Abstract
Paget’s disease of bone (PDB) is a common disorder with a strong genetic component characterised by focal increases in bone turnover which in some cases is caused by SQSTM1 mutations. To identify additional susceptibility genes we performed a genome wide association study in 750 PDB cases without SQSTM1 mutations and 1002 controls and identified three candidate loci for the disease which were replicated in an independent set of 500 cases and 535 controls. The strongest signal was with rs484959 on 1p13 close to the CSF1 gene (P = 5.38 × 10−24) and significant associations were also observed with rs1561570 on 10p13 within the OPTN gene (P = 6.09 × 10−13) and with rs3018362 on 18q21 close to the TNFRSF11A gene (P = 5.27 × 10−13). These studies provide new insights into the pathogenesis of PDB and identify OPTN, CSF1 and TNFRSF11A as novel candidate genes for disease susceptibility.
Paget’s disease of bone (PDB) is a common disorder of the skeleton that affects up to 2% of Caucasians aged 55 years and above1. The disease is characterised by focal areas of increased and disorganised bone remodelling which can cause bone pain, bone deformity, pathological fracture, deafness and secondary osteoarthritis2. Genetic factors are important in PDB and between 15%-40% of individuals have an affected first degree relative3. Mutations affecting the ubiquitin associated domain of the SQSTM1 gene have been identified in about 10% of patients with “sporadic” PDB and 40% of patients with a familial PDB4,5. Despite extensive research efforts, and the identification of several susceptibility loci by linkage analysis6-8, the remaining genes that predispose to PDB remain to be identified.
In this study we sought to identify novel genetic variants that predispose to PDB in patients without SQSTM1 mutations using a genome wide association approach. In the discovery sample (stage 1) we genotyped 750 PDB patients and 1002 controls9 using Illumina arrays. In the replication sample (stage 2), we genotyped the most significant SNPs identified from stage 1 in an independent set of 500 cases and 535 controls using the Sequenom MassARRAY iPLEX platform. Details of the subjects used in the discovery and replication stages of the study are provided in the online methods section.
We used multidimensional scaling analysis of identity-by-state (IBS) sharing matrix of all individuals combined with HapMap project samples to assess population ancestry (Supplementary Fig. 1). After application of quality control measures and exclusion of subjects with non-European ancestry, genotypic data in the discovery sample were available for 294,663 SNPs in 692 PDB cases and 1001 controls. Association testing of the discovery stage data was performed using a stratified Cochran-Mantel-Haenszel (CMH) test in which samples were stratified according to their genome-wide IBS sharing similarity. A quantile-quantile plot comparing the observed and expected distribution of −log10 P showed some evidence for inflation of the test statistics given the multiple origin of PDB cases (Genomic inflation factor10 λ = 1.096, Supplementary Fig. 2). To exclude the possibility of false positive association due to hidden population substructure, the observed test statistics were corrected using the genomic control method10. Six SNPs showed genome-wide significant associations with PDB after Bonferroni correction for multiple testing (Fig. 1). Three SNPs were located within a 14kb region of chromosome 1p13.3 (rs10494112, rs499345, and rs484959); one SNP was located on chromosome 10p13 (rs1561570); and two SNPs were located within a 22kb region of chromosome 18q21.33 (rs2957128 and rs3018362; Table 1, Supplementary Table 1 and Fig. 2). We then imputed SNPs located within 1.0Mb of the six SNPs reaching genome wide significance using genotype data from the HapMap project. Association testing of imputed SNPs revealed several variants with genome wide significance close to the six genotyped SNPs identified from stage 1 (Fig. 2). The imputed SNPs showed a slightly stronger association signal than the genotyped markers due to the conservative nature of the stratified CMH test used to analyse genotyped markers compared to the regression methods used to test imputed markers.
Figure 1. Loci for susceptibility to PDB detected by genome wide association study.
Manhattan plot of association test results from the discovery cohort showing chromosomal positions of the 294,633 SNPs passing quality control plotted against genomic-control adjusted −log10 P. Association with Paget’s disease was tested using stratified Cochran-Mantel-Haenszel test. The red horizontal line indicates the threshold for genome wide significance (P < 1.7 × 10−7).
Table 1.
