulated by an unknown receptor at the cell surface whose localization or activity is controlled by interaction with Mer and Ex. This notion is consistent with the fact that both Mer and Ex have FERM domains, which are known to interact with the cytoplasmic tails of transmembrane proteins (Bretscher et al., 2002). Previous studies have suggested that Ex interacts with the transmembrane protein Crumbs (Ling et al., 2010), though the mechanistic significance of this interaction is unclear. It is not currently known whether Drosophila Merlin has transmembrane binding partners.
Two additional ideas related to Tao-1 function are suggested by our data. In S2 cells, Tao-1 kinase activity is required for normal levels of Hpo phosphorylation at T195 in the kinase activation loop, suggesting that Tao-1 could function to maintain constant, low levels of pathway activation. In turn, this low level of Hpo activation might be necessary so that other, regulated inputs into HSW activity can quickly transition cells away from actively dividing and into a differentiated state following periods of growth. Alternatively, it is possible that Tao-1 activity itself is dynamically regulated during development, allowing it to rapidly alter levels of HSW pathway activity via its effect on Hpo phosphorylation. In either case, phosphorylation by Tao-1 at T195 is likely to promote Hpo’s known ability to undergo autophosphorylation (Colombani et al., 2006), thus amplifying the effect of even a small change in Tao-1 activity. Further studies will be required to answer these questions and to determine if, and how, Tao-1 activity is regulated.
An interesting aspect of our discovery that Tao-1 regulates HSW signaling is that Tao-1, and its mammalian orthologues TAOK1–3, previously have been shown to regulate MT stability (Mitsopoulos et al., 2003; Timm et al., 2003; Liu et al., 2010). Our results indicate that this effect on MT stability is not mediated through HSW signaling, since mutations in other HSW pathway components do not display similar MT phenotypes (data not shown). However, it is interesting to speculate that Tao-1’s association with MTs might affect its ability to regulate HSW pathway activation. More work will be required to determine whether the function of Drosophila Tao-1 in HSW signaling is entirely independent of its role in microtubule dynamics, though a recent study in mammalian cultured cells found that microtubule disruption did not affect localization of Yap, a mammalian Yki orthologue (Dupont et al., 2011), suggesting that in mammalian cells these roles might be independent.
An additional possible mechanistic link between Tao-1 and HSW signaling is suggested by studies in flies and in mammalian cells indicating that Par-1, a polarity protein, is positively regulated by Tao-1 (Timm et al., 2003; King et al., 2011). Par-1 has been shown to promote basolateral polarity in the Drosophila follicular epithelium and to regulate the stability and organization of MTs in these cells (Doerflinger et al., 2003). Recent studies have implicated components of both apical and basolateral polarity in the regulation of HSW signaling (reviewed in Halder and Johnson, 2011). Conversely, HSW signaling also seems to feed back onto Crumbs, an apical determinant, and perhaps other components to regulate apical-basal polarity (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010). Whether Tao-1 plays a role in the linkage between cell polarity and growth control remains to be established, but the ability to both directly activate Hpo function through phosphorylation and control cytoskeletal organization and cell polarity through microtubule organization potentially places Tao-1 in a unique position to coordinate these important cellular processes.
EXPERIMENTAL PROCEDURES
Drosophila Genetics
All crosses were done at 25°C unless otherwise noted. We used the GAL4/UAS system throughout these experiments (Brand and Perrimon, 1993). Transgenic RNAi stocks were provided by the Vienna collection and include Tao-1 RNAi (3) (ID 17432), Tao-1 RNAi (2) (ID 107645), hpo RNAi (ID 104169), and yki RNAi (ID 104523) (Dietzl et al., 2007). dpp>Tao-1 RNAi wing area was measured using the AxioVision software (Zeiss) and compared to the average wing area of wild-type flies.
Immunofluorescence
Dissections and antibody staining were carried out as previously described (Maitra et al., 2006) with the following exceptions: For anti-Crb staining, discs were dissected in PBS and treated with methanol for 10 minutes following fixation. For BrdU incorporation, eye discs were dissected in 50 ug/mL BrdU in Ephrussi and Beadle’s Ringers (4.7mM KCl, 128mM NaCl, 1.5 mM CaCl2) within 10 minutes and incubated for a total of 30 minutes. Following one quick rinse in PBS with 0.1% Triton X-100 (PBST), discs were fixed in 3:1 EtOH:Glacial Acetic Acid for 15 min, then rehydrated through a 75%, 50%, 25%, and 10% EtOH (in H2O) series. Three rinses in PBST were followed by a 20 min treatment in 2N HCl and three more rinses in PBST. Finally, discs were incubated in PBST plus 0.1% NGS prior to antibody staining.
Fluorescent secondary antibodies were diluted to 1:1,000 (Jackson Immunoresearch Laboratories). Tissue samples were mounted in ProLong Antifade (Invitrogen). Confocal images were acquired on a laser-scanning confocal microscope (LSM510, Zeiss) using LSM software. The following objectives were used: 20X NA 0.8 Plan-Apochromat, 40X NA 1.3 EC Plan-NeoFluar and 63X NA 1.4 Plan-Apochromat. Images were compiled using Photoshop CS3 (Adobe, Inc.).