SNPs showing genome-wide significant association with Paget’s disease of bone.
| Discovery |
Replication |
Combined |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Position | MA | MAF PDB | MAF Cont | P a | OR (95%CI) | MAF PDB | MAF Cont | P a | OR (95%CI) | P a | OR (95%CI) |
| 1 | rs10494112 | 110,154,000 | G | 0.312 | 0.200 | 9.14 × 10−12 | 1.80 (1.53 – 2.12) | 0.277 | 0.210 | 4.34 × 10−4 | 1.45 (1.18 – 1.77) | 1.67 × 10−14 | 1.65 (1.45 – 1.88) |
| 1 | rs499345 | 110,163,205 | A | 0.396 | 0.284 | 1.13 × 10−10 | 1.68 (1.44 – 1.95) | 0.400 | 0.293 | 5.15 × 10−7 | 1.61 (1.33 – 1.94) | 3.02 × 10−16 | 1.65 (1.47 – 1.85) |
| 1 | rs484959 | 110,167,606 | A | 0.355 | 0.489 | 1.15 × 10−13 | 0.56 (0.49 – 0.65) | 0.340 | 0.491 | 7.25 × 10−12 | 0.53 (0.45 – 0.64) | 5.38 × 10−24 | 0.55 (0.49 – 0.62) |
| 10 | rs1561570 | 13,195,732 | C | 0.346 | 0.451 | 1.11 × 10−8 | 0.64 (0.56 – 0.74) | 0.366 | 0.463 | 1.19 × 10−5 | 0.67 (0.56 – 0.80) | 6.09 × 10−13 | 0.65 (0.58 – 0.73) |
| 18 | rs2957128 | 58,211,715 | A | 0.499 | 0.384 | 4.21 × 10−10 | 1.61 (1.40 – 1.86) | 0.463 | 0.408 | 1.35 × 10−2 | 1.25 (1.05 – 1.49) | 1.86 × 10−11 | 1.46 (1.30 – 1.63) |
| 18 | rs3018362 | 58,233,073 | A | 0.458 | 0.341 | 1.35 × 10−10 | 1.64 (1.42 – 1.89) | 0.415 | 0.343 | 9.81 × 10−4 | 1.36 (1.13 – 1.62) | 5.27 × 10−13 | 1.52 (1.36 – 1.70) |
Genomic-control adjusted P values from association testing using stratified Cochran-Mantel-Haenszel test. MA, minor allele; MAF, minor allele frequency; PDB, Paget’s disease of bone subjects; Cont, Control subjects; OR, odds ratio for the minor allele and CI is confidence interval.
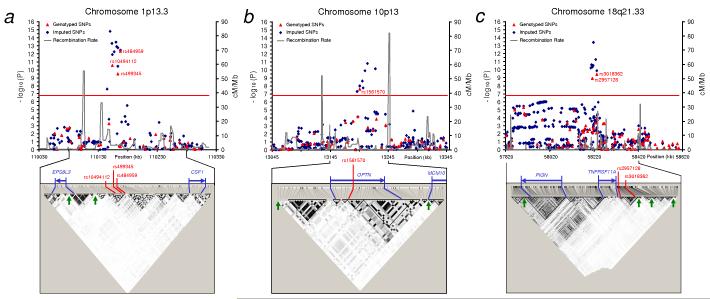
Figure 2. Details of loci associated with Paget’s disease.
Association and linkage disequilibrium (LD) plots of regions showing genome wide significant association with Paget’s disease located on (a) chr 1p13, (b) chr 10p13 and (c) 18q21. The chromosomal position (based on NCBI human genome Build 36) of SNPs is plotted against genomic-control adjusted −log10 P. Genotyped SNPs are shown as red triangles and imputed SNPs as blue diamonds. The estimated recombination rates (cM/Mb) from HapMap CEU release 22 are shown as grey lines and the red horizontal line indicates genome wide significance threshold (P < 1.7 × 10−7). Genotyped SNPs were tested using stratified Cochran-Mantel-Haenszel test and imputed SNPs were tested using regression analysis based on imputed allelic dosage and adjusting for population clusters. SNPs reaching genome wide significance are shown with red text. LD plots for the indicated regions are based on HapMap CEU release 22 showing LD blocks depicted for alleles with MAF > 0.05 using the r2 colouring scheme of Haploview37. The blue arrows indicate known genes in the region and possible recombination hotspots (> 20 cM/Mb) are shown as green arrows on the LD plots.
In total, 76 SNPs with P-values of 1 × 10−4 or less were identified in the discovery sample (Supplementary Table 2). From these SNPs we selected those with P < 1.0 × 10−6 and those with P < 1.0 × 10−5 in which another SNP within 50kb attained a P-value of 1.0 × 10−3 or less for further analysis in the replication sample. Following application of quality control measures on the replication dataset, genotype data were obtained in 481 cases and 520 controls for the 16 selected SNPs (Supplementary Table 3). Eight SNPs showed significant association with PDB in the replication stage after correction for multiple testing (P < 3 × 10−3), resulting in the identification of 8 SNPs where P-values attained genome wide significance in the combined dataset (Supplementary Table 3). The distribution of minor alleles and the direction of associations were similar in both the discovery and replication samples (Table 1 and Supplementary Table 3). Whilst all samples used in the replication stage were of European ancestry, confounding due to population substructure is possible, given the multiple nationalities of the replication cohorts. To address this issue we tested for association only in replication samples of British descent (256 PDB cases and 488 controls) using the CMH test. This yielded results that were qualitatively similar to those obtained from the whole replication cohort (Supplementary Table 4). Linkage disequilibrium (LD) patterns in the associated regions were also similar across the study samples and to those observed in HapMap CEU samples (Supplementary Fig. 3). Furthermore, the distribution of allele frequencies for SNPs showing genome wide significant association was broadly similar across individuals when grouped by origin (Supplementary Table 5) and the replicated hits were not located in genomic regions with known geographic variations in European population11,12 indicating that the association reported here is unlikely to be confounded by population substructure.
A significant association with PDB was observed on chromosome 1p13.3 for three SNPs (rs10494112, rs499345, and rs484595) but the strongest signal was with rs484959 (combined P = 5.38 × 10−24; Table 1 and Supplementary Table 1). These SNPs were poorly correlated (r2 < 0.36) with other genotyped SNPs and are located in a 14 kb LD block 87kb upstream of the CSF1 gene (Fig. 2). Another gene (EPS8L3) is located 47kb further upstream but is separated by two recombination hotspots (Fig. 2) making EPS8L3 a less likely candidate. Stepwise regression analysis adjusting for population clusters and accounting for the genotypic additive effect of the three SNPs showed strong evidence for independent association with rs484959 (P = 4.7 × 10−10) and rs10494112 (P = 9.28 × 10−3) compared to rs499345 (P = 0.80; Supplementary Table 6). This is not surprising since rs499345 is correlated with rs10494112 (r2 = 0.61) but rs484959 and rs10494112 are not (r2 = 0.21; Supplementary Fig. 3). Analysis of haplotypes formed by the three SNPs did not show a stronger association than analysis of single-SNP (Supplementary Table 7). The CSF1 gene encodes macrophage colony stimulating factor which is an extremely strong functional candidate for susceptibility to PDB since it plays a critical role in osteoclast formation and survival13,14. Furthermore, loss of function mutations in rat Csf1 causes osteopetrosis due to failure of osteoclast differentiation14,15, whereas clinical studies have shown that patients with PDB have increased serum levels of CSF-116,17.
A second locus showing significant association with PDB was situated on chromosome 10p13. Three SNPs (rs1561570, rs825411 and rs2095388) all located within a 30kb region were analysed in both stages of the study and the strongest signal was observed for rs1561570 (combined P = 6.09 × 10−13; Table 1 and Supplementary Table 3). These three SNPs are poorly correlated with other genotyped SNPs (r2 < 0.37). The rs825411 is not in LD with rs1561570 (r2 = 0.04, D′ = 0.21; Supplementary Fig. 3) but attained borderline genome wide significance (P = 7.82 × 10−8) and appeared to have an independent association with PDB as revealed by regression analysis accounting for the genotypic additive effect of rs1561570 (P = 2.23 × 10−9) and rs825411 (P = 5.15 × 10−6; Supplementary Table 6). Haplotype analysis showed a stronger association signal for alleles formed by rs1561570 and rs825411 combined, compared to single-SNP analysis with the risk haplotype “TC” showing the strongest association (P = 2.50 × 10−17; OR = 1.67; Supplementary Table 7). The third SNP (rs2095388) showed no significant association with PDB in the combined analysis (P = 3.08 × 10−6; Supplementary Table 3) and had no independent effect after accounting for rs1561570 and rs825411 (P = 0.14; Supplementary Table 6). The 10p13 locus is marked by two recombination hotspots and contains only one known gene (OPTN; Fig. 2). It is interesting to note that this region of chromosome 10p13 has been previously