Cell Culture and In Vitro Kinase Assays
Schneider-2 (S2) cell maintenance and transfection were performed as previously described (Neisch et al., 2010). Cells were lysed in lysis buffer containing 150 mM NaCl, 0.5% NP-40, 50 mM NaF, 100 mM Tris pH 8.0 and protease inhibitors (Roche). Immunoblots were imaged using an Odyssey scanner with version 2.1 software (LI-COR Biosciences).
For RNAi knockdown experiments, T7-primers corresponding to non-conserved coding regions were used to generate 450–700 bp PCR products and dsRNA was transcribed from these templates using the MEGAscript High Yield Transcription Kit (Applied Biosystems).
For in vitro kinase assays, S2 cells expressing Flag-tagged Tao-1, Tao-1KD or TAOK3 were lysed in lysis buffer (see above) and immunoprecipitated with M2 anti-Flag agarose beads (Sigma-Aldrich), washed in PBS and incubated with recombinant GST protein in kinase buffer containing 25 mM HEPES pH 7.2, 25 mM MgCl2, 50 mM β-glycerol phosphate, 2 mM dithiothreitol, 0.5 mM sodium vanadate, 10 µM ATP and 10 µCi/ml gamma-32P-ATP at 30°C for 40 min.
Expression Constructs
The kinase domains of Hpo (kinase dead), MST1 and MST2 consisting of amino acids 1–367, 1–320 and 1–316, respectively were cloned into the pGEX expression vector and transformed into E. coli (BL21) electro-competent cells. Protein expression was induced with 0.8 mM IPTG at 25°C for 3hr and GST-tagged protein was purified using glutathione Sepharose beads (Sigma-Aldrich). Flag-tagged Tao-1 constructs for expression in S2 cells were gifts from S. Kobayashi’s lab in Japan (Sato et al., 2007). Myc-tagged Hpo, V5/His-tagged Wts and Myc-tagged Kibra were gifts from D.J. Pan’s lab (Wu et al., 2003; Yu et al., 2010). Ex-HA and Mer- HA were gifts from G. Halder’s lab (Hamaratoglu et al., 2006). Human TAOK3 cDNA was obtained from Thermo Scientific Inc. and subcloned into the pTFW destination vector of the Gateway System (Invitrogen) for expression in Drosophila S2 cells.
HIGHLIGHTS.
-
⊙
Tao-1 restricts cell proliferation and regulates HSW target gene expression
-
⊙
Tao-1 genetically interacts with HSW genes and promotes pathway activation
-
⊙
Tao-1 directly phosphorylates Hippo at T195 in the kinase activation loop
-
⊙
Effects of Tao-1 on the HSW pathway are strictly Hippo dependent
Supplementary Material
ACKNOWLEDGMENTS
We thank D.J. Pan, K. Irvine, G. Halder, N. Tapon, H. McNeill, U. Heberlein, B. Baum, S. Kobayashi, the Bloomington and Vienna stock centers and the Developmental Studies Hybridoma Bank for fly stocks, antibodies and bacterial plasmids. We are grateful to Y. Wang for help with the RNAi screen, and S. Morillo for assistance with the in vitro kinase assay. We also thank I. Rebay and members of the Fehon and Rebay laboratories for critical comments throughout the course of this work, and S. Horne-Badovinac and M. Glotzer for comments on the manuscript. PJV was supported by postdoctoral fellowships from the National Institutes of Health (T32 CA009594) and the CDMRP (W81XWH-10-1-0610) and JCB was supported by National Institutes of Health training grants (T32 HD055164 and T32 GMO7197). This research was funded by a National Institutes of Health grant (NS034783) to RGF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Colombani J, Polesello C, Josue F, Tapon N. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Curr Biol. 2006;16:1453–1458. doi: 10.1016/j.cub.2006.05.059. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Doerflinger H, Benton R, Shulman JM, St Johnston D. The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development. 2003;130:3965–3975. doi: 10.1242/dev.00616. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci USA. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hughes SC, Fehon RG. Phosphorylation and activity of the tumor suppressor Merlin and the ERM protein Moesin are coordinately regulated by the Slik kinase. J Cell Biol. 2006;175:305–313. doi: 10.1083/jcb.200608009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I, Tsai LT, Pflanz R, Voigt A, Lee S, Jackle H, Lu B, Heberlein U. Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J Neurosci. 2011;31:1139–1148. doi: 10.1523/JNEUROSCI.4416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Rohn JL, Picone R, Kunda P, Baum B. Tao-1 is a negative regulator of microtubule plus-end growth. J Cell Sci. 2010;123:2708–2716. doi: 10.1242/jcs.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The Neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- Mitsopoulos C, Zihni C, Garg R, Ridley AJ, Morris JD. The prostate-derived sterile 20-like kinase (PSK) regulates microtubule organization and stability. J Biol Chem. 2003;278:18085–18091. doi: 10.1074/jbc.M213064200. [DOI] [PubMed] [Google Scholar]
- Neisch AL, Speck O, Stronach B, Fehon RG. Rho1 regulates apoptosis via activation of the JNK signaling pathway at the plasma membrane. J Cell Biol. 2010;189:311–323. doi: 10.1083/jcb.200912010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Hayashi Y, Ninomiya Y, Shigenobu S, Arita K, Mukai M, Kobayashi S. Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc Natl Acad Sci U S A. 2007;104:7455–7460. doi: 10.1073/pnas.0610052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Timm T, Li XY, Biernat J, Jiao J, Mandelkow E, Vandekerckhove J, Mandelkow EM. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 2003;22:5090–5101. doi: 10.1093/emboj/cdg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.