linked to familial PDB but the causal gene has not been identified6. It is therefore possible that the risk haplotype could be tagging rare allele(s) within OPTN that markedly increase susceptibility to PDB. In this regard there have been other reports where GWAS studies have identified common variants associated with diseases within regions previously mapped by linkage analysis. Examples include amyotrophic lateral sclerosis18 and Crohn’s disease19. Alternatively the risk haplotype could be tagging another common susceptibility variant within the gene but further studies will be required to investigate these possibilities. The OPTN gene, which encodes Optineurin is a novel candidate gene for PDB. Mutations in OPTN have been linked to glaucoma20, but until now, OPTN has not been implicated in the regulation of bone metabolism. Optineurin is a ubiquitously expressed cytoplasmic protein21 which contains a ubiquitin binding domain, similar to that present in NEMO. Optineurin negatively regulates TNF-α induced NFκB activation by interacting with ubiquitylated RIP22. Furthermore, a putative NFκB binding site has been identified in OPTN promoter23 and studies have shown that Optineurin interacts with myosin VI suggesting a role in vesicular trafficking between the Golgi apparatus and plasma membrane24. This is of interest since mutations affecting the VCP protein which is also involved in vesicular trafficking cause the syndrome of inclusion body myopathy with early-onset Paget’s disease and frontotemporal dementia (IBMPFD)25. Taken together these data indicate that optineurin may well play a hitherto unrecognised role in the regulation of bone metabolism through its effects on NFκB signaling and/or vesicular trafficking, but further studies will be required to investigate this issue.
The third region showing a significant association with PDB was located on chromosome 18q21.33 close to the TNFRSF11A gene which encodes receptor activator of NFκB (RANK). Four SNPs within a 300kb region reached genome wide significance in the combined analysis (rs663354, rs2980996, rs2957128, and rs3018362). Regression analysis accounting for the genotypic additive effect of the four SNPs showed that only rs2957128 (P = 0.047) and rs3018362 (P = 0.022) had independent effects (Supplementary Table 6). Analysis of haplotypes formed by alleles of rs2957128 and rs3018362 showed that a risk haplotype “AA” was consistently over-represented in PDB cases as compared with controls in the combined sample of cases and controls (P = 8.71 × 10−14; OR = 1.55; Supplementary Table 7). These two SNPs are moderately correlated (r2 = 0.55) and are located in adjacent LD blocks about 5kb downstream of TNFRSF11A (Fig.2c).
The TNFRSF11A gene product RANK plays a critical role in osteoclast differentiation and function. Mice with targeted disruption of TNFRSF11A exhibit severe osteopetrosis due to complete absence of osteoclasts26 and loss of function mutations in TNFRSF11A cause osteoclast-poor osteopetrosis in humans27. Mutations affecting the signal peptide region of RANK cause the PDB-like syndromes of Familial Expansile Osteolysis, early onset familial PDB and expansile skeletal hyperphosphatasia28-30. Mutations of TNFRSF11A have not so far been identified in patients with classical PDB28,31, although this region of chromosome 18q22 has been linked to PDB in some families32. It is also of interest to note that rs3018362 and rs884205 located downstream of TNFRSF11A have recently been associated with bone mineral density (BMD) and fracture33-35. The allele of rs3018362 that was associated with PBD was also associated with reduced BMD raising the possibility that this allele may be associated with increased bone turnover but further studies will be required to confirm this. The rs884205 was not directly genotyped in our study but it is moderately correlated with both rs3018362 (r2 = 0.53) and rs2957128 (r2 = 0.52). Imputation analysis showed evidence for association of rs884205 with PDB (Imputed P value = 5.93 × 10−11) confirming the importance of TNFRSF11A in the genetic regulation of bone metabolism.
The three loci on chromosomes 1p13, 10p13 and 18q21 identified in this study appear to have an independent role, since we found no evidence to suggest that the associated SNPs within these loci interacted to regulate susceptibility to PDB (P > 0.33 for all inter-locus pair wise interactions; Supplementary Table 8). These data are consistent with a multiplicative model for association with PDB. The cumulative population attributable risk for the SNPs showing independent association with PDB was 70% indicating substantial contribution of the identified loci to PDB development. Additionally the risk of PDB increased with increasing number of risk allele scores (ORper-riskallele = 1.34, 95% CI = 1.29 – 1.40, P = 5.81 × 10−45) with individuals carrying 10 or more risk alleles having six fold increase in PDB risk compared to those with median number of risk alleles (Table 2).
Table 2.
The cumulative contribution of identified variants to PDB risk.
| Risk Allele Scorea |
Allele score frequency (%) |
ORb (95% CI) | P | |
|---|---|---|---|---|
| Cases | Controls | |||
| 0,1 | 0.94 | 3.02 | 0.41 (0.21 - 0.82) | 9.00 ×10−3 |
| 2 | 1.71 | 6.57 | 0.34 (0.21 - 0.58) | 2.81 × 10−5 |
| 3 | 6.99 | 12.49 | 0.74 (0.54 - 1.02) | 0.07 |
| 4 | 11.59 | 18.67 | 0.83 (0.63 - 1.09) | 0.17 |
| 5 | 15.17 | 20.18 | 1.00 (0.77 - 1.30) | 1.00 |
| 6 | 18.33 | 16.90 | 1.44 (1.11 - 1.87) | 5.00 × 10−3 |
| 7 | 19.10 | 11.18 | 2.27 (1.73 - 2.98) | 2.25 × 10−9 |
| 8 | 13.73 | 6.51 | 2.81 (2.05 - 3.83) | 4.42 × 10−11 |
| 9 | 6.39 | 3.16 | 2.69 (1.79 - 4.05) | 1.07 × 10−6 |
| 10 | 4.43 | 0.99 | 5.98 (3.27 - 10.93) | 1.60 × 10−10 |
| 11,12 | 1.62 | 0.33 | 6.55 (2.40 - 17.85) | 3.06 × 10−5 |
Risk allele score of the 6 SNPs showing independent association with PDB in the combined dataset (chr1:rs10494112, rs484959; chr10:rs1561570, rs825411; chr18:rs2957128, rs3018362). Allele scores were normally distributed in cases and controls. Individuals carrying low frequency scores (0,1 and 11,12) were combined together.
Odds ratios (OR) are relative to the median number of risk alleles in the controls (5 risk alleles).
It is likely that other genomic regions might also contribute to PDB since the present study was powered only to detect variants with a moderate effect size (risk allele OR > 1.6). A quantile-quantile plot showing the distribution of P values after removal of all genome wide significant SNPs and correlated markers showed excess in the number of SNPs with small P values compared to what is expected by chance (Supplementary Fig. 2). For example, we observed 19 SNPs with P < 1 × 10−5 compared to 3 expected SNPs suggesting that other risk variants with modest effect remain to be identified. Among these are variants located on chromosome 3p24, 8q22, 10q24, and 14q32 which didn’t reach genome wide significance but could be considered suggestively associated with PDB (combined P < 1 × 10−5 ; Supplementary Table 3). Of particular interest is the 14q32 locus containing RIN3 (Ras interaction/interference protein 3) gene which is involved in vesicular trafficking36 and could be important in osteoclast function.
In summary we have demonstrated that common genetic variants at loci close to the CSF1, OPTN, and TNFRSF11A genes are independently associated with PDB. Further studies are now warranted to explore the mechanisms responsible for these associations.
ONLINE METHODS
Study subjects
The genome wide association study was conducted in a discovery sample of 750 patients of predominantly British descent with clinical and radiological evidence of PDB in whom mutations of the SQSTM1 gene had been excluded by DNA sequencing. These comprised subjects who had participated in the PRISM study (n=597) a randomised trial of two different treatment strategies for PDB38; clinic-based patients from the UK with sporadic PDB (n=55); and patients with a family history of PDB derived from the UK (n=20), Australia (n=66), New Zealand (n=8) and Italy (n=4). Details of the 1002 control subjects have previously been described 9, but in brief, they comprised healthy subjects of Scottish descent with no clinical evidence of PDB. For the replication study we conducted genotyping in an additional 500 PDB patients without SQSTM1 mutations diagnosed according to standard techniques. These comprised subjects with sporadic PDB who had been recruited from hospital clinics in the UK (n=226), Italy (n=20) and Spain (n=200); patients with sporadic PDB who had participated in the PRISM study (n=43) and subjects with a positive family history of PDB who had been recruited from hospital clinics in Australia (n=10), and the UK (n=1). The 535 replication controls comprised subjects from the UK who had been referred for investigation of osteoporosis but who had been found to have normal bone density on examination by dual energy x-ray absorptiometry (n=248); spouses of participants of the PRISM study who were not known to be affected by PDB (n=252) and clinic based controls from Spain (n=35). All study participants were of European descent and they provided informed consent. The discovery sample had 96 % power to detect disease associated allele with MAF = 0.2 and genotype relative risk of 1.6 assuming a multiplicative model and a disease with population prevalence of 2%.
Stage 1 genotyping and quality control
Genotyping of PDB cases was performed at the genetics core of the Wellcome Trust Clinical Research Facility (Edinburgh, UK) using Illumina HumanHap300 BeadChip version 2 duo. Genotyping of the controls had been previously performed by Illumina Inc. (San Diego) using HumanHap300 v1 and HumanHap240S arrays9. Genotypes for cases and controls were called using BeadStudio v3.2 (Illumina, Inc.) by following the manufacturer’s recommended protocol. Genotype data for control subjects were provided after applying the quality control (QC) measures described previously9. For the cases, we used a no-call threshold of 0.15 in BeadStudio and quality control metrics such as cluster separation, AB T mean and AB R mean to exclude badly performing SNPs. Samples with a call rate of less than 90% were excluded (n = 30). The data were then subjected to further QC measures using PLINK39 to exclude SNPs with a call rate of less than 95%, those with Hardy-Weinberg equilibrium (HWE) P-values of less than 1.0 × 10−4 in controls, and those with a minor allele frequency of less than 1%. This left a total of 294,663 SNPs common to cases and controls with at least 95% call rates in each set. Samples with excess heterozygosity (1 case), non-European ancestry (21 cases and 1 control), and related subjects (6 cases) were excluded before analysis leaving a final total of 692 cases and 1001 controls with an average (± SD) genotype call rate of 99.63 ± 1.0. The genotype cluster plots for all SNPs showing association with PDB at P < 1.0 × 10−4 were visually inspected in BeadStudio. Population ancestry was determined using multidimensional scaling analysis of identity-by-state (IBS) distances matrix of all individuals after combining genotype data from the HapMap project (release 22) samples of European (CEU), Asian (CHB and JPT), and African (YRI) ancestry. For this analysis we first removed SNPs in areas of extended linkage disequilibrium (LD) (Chr2: 134.0 -138.0, Chr6: 25.0 - 34.0, Chr8: 8.0 – 12.0, Chr11: 45.0 – 57.0)40, and those with r2 > 0.2 within a 150 SNP window. SNPs with call rate < 99%, MAF < 5%, and HWE P < 1.0 × 10−4 in cases or controls were also excluded leaving a total of 63,528 SNPs. The genome wide average IBS distances matrix for all pair of individuals was then calculated based on the 63,528 SNPs using PLINK and then used for multidimensional scaling analysis. Supplementary Fig 1 is a plot of the first two component of the multidimensional scaling analysis showing three clusters corresponding to CEU, CHB+JPT, and YRI samples with the majority of cases and controls located within the European CEU cluster. We identified 21 cases and 1 control as outliers from the CEU cluster and these were excluded from further analysis. Based on genome wide IBS distance, we identified 5 identical pairs (IBS distance > 99%) and 1 related pair (IBS distance > 85%) of samples from the cases cohort; the sample with the lowest call rate was excluded from each pair before further analysis.
Stage 2 genotyping and quality control
Genotyping of replication samples was performed by Sequenom (Hamburg, Germany) using the MassARRAY iPLEX platform. DNA from cases and controls were distributed into 384 well plates so that each plate had the same number of cases and controls to minimize genotyping bias due variations between runs. We included 100 samples from stage 1 as a quality control measure. The concordance rate between Illumina and Sequenom platforms was > 99.9%. Replication samples with call rate < 95% were excluded (19 cases and 15 controls) leaving a total of 481 cases and 520 controls with an average genotype call rate of 99.61%. The call rate of all genotyped SNPs was >95%.
Imputation
Genotypes were imputed for untyped variants located within 2.0 Mb of SNPs identified in stage 1 with genome wide significant association with PDB using MACH41. The HapMap European (CEU) genotype data from release 22 were used as a reference. To minimize end effects, only data from the middle 2 Mb of each 4 Mb imputed segment was used for further analysis. We used 200 rounds of Markov chain iterations to estimate allele dosage and best guess genotypes in stage 1 data. Imputation quality was assessed by estimating the correlation (r2) between imputed and true genotypes. SNPs with r2 < 0.3 were excluded before further analysis. Analysis of imputed data was performed using logistic regression implemented in mach2dat42 in which the imputed allelic dosage was used to account for uncertainty in imputed genotypes.
Statistical analysis
Statistical analyses were performed using PLINK (Version 1.07) 39. In stage 1, genotyped SNPs were tested for association with PDB using a stratified Cochran-Mantel-Haenszel (CMH) test. Samples were stratified based on their genome-wide IBS similarity so that individuals assigned to one cluster were not genetically different (P > 0.001 from pair wise population concordance test). The quantile-quantile plot and genomic control λ were used to assess overdispersion of the test statistics and were calculated using the statistical package R version 2.7.2 based on the 90% least significant SNPs as described previously10. Stepwise logistic regression was used to test for independent effects of SNP where the allelic dosage of the conditioning SNP was entered as a covariate in the regression model along with the population clusters identified by IBS sharing analysis described above to adjust for population substructure. Haplotype analysis was performed by logistic regression looking at the presence or absence of the test haplotype and including the population clusters as a covariate in the model. Haplotypes were phased using the E-M algorithm implemented in PLINK and only haplotypes with a frequency of ≥ 1% were analyzed. The cut off point for genome wide significance was set as P < 1.7 × 10−7 (0.05/294,663) for stage 1, and P < 3 × 10−3 (0.05/16) for the replication stage. For the combined analysis we set the threshold for significance as P < 5 × 10−8 as recently proposed43. The replication and combined data sets were analysed as described above except that the replication dataset was considered as a separate cluster when population clusters were used in a stratified CMH test or as a covariate in logistic regression models. The population attributable risk (PAR) for markers showing association with PDB was calculated according to the following formula: PAR = p(OR-1)/[p(OR-1)+1]; where p is the frequency of the risk allele in controls and OR is the risk allele odds ratio. The cumulative PAR was calculated as follows: Cumulative PAR = 1- (π1→n (1-PARi)); where n is the number of variants and PARi is the individual PAR for the ith SNP.
Supplementary Material
ACKNOWEDGEMENTS
The authors would like to acknowledge the contribution of the many participants who provided samples for the analysis. Dr Lee Murphy and Ms Angie Fawkes of the Wellcome Trust Clinical Research Facility for technical support with the Illumina genotyping. Mrs Aman Khatib for her assistance in data management. The study was supported in part by grants to SHR from the Arthritis Research Campaign (grants 13724, 17646 & 15389) and a grant to OMEA from the National Association for Relief of Paget’s disease.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
References
- 1.Cooper C, et al. The epidemiology of Paget’s disease in Britain: is the prevalence decreasing? J.Bone Miner.Res. 1999;14:192–197. doi: 10.1359/jbmr.1999.14.2.192. [DOI] [PubMed] [Google Scholar]
- 2.Siris ES. Paget’s disease of bone. J.Bone Miner.Res. 1998;13:1061–1065. doi: 10.1359/jbmr.1998.13.7.1061. [DOI] [PubMed] [Google Scholar]
- 3.Morales-Piga AA, Rey-Rey JS, Corres-Gonzalez J, Garcia-Sagredo JM, Lopez-Abente G. Frequency and characteristics of familial aggregation of Paget’s disease of bone. J.Bone Miner.Res. 1995;10:663–670. doi: 10.1002/jbmr.5650100421. [DOI] [PubMed] [Google Scholar]
- 4.Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am.J.Hum.Genet. 2002;70:1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hocking LJ, et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s disease. Hum.Mol.Genet. 2002;11:2735–2739. doi: 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- 6.Lucas GJ, et al. Identification of a major locus for Paget’s disease on chromosome 10p13 in families of British descent. J.Bone Miner.Res. 2008;23:58–63. doi: 10.1359/jbmr.071004. [DOI] [PubMed] [Google Scholar]
- 7.Hocking LJ, et al. Genomewide search in familial Paget disease of bone shows evidence of genetic heterogeneity with candidate loci on chromosomes 2q36, 10p13, and 5q35. Am.J.Hum.Genet. 2001;69:1055–1061. doi: 10.1086/323798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurin N, et al. Paget disease of bone: mapping of two loci at 5q35-qter and 5q31. Am.J.Hum.Genet. 2001;69:528–543. doi: 10.1086/322975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat.Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat.Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 11.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsurukai T, Udagawa N, Matsuzaki K, Takahashi N, Suda T. Roles of macrophage-colony stimulating factor and osteoclast differentiation factor in osteoclastogenesis. J.Bone Miner.Metab. 2000;18:177–184. doi: 10.1007/s007740070018. [DOI] [PubMed] [Google Scholar]
- 14.Bouyer P, et al. Colony-stimulating factor-1 increases osteoclast intracellular pH and promotes survival via the electroneutral Na/HCO3 cotransporter NBCn1. Endocrinology. 2007;148:831–840. doi: 10.1210/en.2006-0547. [DOI] [PubMed] [Google Scholar]
- 15.Van Wesenbeeck L, et al. The osteopetrotic mutation toothless (tl) is a loss-of-function frameshift mutation in the rat Csf1 gene: Evidence of a crucial role for CSF-1 in osteoclastogenesis and endochondral ossification. Proc.Natl.Acad.Sci.U.S.A. 2002;99:14303–14308. doi: 10.1073/pnas.202332999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roodman GD, et al. Interleukin 6. A potential autocrine/paracrine factor in Paget’s disease of bone. J.Clin.Invest. 1992;89:46–52. doi: 10.1172/JCI115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neale SD, Schulze E, Smith R, Athanasou NA. The influence of serum cytokines and growth factors on osteoclast formation in Paget’s disease. QJM. 2002;95:233–240. doi: 10.1093/qjmed/95.4.233. [DOI] [PubMed] [Google Scholar]
- 18.van Es MA, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat.Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 19.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat.Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rezaie T, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 21.Rezaie T, Waitzman DM, Seeman JL, Kaufman PL, Sarfarazi M. Molecular cloning and expression profiling of optineurin in the rhesus monkey. Invest Ophthalmol.Vis.Sci. 2005;46:2404–2410. doi: 10.1167/iovs.04-1243. [DOI] [PubMed] [Google Scholar]
- 22.Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFalpha- induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr.Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 23.Sudhakar C, Nagabhushana A, Jain N, Swarup G. NF-kappaB mediates tumor necrosis factor alpha-induced expression of optineurin, a negative regulator of NF-kappaB. PLoS.One. 2009;4:e5114. doi: 10.1371/journal.pone.0005114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahlender DA, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J.Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat.Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 26.Li J, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc.Natl.Acad.Sci.U.S.A. 2000;97:1566–1571. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villa A, Guerrini MM, Cassani B, Pangrazio A, Sobacchi C. Infantile malignant, autosomal recessive osteopetrosis: the rich and the poor. Calcif.Tissue Int. 2009;84:1–12. doi: 10.1007/s00223-008-9196-4. [DOI] [PubMed] [Google Scholar]
- 28.Hughes AE, et al. Mutations in TNFRSF11A, affecting the signal peptide of RANK, cause familial expansile osteolysis. Nat.Genet. 2000;24:45–48. doi: 10.1038/71667. [DOI] [PubMed] [Google Scholar]
- 29.Nakatsuka K, Nishizawa Y, Ralston SH. Phenotypic characterization of early onset Paget’s disease of bone caused by a 27-bp duplication in the TNFRSF11A gene. J.Bone Miner.Res. 2003;18:1381–1385. doi: 10.1359/jbmr.2003.18.8.1381. [DOI] [PubMed] [Google Scholar]
- 30.Whyte MP, Hughes AE. Expansile skeletal hyperphosphatasia is caused by a 15-base pair tandem duplication in TNFRSF11A encoding RANK and is allelic to familial expansile osteolysis. J Bone Miner Res. 2002;17:26–29. doi: 10.1359/jbmr.2002.17.1.26. [DOI] [PubMed] [Google Scholar]
- 31.Wuyts W, et al. Evaluation of the role of RANK and OPG genes in Paget’s disease of bone. Bone. 2001;28:104–107. doi: 10.1016/s8756-3282(00)00411-7. [DOI] [PubMed] [Google Scholar]
- 32.Hocking L, et al. Familial Paget’s disease of bone: patterns of inheritance and frequency of linkage to chromosome 18q. Bone. 2000;26:577–580. doi: 10.1016/s8756-3282(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 33.Styrkarsdottir U, et al. Multiple genetic loci for bone mineral density and fractures. N.Engl.J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 34.Rivadeneira F, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat.Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Styrkarsdottir U, et al. New sequence variants associated with bone mineral density. Nat.Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 36.Saito K, et al. A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J.Biol.Chem. 2002;277:3412–3418. doi: 10.1074/jbc.M106276200. [DOI] [PubMed] [Google Scholar]
- 37.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 38.Langston AL, et al. Randomised Trial of Intensive Bisphosphonate Treatment Versus Symptomatic Management in Paget’s Disease of Bone. J.Bone Miner.Res. 2009 doi: 10.1359/jbmr.090709. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am.J.Hum.Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am.J.Hum.Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Abecasis GR. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am.J.Hum.Genet. 2006;S79:2290. [Google Scholar]
- 42.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu.Rev.Genomics Hum.Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet.Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.